OCT4 and NANOG are required for maintenance of pluripotent cells. Do et al. find that STAT3 becomes phosphorylated and translocates to the nucleus in the four-cell and later stage embryos. IL-6 acts in an autocrine fashion to ensure STAT3 phosphorylation, mediated by JAK1. STAT3 is essential for the maintenance of inner cell mass (ICM) lineages but not for ICM and trophectoderm formation. STAT3 directly binds to the Oct4 and Nanog distal enhancers, modulating their expression to maintain pluripotency of mouse embryonic and induced pluripotent stem cells.
Keywords: embryogenesis, inner cell mass, STAT3, OCT4, NANOG, embryonic stem cell
Abstract
Although it is known that OCT4–NANOG are required for maintenance of pluripotent cells in vitro, the upstream signals that regulate this circuit during early development in vivo have not been identified. Here we demonstrate, for the first time, signal transducers and activators of transcription 3 (STAT3)-dependent regulation of the OCT4–NANOG circuitry necessary to maintain the pluripotent inner cell mass (ICM), the source of in vitro-derived embryonic stem cells (ESCs). We show that STAT3 is highly expressed in mouse oocytes and becomes phosphorylated and translocates to the nucleus in the four-cell and later stage embryos. Using leukemia inhibitory factor (Lif)-null embryos, we found that STAT3 phosphorylation is dependent on LIF in four-cell stage embryos. In blastocysts, interleukin 6 (IL-6) acts in an autocrine fashion to ensure STAT3 phosphorylation, mediated by janus kinase 1 (JAK1), a LIF- and IL-6-dependent kinase. Using genetically engineered mouse strains to eliminate Stat3 in oocytes and embryos, we firmly establish that STAT3 is essential for maintenance of ICM lineages but not for ICM and trophectoderm formation. Indeed, STAT3 directly binds to the Oct4 and Nanog distal enhancers, modulating their expression to maintain pluripotency of mouse embryonic and induced pluripotent stem cells. These results provide a novel genetic model of cell fate determination operating through STAT3 in the preimplantation embryo and pluripotent stem cells in vivo.
One of most challenging questions in developmental biology is understanding how a totipotent zygote differentiates into an embryo containing all cell lineages of the developing organism. The limited amount of mammalian embryonic material has hampered the molecular study of lineage commitment, but the discovery, isolation, and culture of embryonic stem cells (ESCs) (Evans and Kaufman 1981; Martin 1981) enabled biochemical studies, which led to the understanding of gene circuitries that control pluripotency (for review, see Jaenisch and Young 2008).
OCT4, a homeobox-containing transcription factor, was originally shown to be one of the essential factors regulating pluripotency in vitro and in vivo (Scholer et al. 1989a,b; Nichols et al. 1998). OCT4 is a master regulator of a gene circuit that includes NANOG, SOX2, and other genes that coordinate formation of early embryonic lineages (Koutsourakis et al. 1999; Avilion et al. 2003; Mitsui et al. 2003; Strumpf et al. 2005; Yagi et al. 2007). OCT4 and Caudal-related homeobox 2 (CDX2) are major transcription factors regulating inner cell mass (ICM) and trophectoderm (TE) lineage, respectively (Nichols et al. 1998; Strumpf et al. 2005). In implanting embryonic day 4.5 (E4.5) embryos, NANOG and GATA4/6 are key determinants for the epiblast (EPI) and primitive endoderm (PE) fate, respectively (Koutsourakis et al. 1999; Mitsui et al. 2003). However, the upstream signals that regulate Oct4 expression and its gene circuitry during early embryogenesis are not well documented.
Many initial attempts to identify the upstream regulators were undertaken in vivo (Rothstein et al. 1992; Ko et al. 2000; Guo et al. 2010; Tang et al. 2010), yet it was from in vitro work that leukemia inhibitory factor (LIF) was found to be the key growth factor for the culture and maintenance of mouse ESCs (mESCs) (Smith et al. 1988; Williams et al. 1988). LIF, a member of the interleukin 6 (IL-6) family of cytokines, binds to glycoprotein 130 (gp130) in association with a ligand-specific receptor subunit, LIF receptor (LIFR). The binding of LIF to the LIFR results in activation of receptor-associated janus kinases (JAKs) in the phosphorylation of receptor docking sites and, finally, in the recruitment of the Src homology-2 (SH2) domain and phosphorylation on Tyr705 residues of signal transducers and activators of transcription (STAT), a gene family originally described in the interferon-induced regulatory pathways (Fu et al. 1990, 1992; Schindler et al. 1992; Darnell et al. 1994). The STAT proteins are well conserved through evolution (L Zhang, CPK Patro, CY Ung, TP Phan, PJ Lin, Y Zheng, G Song, A Jean, JC Tong, YE Chin, et al., unpubl.), regulating gene expression in response to external signals (Darnell 1997). STAT3, first identified as a transcription factor for the IL-6 family of cytokines (Akira et al. 1994; Zhong et al. 1994), was subsequently found to be crucial for ESC pluripotency (Boeuf et al. 1997; Niwa et al. 1998; Raz et al. 1999; Ying et al. 2003). However, it is not clear how LIF/STAT3 signaling interacts with the core set of pluripotency factors for ESCs; namely, Oct4, Nanog, and Sox2 (Boyer et al. 2005; Loh et al. 2006). Using a genome-wide approach, STAT3 was initially implicated in the OCT4-mediated gene circuitry in cultured ESCs (Chen et al. 2008); other studies showed direct regulation of Klf4, which is also a member of this circuitry (Hall et al. 2009; Niwa et al. 2009).
Conventional knockout of Stat3 in mice results in embryonic lethality at E6.5 (Takeda et al. 1997). However, the ability of zygotic Stat3 knockout embryos to undergo apparently normal EPI expansion is in conflict with the central role of STAT3 in maintaining ESCs (Boeuf et al. 1997; Niwa et al. 1998; Raz et al. 1999; Ying et al. 2003). To resolve this paradox, we investigated the role of STAT3 in preimplantation embryos and pluripotent stem cells. By eliminating Stat3 in mouse oocytes and embryos, we found STAT3 to be essential for the maintenance of ICM lineages but not for blastocyst formation or TE maintenance. By working with mouse ESCs and induced pluripotent stem cells (iPSCs), we found that STAT3 directly binds and regulates Oct4 (Pou5F1) expression to maintain their pluripotency. These results lead us to propose that STAT3 has an essential role in ICM lineage specification and maintenance and pluripotent stem cell identity.
Results
Presence of activated STAT3 in preimplantation embryos
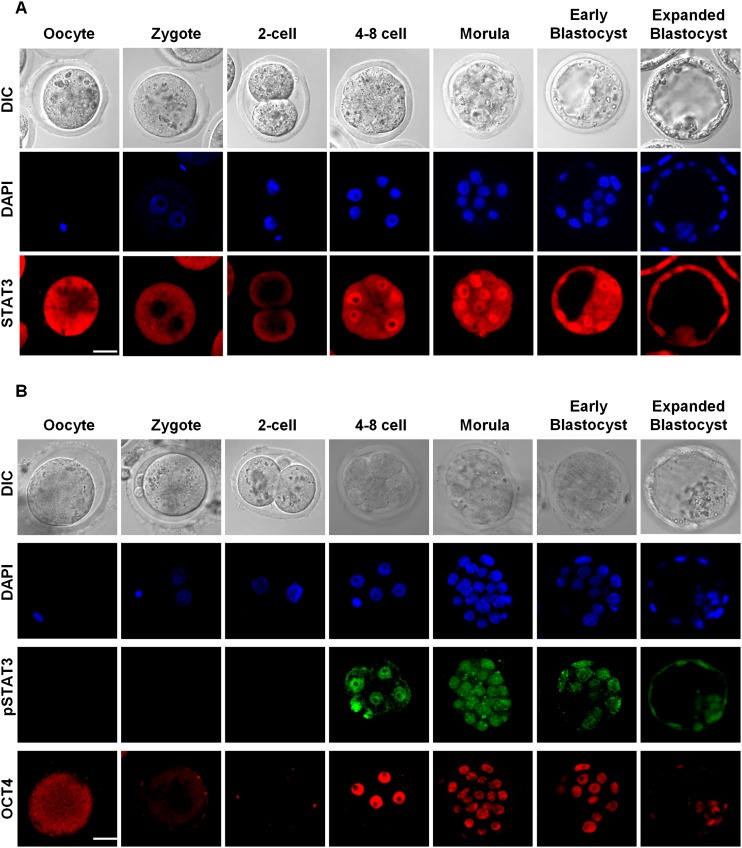
STAT3 is present exclusively in the cytoplasm of oocytes, zygotes, and two-cell stage embryos but strikingly translocates to the nucleus specifically at the four-/eight-cell stage (Fig. 1A). Using an antibody against phosphorylated tyrosine-specific (pY705)-STAT3, we confirmed that nuclear STAT3 at the four-cell stage embryo is indeed tyrosine-phosphorylated (Fig. 1B). We also observed OCT4 colocalization with nuclear, phosphorylated STAT3 (pY705; pSTAT3) in most four-cell stage embryos. These results indicate that STAT3, while present in earlier stage embryos, is only functionally activated at the four-cell stage.
Figure 1.
Expression of STAT3 during preimplantation mouse development. (A) Immunodetection with pan-STAT3 antibody indicates nuclear localization of STAT3 starting from the four-cell stage. (B) Immunodetection with antibody specific to phosphorylated tyrosine (pY705)-STAT3 and OCT4 show nuclear STAT3 is tyrosine-phosphorylated and correlates with re-expression of OCT4 in the four-cell embryos. Bar, 20 μm.
STAT3 phosphorylation is dependent on LIF, IL-6, and JAK1 in preimplantation mouse embryos
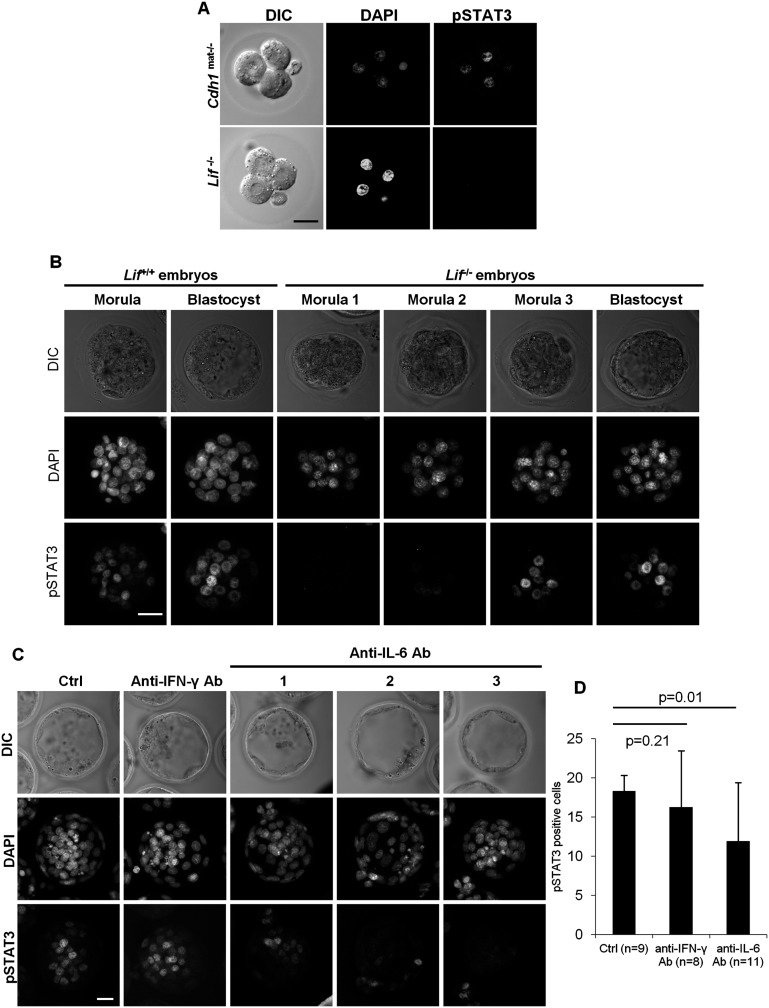
To identify the cytokines responsible for the STAT3 activation in the four-cell embryos, we examined STAT3 phosphorylation in Lif-null and E-cadherin (Cdh1)-null preimplantation embryos. STAT3 is known to be phosphorylated by LIF (Akira et al. 1994; Zhong et al. 1994), and Cdh1-null ESCs exhibit loss of tyrosine-phosphorylated STAT3 (Hawkins et al. 2012). We found no discernible tyrosine-specific phosphorylated STAT3 in Lif-null four-cell embryos, while STAT3 phosphorylation is present in Cdh1-null embryos (Fig. 2A). This suggests that LIF is the cytokine responsible for STAT3 phosphorylation in four-cell embryos, and in contrast to ESCs, this is E-cadherin-independent in vivo. Interestingly, when we examined the STAT3 phosphorylation in later stage embryos, we found that active STAT3 (pY705; pSTAT3) is nontheless present at the late morula and blastocyst stages (Fig. 2B). In addition, tyrosine-phosphorylated STAT3 is present in blastocysts derived from zygotes cultured in KSOM (Fig. 2C). These results suggest that cytokines other than LIF produced by the blastocysts act in an autocrine manner to stimulate STAT3 phosphorylation. Since IL-6 and IFN-γ are responsible for STAT3 phosphorylation (Akira et al. 1994; Zhong et al. 1994) and appear to be expressed in blastocysts (Rothstein et al. 1992), we examined their role in STAT3 phosphorylation in the blastocyst. While treatment of zygotes with a neutralizing antibody against IFN-γ showed no significant change in STAT3 phosphorylation, embryos treated with a neutralizing antibody against IL-6 showed a significant reduction in phosphorylated STAT3 expression (Fig. 2C,D). Taken together, these results suggest that IL-6 is the additional cytokine produced by the blastocysts and responsible for STAT3 phosphorylation in an autocrine manner in the absence of LIF.
Figure 2.
STAT3 is phosphorylated by LIF-dependent and -independent pathways during preimplantation mouse development. Immunostaining of (pY705)-STAT3 in Cdh1mat−/− and Lif−/− four-cell embryos (A), (pY705)-STAT3 in Lif+/+ and Lif−/− morulas and early blastocysts (B), and (pY705)-STAT3 in control embryos and embryos treated with neutralizing antibodies against IFN-γ and IL-6 cytokines (C). Bar, 20 μm. (D) Bar graph showing the average of pSTAT3-positive cells in controls and in embryos treated with anti-IFN-γ and anti-IL-6 antibodies.
The JAKs TYK2, JAK1, JAK2, and JAK3 are responsible for the activation of STAT proteins; TYK2, JAK1, and JAK2 are activated by IL-6 (Schindler and Darnell 1995). We therefore examined RNA sequencing (RNA-seq) data obtained from single preimplantation mouse embryos for expression of these kinases during preimplantation mouse development. We found that Jak2 and Tyk2 mRNAs are highly expressed in the four-cell stage, whereas Jak1 is highly expressed in the blastocyst stage (Supplemental Fig. S1A). To determine whether JAK1 is a major kinase responsible for STAT3 phosphorylation in the blastocyst, we cultured zygotes in the presence of different JAK inhibitors: JAK inhibitor I (JAKi), an inhibitor for all four JAK kinases; AG490, an inhibitor for JAK2 and JAK3; and ruxolitinib, an inhibitor for JAK1 and JAK2. JAKi- and ruxolitinib-treated blastocysts showed no discernible expression of tyrosine-phosphorylated STAT3, while AG490-treated embryos were positive for STAT3 phosphorylation, suggesting that JAK1 is responsible for STAT3 phosphorylation in the blastocysts (Supplemental Fig. S1B). In addition, OCT4 expression was reduced in JAKi-treated blastocysts compared with controls (Supplemental Fig. S1C), suggesting that embryos treated with JAK inhibitors recapitulate the loss of OCT4 expression in maternal/zygotic Stat3-null expanded blastocysts (see below).
Maternal and paternal Stat3 knockout E4.5 embryos exhibit loss of the EPI and PE lineages
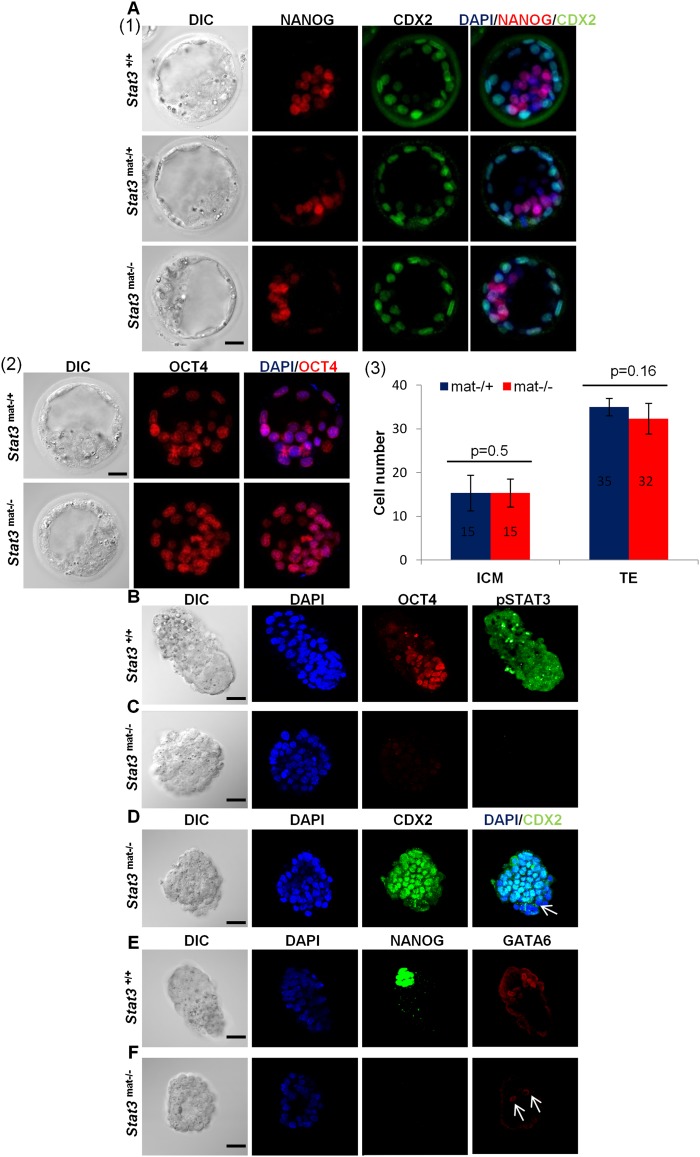
Since STAT3 is present in oocytes (Fig. 1A), conventional zygotic knockout (Stat3−/−) embryos may be able to use this maternal protein during early embryogenesis. We therefore established maternal Stat3 knockout oocytes using a Zp3-Cre mating strategy (De Vries et al. 2004), rendering STAT3 absent in all eggs. However, when fertilized by wild-type males, the maternal deletion is rescued by expression from the paternal allele at the morula stage. Preimplantation embryos are normal, and pups derived from these matings are viable, indicating that maternal STAT3 is not essential for oogenesis and preimplantation development. In contrast, if maternal-null Stat3 oocytes (Stat3mat−) are fertilized by sperm from heterozygous males (Stat3+ or Stat3−), no viable pups or E5.5 post-implantation maternal/zygotic Stat3-null (Stat3mat−/−) embryos were found, although normal Stat3mat−/+ embryos are present (Supplemental Fig. S2A). However, Stat3mat−/− E3.5 blastocysts show normal morphology; express the ICM/TE markers OCT4, NANOG, and CDX2, respectively; and have ICM and TE cell numbers comparable with their Stat3mat−/+ littermates (Fig. 3A). These observations show that STAT3 is not required for the first lineage specification in the preimplantation embryo.
Figure 3.
Maternal and paternal Stat3 knockout embryos isolated at E4.5 exhibit loss of the EPI and PE lineages but not TE. (A) (Panel 1) Immunodetection of NANOG and CDX2 in Stat3+/+, Stat3mat−/+, and Stat3mat−/− early blastocysts isolated at E3.5. (Panel 2) Immunodetection of OCT4 in Stat3mat−/+ and Stat3mat/− E3.5 embryos. (Panel 3) Bar graphs showing the average cell number in ICM and TE lineages of Stat3mat−/+ (n = 3) and Stat3mat−/− (n = 3) early blastocysts isolated at E3.5. ICM and TE cells are identified as NANOG+/CDX2− and CDX2+ cells, respectively. (B,C) Immunodetection of pSTAT3 and OCT4 in Stat3+/+ and Stat3mat/− E4.5 embryos. (D) Immunodetection of CDX2 in Stat3mat−/− E4.5 embryos. White arrow indicates the presence of CDX2-negative cells in the knockout embryos. (E,F) Immunodetection of NANOG and GATA6 in Stat3+/+ and Stat3mat−/− E4.5 embryos. White arrows indicate the presence of remaining GATA6-positive cells in the knockout embryos. Bar, 20 μm.
Interestingly, when we compared Stat3+/+ implanting E4.5 embryos (Fig. 3B) with Stat3mat−/− embryos, we found severely morphologically challenged embryos, which expressed little to no OCT4 (Fig. 3C). To confirm that these defects are primarily in the ICM lineages, we immunostained E4.5 embryos for the TE marker CDX2 and found almost all cells to be CDX2-positive (Fig. 3D), suggesting that maternal/zygotic deletion of Stat3 causes specific loss of OCT4-positive ICM but not CDX2-positive TE cells. Since there were few CDX2-negative cells in Stat3mat−/− E4.5 embryos, we performed immunostaining of NANOG and GATA6 to detect cells of the EPI and PE lineages, respectively. While ICMs of control E4.5 embryos have two normal and distinct populations of NANOG- and GATA6-positive cells (Fig. 3E), we found no NANOG-positive cells and few to no discernible GATA6-positive cells in ICMs of Stat3mat−/− embryos (Fig. 3F; Supplemental Fig. S2B). In addition, while there are no active Caspase3 (CASP3)-positive cells in control E4.5 embryos (Supplemental Fig. S2C), we found active CASP3-positive cells in Stat3mat−/− mutants, indicating loss of ICM cells, at least in part, through apoptosis (Supplemental Fig. S2D). Thus, using this genetic approach, we found STAT3 to be essential for ICM but not TE lineage maintenance in the implanting mouse embryo.
Loss of maternal/zygotic Stat3 results in random lineage marker expression in ICM cells
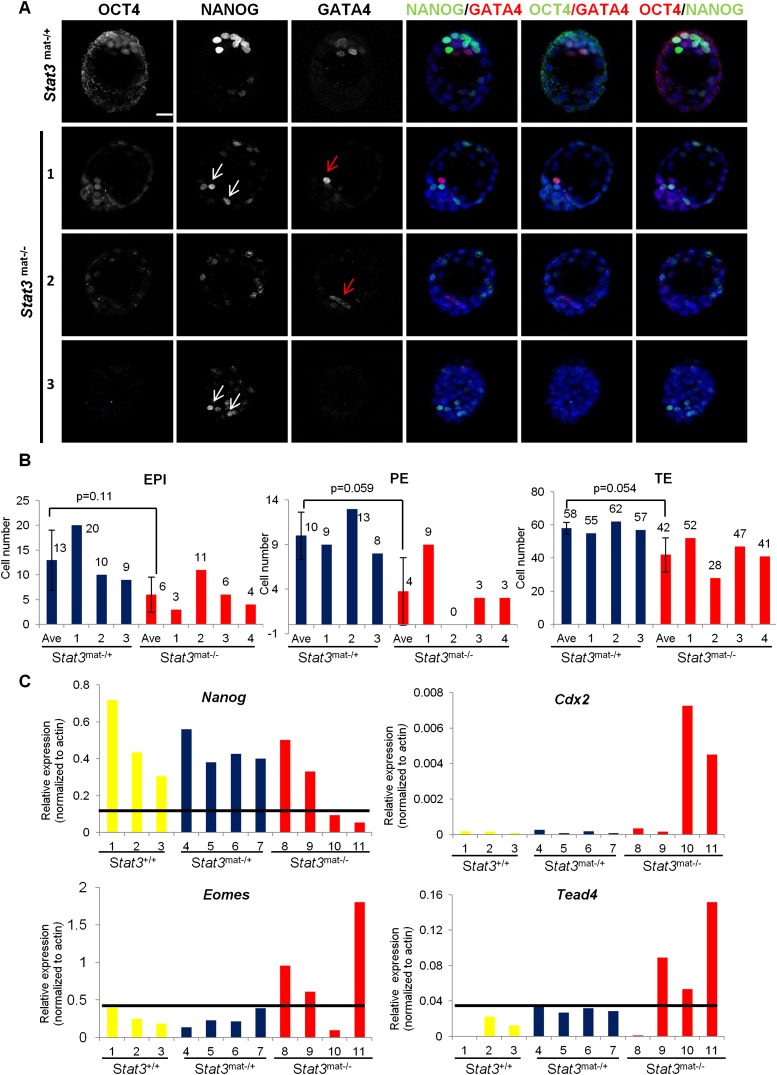
While maternal/zygotic null E3.5 embryos show appropriate lineage allocation and marker gene expression, E4.5 embryos are morphologically deranged, and ICM cells are lost. To further understand the nature of this rapid, progressive loss of lineage allocation and maintenance, we performed coimmunostaining of OCT4, NANOG, and GATA4 in expanded blastocysts at E4.0. In control blastocysts, OCT4, NANOG, and GATA4 were appropriately expressed in the ICM domain, with NANOG and GATA4 labeling two intermingled cell populations in a “salt-and-pepper-like” fashion (Fig. 4A, top row). In contrast, we found an abnormal phenotype in the ICM lineages of Stat3mat−/− embryos, showing either highly variable ICM marker gene expression or none at all (Fig. 4A; Supplemental Fig. S3A,B). For instance, few ICM cells in embryo 1 are positive for OCT4 and NANOG (Fig. 4A, white arrows) or OCT4 and GATA4 (Fig. 4A, red arrows), while others are OCT4-, NANOG-, and GATA4-negative. This variability is pronounced, with extreme cases not showing any NANOG-positive (embryo 2) or GATA4-positive (embryo 3) ICM cells (Fig. 4A). To confirm that these defects are primarily in the ICM, we immunostained Stat3mat−/− expanded blastocysts with CDX2 antibody and found the CDX2-positive TE lineage intact (Supplemental Fig. S3B). To further characterize the defect in ICM lineage formation in Stat3mat−/− expanded blastocysts, we counted the number of the EPI-committed (NANOG+/CDX2−), PE-committed (GATA4+/CDX2−), and TE-committed (CDX2+) in mutant embryos. Consistent with the immunostaining results, we found highly variable cell numbers for the EPI and PE lineages but not the TE lineage (Fig. 4B). In addition, the average cell numbers in the EPI and PE lineages were reduced, but this was not the case in the TE lineage when Stat3mat−/− expanded blastocysts were compared with Stat3mat−/+ embryos. However, because of the high variation among individual mutant embryos, this reduction is not statistically significant (Fig. 4B). Consistent with the protein expression results, quantitative real-time PCR (qRT-PCR) also showed variable mRNA expression of ICM and TE marker genes in individual immunosurgically isolated ICMs of Stat3mat−/− E3.5 blastocysts (Fig. 4C). Taken together, our results suggest that STAT3 is required for ICM lineage maintenance in the expanded blastocyst after initial lineage segregation, most likely by modulating ICM gene expression.
Figure 4.
Maternal and paternal Stat3 knockout, expanded blastocysts exhibit a variable abnormal phenotype. (A) Immunodetection of OCT4, NANOG, and GATA4 in Stat3mat−/+ and Stat3mat−/− expanded blastocysts. Embryo 1 has NANOG-positive (white arrows) and GATA4-positive (red arrow) cells in ICM. Embryo 2 has GATA4-positive cells (red arrow) but no OCT4- and NANOG-positive cells in the ICM. Embryo 3 has Nanog-positive cells in either the ICM or TE (white arrow) and no OCT4- and GATA4-positive cells. Bar, 20 μm. (B) The bar graphs show the average and individual cell number in ICM, PE, and TE lineages in the Stat3mat−/+ (blue bars) and Stat3mat−/− (red bars) expanded blastocysts. The EPI, PE, and TE cells are identified as NANOG+/CDX2−, GATA4+/CDX2−, and CDX2+ cells, respectively. (C) Real-time PCR shows mRNA expression of Nanog, Cdx2, Eomes, and Tead4 in individual ICMs that were isolated by immunosurgery from Stat3+/+ (yellow bars), Stat3mat−/+ (blue bars), and Stat3mat−/− (red bars) early blastocysts.
STAT3 directly regulates Oct4 and Nanog expression to maintain ESC pluripotency
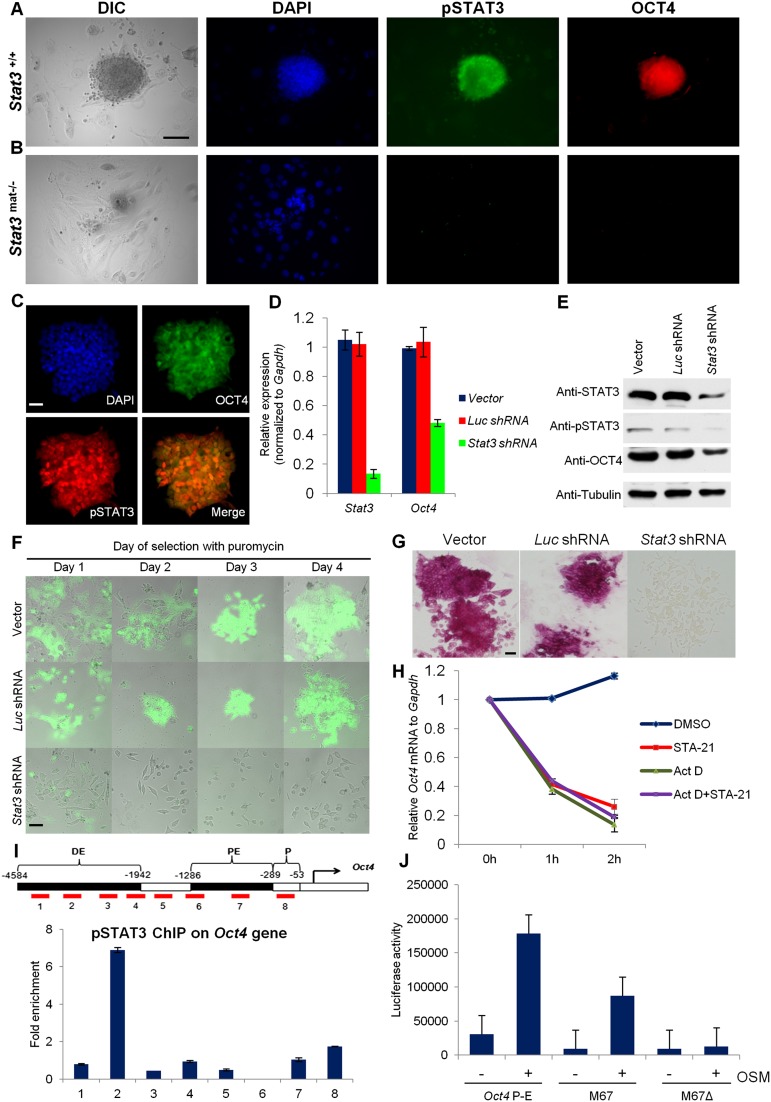
Although Stat3mat−/− early blastocysts present a normal morphology, these embryos fail to form typical ICM-derived, OCT4-positive outgrowths (Fig. 5A,B), and ESCs cannot be derived. To examine a possible molecular link between STAT3 and OCT4, we used two pluripotent in vitro cell systems: mESCs carrying an Oct4-GFP reporter (Yamagata et al. 2010) and mouse iPSCs (Takahashi and Yamanaka 2006). As in preimplantation embryos, the presence of tyrosine-phosphorylated (pY705) STAT3 correlated with GFP expression (i.e., Oct4) in ESCs (Fig. 5C) and iPSCs (Supplemental Fig. S4A). When we reduced Stat3 transcript levels by using Stat3 shRNA knockdowns, Oct4-Gfp mRNA expression and OCT4 levels in ESCs were reduced (Fig. 5D–F). Similarly, Oct4 and Nanog mRNA and protein levels were reduced in iPSCs after Stat3 knockdown (Supplemental Fig. S4C–F). Furthermore, Stat3 knockdown in either ESCs or iPSCs resulted in failure to form distinct alkaline phosphatase-positive colonies (Fig. 5G; Supplemental Fig. S4B), although differentiated cells were observed (Fig. 5G; Supplemental Fig. S4B). These results suggest that the STAT3–OCT4–NANOG link is important for maintenance of pluripotent, self-renewing cells.
Figure 5.
STAT3 directly regulates Oct4 expression to maintain ESC pluripotency. (A,B) Immunodetection of pSTAT3 and OCT4 in 5-d in vitro blastocyst outgrowths of Stat3+/+ and Stat3mat−/−. Bar, 100 μm. (C) Nuclear expression of pSTAT3 and OCT4-GFP in ESCs. Bar, 20 μm. (D) Transfection of Stat3 shRNA results in down-regulation of Stat3 and Oct4 mRNA levels. (E) Stat3 knockdown in Oct4-Gfp ESCs results in decreased levels of STAT3, pSTAT3, and OCT4 by Western blot analysis. (F) Transfection of Stat3 shRNA into Oct4-Gfp ESCs leads to down-regulation of OCT4-GFP. Bar, 50 μm. (G) Reduced alkaline phosphatase detection in Stat3 knockdown Oct4-Gfp ESCs. Bar, 50 μm. (H) Treatment of iPSCs with STAT3 inhibitor STA-21 results in down-regulation of Oct4 mRNA. Treatment with actinomycin D or with actinomycin D plus a STAT3 inhibitor does not produce any additional effects. (I) STAT3 binds to the distal enhancer of Oct4 in mouse iPSCs. ChIP was performed on sonicated chromatin from iPSCs using pSTAT3 antibody. Immunoprecipitated DNA was analyzed by qRT-PCR with primer sets designed to detect ChIP-enriched DNA fragments (red boxes), shown within the context of the genomic structure of mouse Oct4. Amplicons are numbered in order, relative to their sites along the gene. Fold enrichment is the relative abundance of DNA fragments at the indicated regions over control region as quantified by real-time PCR. Standard deviations were calculated from biological replicates of qPCR data. (J) Luciferase results show that STAT3 could activate gene expression driven by the Oct4 promoter enhancer (P-E) in STAT3-overexpressing 293T cells in response to OSM. Oct4 P-E is the pGL3 luciferase vector containing the 4.6-kb upstream (P-E) region of Oct4. M67 and M67Δ are control luciferase vectors with the STAT3- and mutated STAT3-binding sites, respectively.
To determine whether STAT3 regulates Oct4 directly, we treated iPSCs with the STAT3 inhibitor STA-21, which resulted in a significant reduction of Oct4 mRNA after 1 h but did not change the mRNA expression of Sox2 and Tbx3 (STAT3 nontarget genes) (Fig. 5H; Supplemental Fig. S5). Treatment with actinomycin D or actinomycin D plus STA-21 did not produce any additional effects. This indicates that STAT3 directly regulates Oct4 expression without affecting mRNA stability (Fig. 5H). To examine whether there is direct binding of phosphorylated STAT3 to either the Oct4 or Nanog promoter, including the proximal enhancer or distal enhancer region (Fig. 5I, PE and DE, respectively; Supplemental Fig. 3SG; Chen et al. 2008), we performed chromatin immunoprecipitation (ChIP) assays using the pY705-STAT3-specific antibody. With Oct4, the highest enrichment of STAT was found in a region (−3161 base pairs [bp]/−3010 bp) of the distal enhancer (region 2), and less was found in the promoter region (region 8 in Fig. 5I). In addition, with Nanog ChIP, the highest enrichment of pSTAT3 was found in a region (−4320 bp/−4116 bp) of the distal enhancer (region 1), with less binding in the promoter region (region 7 in Supplemental Fig. S3G). ChIP using a STAT1-specific antibody shows no binding of STAT1 at the STAT3-binding sites of either Oct4 or Nanog (Supplemental Fig. S6). These results show that STAT3 directly and specifically interacts with the regulatory regions of Oct4 and Nanog. Finally, to assess the transcriptional activation of the Oct4 promoter/enhancer, we prepared an Oct4-luciferase reporter construct containing the 4.6-kb upstream promoter enhancer region of Oct4. For controls, we used luciferase reporter constructs M67 and M67Δ, containing STAT3- and mutated STAT3-binding sites, respectively. STAT3-overexpressing 293T cells (Supplemental Fig. S7) transfected with the Oct4 promoter/enhancer luciferase construct displayed high levels of luciferase activity after treatment of oncostatin M (OSM), a STAT3 activator (Fig. 5J). The specificity of this activation was confirmed by transfection of STAT3-overexpressing 293T cells with the M67 construct, which produced high luciferase activity after OSM treatment, while those transfected with M67Δ did not. Taken together, the evidence from mutant embryos, shRNA knockdowns in ESCs/iPSCs, STAT3 inhibitors, ChIPs, and luciferase assays demonstrates that pSTAT3 directly and positively regulates Oct4 and Nanog expression in pluripotent cells in vivo and in vitro.
Discussion
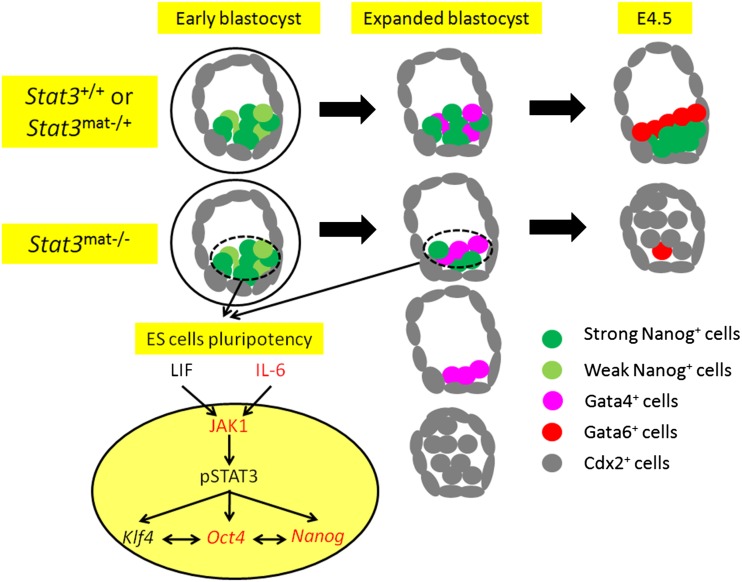
In this study, we investigated the molecular mechanisms of STAT3 action and whether STAT3-targeted genes are involved in both the regulatory circuitry of preimplantation embryos and pluripotent stem cells. Strikingly, we found that STAT3 is highly expressed in mouse oocytes, becomes tyrosine-phosphorylated, and translocates to the nucleus in the four-cell stage embryo, where it is coexpressed with OCT4 in the four- to eight-cell stage. Using Lif-null embryos, we found that STAT3 tyrosine phosphorylation is dependent on LIF in the four-cell embryos, while IL-6, one of the cytokines produced by the blastocyst, acts in an autocrine fashion to ensure STAT3 phosphorylation. We further identified JAK1 as the IL-6 downstream kinase and responsible for STAT3 tyrosine phosphorylation in the blastocyst stage. Using genetically engineered mouse strains designed to eliminate Stat3 in the oocyte and embryo, we demonstrate that STAT3 is essential to maintain the ICM lineages but not TE in late blastocysts (Fig. 6). We further showed that STAT3 directly binds and regulates Oct4 (Pou5F1) and Nanog gene expression to maintain pluripotency of mouse ESCs and iPSCs.
Figure 6.
Proposed model showing that STAT3 is required for maintenance of the embryonic EPI and preservation of pluripotent stem cell identity. Stat3mat−/− E4.5 blastocysts are collapsed with no OCT4- or NANOG-positive cells and reduced GATA6-positive cells in the ICM. Stat3mat−/− E4.0 expanded blastocysts show a variable, abnormal phenotype with reduction or loss of NANOG- and GATA4-positive cells. There are consistent changes in expression of pluripotent and TE marker genes in the ICMs of Stat3mat−/− blastocysts. IL-6, a cytokine produced by the blastocyst, acts in an autocrine fashion to ensure STAT3 phosphorylation. JAK1, a LIF- and IL-6-dependent downstream kinase, is responsible for STAT3 phosphorylation in the blastocyst. Stat3mat−/− early blastocysts are morphologically normal, yet these embryos fail to form ICM-derived structures after in vitro outgrowth, and ESCs cannot be established. In vitro studies using ESCs and iPSCs indicate that STAT3 directly regulates the pluripotency gene network (Oct4, Nanog, and Klf4).
The complexity of STAT3 signaling in preimplantation embryos
Studies on knockout mouse models indicate that the LIF signaling pathway is dispensable during preimplantation development. For instance, the Lif-null embryos develop normally into adulthood, but maternal Lif-null blastocysts cannot implant due to the absence of LIF production by the uterus (Stewart et al. 1992). Lifr mutant embryos develop to term but have severe deficits in motor neuron and glia cell populations (Li et al. 1995; Ware et al. 1995); gp130−/− embryos develop to blastocysts and implant but die by E12 with placental, cardiac, hematopoietic, and neuronal defects (Yoshida et al. 1996; Nakashima et al. 1999). Jak1-null mice die within the first 24 h after birth, and Jak2-null mice die around E12.5 (Neubauer et al. 1998; Rodig et al. 1998). However, no evidence for early embryo loss has been reported from any of these mutants. In contrast, mESC derivation and maintenance is dependent on LIF (Smith et al. 1988; Williams et al. 1988), LIFR (Ware et al. 1995), and gp130 (Nichols et al. 2001), suggesting that alternative pathways might be involved in maintaining pluripotency in vivo and in vitro. Our results, showing that phosphorylated STAT3 is present in E3.5 despite the absence of LIF, support this suggestion. We further provide a possible explanation for the differential phenotypes of maternal/zygotic Lif- and Stat3-null embryos, showing IL-6 to be involved in STAT3 tyrosine phosphorylation in the blastocyst. This is consistent with the fact that the combination of IL-6 and soluble IL-6 receptor can be used to derive and maintain ESCs without involvement of LIFR (Nichols et al. 1994; Yoshida et al. 1994). Il6-deficient mice can develop normally (Kopf et al. 1994), suggesting that either LIF or additional, redundant cytokines such as IL-6 are required for STAT3 phosphorylation in vivo. Taken together, our results suggest that multiple cytokines are used for STAT3 tyrosine phosphorylation to ensure proper ICM lineage formation in the blastocyst in vivo. In this light, it would be interesting to examine STAT3 phosphorylation and the developmental capacity of embryos deficient in both Lif and Il-6.
Strikingly, we also identified JAK1 as a possible IL-6 and LIF downstream kinase responsible for STAT3 phosphorylation in the blastocyst. Embryos treated with JAK inhibitors in vitro show loss of STAT3 tyrosine phosphorylation and reduced OCT4 expression in the ICM, a phenocopy of the maternal/zygotic Stat3-null embryos in vivo. Zygotic Jak1-null mice develop normally and die within the first 24 h after birth (Rodig et al. 1998), suggesting that maternal JAK1 may play an important role in the development of preimplantation embryos. Overall, we established that an independent, redundant pathway ensuring STAT3 activation is integral to preimplantation development.
Role of maternal STAT3
Absence of STAT3 in the mouse egg appears inconsequential for development, as maternal-null embryos give rise to viable pups. However, maternal/zygotic null embryos display earlier developmental arrest than that reported for zygotic mutants (Takeda et al. 1997), suggesting a maternal gene function revealed only in the absence of zygotic Stat3 expression. A parsimonious explanation for these observations is that transcription from the paternally inherited allele, which occurs at the eight-cell stage in maternal-null embryos, produces sufficient STAT3 for the maintenance of pluripotent cells in the EPI. As a corollary, the presence of maternal STAT3 in zygotic null embryos may be sufficient to support ICM cell maintenance past implantation, yet this protein pool is depleted eventually, causing developmental arrest at E6.5. Finally, in maternal/zygotic null embryos, the complete absence of STAT3 results in early loss of pluripotent cell lineage maintenance, which is somewhat reminiscent of mutations of two other genes essential to the maintenance of pluripotency: Oct4 and Nanog (Nichols et al. 1998; Mitsui et al. 2003; Silva et al. 2009; Messerschmidt and Kemler 2010).
Role of STAT3 in ICM lineages
Our results, showing that maternal/zygotic Stat3 knockout E4.5 embryos exhibit loss of EPI and PE lineages, are consistent with those previously observed from Oct4- and Nanog-null embryos. Oct4 is known to be important in establishing the distinct identity of the ICM in early embryogenesis. Moreover, homozygous Oct4-null embryos, when cultured in vitro, yield outgrowths of trophoblast giant cells but no recognizable ESCs (Nichols et al. 1998). Formation of the EPI is also dependent on NANOG expression; Nanog-null embryos fail to establish an EPI identity due to degeneration of the ICM (Mitsui et al. 2003; Silva et al. 2009). NANOG is also required for PE formation through a non-cell-autonomous mechanism (Messerschmidt and Kemler 2010). Thus, the Stat3mat−/− embryos present phenotypes similar to those from Oct4- and Nanog-null mice, supporting the requirement of STAT3 for development and maintenance of the ICM lineages.
More detailed examination of Stat3-null embryos shows an intact, CDX2-expressing TE epithelium capable of inducing a decidual reaction, an indication that both maternal and zygotic STAT3 is dispensable for TE lineage formation and maintenance (Fig. 6). This finding is consistent with previous studies showing that CDX2 promotes, but OCT4 represses, TE differentiation (Niwa et al. 2005). Stat3mat−/− E3.5 blastocysts appear morphologically normal and contain NANOG-expressing ICM cells. However, 24 h later in the E4.5 embryo, there are very few CDX2-negative cells, none of which expressed pluripotency markers. We speculate that in the absence of STAT3, ICM cells in the expanding blastocysts begin to differentiate inappropriately and fail to proliferate, an effect possibly even potentiated by the absence of STAT3, which has been shown to have anti-apoptotic activity (Hirano et al. 2000).
Molecular interaction of STAT3 with OCT4 and NANOG
It has been reported that STAT3 maintains ESC pluripotency by integrating with the core transcription circuitry OCT4/NANOG through directly regulating Klf4 (Hall et al. 2009; Niwa et al. 2009). It has also been reported that Stat3−/− ESCs are morphologically indistinguishable from wild-type ESCs, expressing both Oct4 and Nanog in 3i medium, whereas when they are transferred to medium containing LIF and serum, they rapidly differentiate (Ying et al. 2008). However, since 3i inhibitors also prevent repression of Tcf3 and Erk, it is unclear whether this shows a direct relationship with the Stat3/Oct4/Nanog circuitry (Nichols and Smith 2012).
We now provide evidence that STAT3 directly regulates Oct4 and Nanog transcription to maintain pluripotency of ESCs and iPSCs. Short-term treatment of cells with a STAT3 inhibitor leads to substantial reduction of Oct4 mRNA, suggesting that Oct4 down-regulation is a direct effect of STAT3 inhibition rather than a consequence of cell differentiation. Furthermore, consistent with the ChIP sequencing (ChIP-seq) data mapping STAT3-binding sites in ESCs (Chen et al. 2008), we now report that STAT3 directly binds to the distal enhancer of Oct4 and Nanog.
Delineation of the role of STAT3 in ICM lineage maintenance in vivo coupled with the demonstration that STAT3 directly interacts with Oct4 and Nanog to maintain pluripotent cells in vitro reveals the essential nature of this regulatory circuit at the outset of mammalian development.
Materials and methods
Cell culture and transfection
iPSCs were generated as described previously (Feng et al. 2009). Stat3 shRNA was cloned into the pSUPER-puro vector with the target sequence GATCCCCGGGCCATCCTAAGCACAAAttcaagagaTTTGTGCTTAGGATGGCCCTTTTTA. Oct4-Gfp ESCs (kindly provided by Dr. Kazuo Yamagata) or iPSCs seeded at a density of 4 × 105 cells per well in six-well plates were transfected with 4 μg of pSUPER.puro vector, shLuciferase-pSUPER-puro, or shSTAT3-pSUPER.puro. Puromycin selection (1.5 μg/mL) was applied for 3–5 d before analysis. Alkaline phosphatase staining was detected using the Alkaline Phosphatase Detection kit (Millipore) according to the manufacturer's recommendations. The specific STAT3 inhibitor STA-21 was used as previously described (Song et al. 2005).
RNA extraction, reverse transcription, and qRT-PCR
RNA was extracted 3 d after transfection, selected in puromycin using TRIzol reagent (Invitrogen), and treated with DNase (Amicon). cDNA synthesis was performed with 1 μg of total RNA and the SuperScript III kit (Invitrogen) according to the manufacturer's instructions. Endogenous mRNA levels were measured by real-time PCR analysis based on SYBR Green detection with an ABI real-time PCR machine. The sequence of real-time PCR primers is provided in Supplement Table 1. Samples were assayed in duplicate and normalized to endogenous Gapdh or Actin.
Protein extraction and Western blotting
Protein extracts were obtained using cell lysis buffer (20 mM HEPES, 400 nM NaCl, 0.5% NP-40, 10% glycerol, 1 mM DTT) with protease inhibitor (Roche) and phosphatase inhibitor. Proteins were separated by SDS-PAGE, transferred to PDVF membrane (Bio-Rad), and probed with specific primary antibody and an appropriate HRP-conjugated secondary antibody. Signals were detected using SuperSignal West Pico chemiluminescent substrate (Pierce). The following antibodies were used for Western blotting: α-STAT3 (1:1000; Santa Cruz Biotehnology, C-20), α-STAT3 (1:1000; Cell Signaling), α-pY705-STAT3 (1:500; Santa Cruz Biotechnology, B-7), α-pY705-STAT3 (1:1000; Cell Signaling), α-OCT4 (1:1000; Abcam, ab19857), α-OCT4 (1:500; Santa Cruz Biotechnology, N-19), α-NANOG (1:1000; Abcam, ab21624), and α-Tubulin (1:2500; Santa CruzBiotechnology, sc-5274).
Immunocytochemistry
Cells were seeded at 2.5 × 104 cells per well in four-well plates on gelatin-coated coverslips and cultured for 3 d. The cells were fixed in 4% paraformaldehyde for 15 min and permeabilized with 0.25% Triton X-100 for 15 min. Blocking was performed with PBS containing 0.05% Tween and 1% BSA for 30 min. Cells were reacted with primary antibody rabbit α-OCT4 (1:1000; Abcam, ab19857), rabbit α-NANOG (1:1000; Abcam, ab21624), and rabbit α-pY705-STAT3 (1:200; Cell signaling) overnight at 4°C, followed by incubation with the appropriate secondary antibodies rabbit IgG Alexa Fluor 488 (1:1000) and rabbit IG FITC (1:200) for 1 h at room temperature. Images were captured with a Zeiss AxioVision 4.7.
ChIP assays
ChIP assays with iPSCs were carried out as described previously (Loh et al. 2006). Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and formaldehyde was neutralized by addition of 0.2 M glycine. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using 5 μg of α-pY705-STAT3 (Cell Signaling). For all ChIP experiments, qRT-PCR analyses were performed using Stratagene and SYBR Green Master Mix.
Mice and genotyping
Stat3flox/flox mice were crossed with Zp3-Cre transgenic mice carrying cre recombinase under the control of the oocyte-specific Zp3 promoter (De Vries et al. 2004) to generate Zp3-Cre; Stat3flox/+ male mice. These mice were then backcrossed to Stat3flox/flox female mice, and Zp3-Cre;Stat3flox/flox females were selected. Oocytes from these females were Stat3-deleted. To generate maternal- and zygotic-deleted embryos, Zp3-Cre;Stat3flox/flox female mice were mated with Stat3+/− males; one-half of their progeny were maternal–zygotic null. Genotyping was done by PCR using DNA extracted from tails tips of 21-d-old mice. The primer pairs used to detect the presence of the Zp3-Cre transgene and the Stat3-floxed alleles were as described (Welte et al. 2003; De Vries et al. 2004). Genotyping of embryos followed a previous method (Nichols et al. 1998).
Zp3-Cre; Cdh1flox/flox female mice were mated with Cdh1+/− to generate maternal- and zygotic-deleted embryos; one-half of their progeny were maternal/zygotic null (De Vries et al. 2004). Lif−/− embryos were flushed from pregnant Lif−/− females after mating with Lif−/− males (Stewart et al. 1992).
Harvesting mouse oocytes and embryos and in vitro culture and treatment of embryos
C57BL/6 female mice were superovulated by injecting pregnant mare's serum gonadotropin (PMSG) followed by human chorionic gonadotropin (hCG). After 48 h, they were mated with male mice. Oocytes and zygotes were harvested from oviducts 17–22 h after hCG injection. Cumulus cells were removed by incubation with 0.3 mg/mL hyaluronidase (Sigma, H4272) in M2 medium. The embryos were recovered in M2 medium, washed in a few drops of KSOM (Millipore), and cultured in microdrops under mineral oil in 5% CO2 at 37°C. The embryos were collected at different stages for immunostaining. Alternatively, embryos from the two- to eight-cell stage were flushed from oviducts at 1.5 and 2.5 d post-coitum (dpc), and embryos from blastocyst stage were flushed from the uterus at 3.5 dpc.
Zygotes were cultured with neutralizing antibodies against IFN-γ and IL-6 cytokine at 2 ng/mL concentration (eBioscience) and fixed as expanded blastocysts for immunostaining. Zygotes were cultured with 5 μM JAKi (Calbiochem,), 5 μM AG490 (Tortis bioscience), and 5 μM Ruxolitinib (InvivoGen) in vitro and fixed as expanded blastocysts for immunostaining.
Immunostaining of oocytes and preimplantation embryos
Embryos were fixed in 2% paraformaldehyde in PBS for 15 min at room temperature or overnight at 4°C. They were washed in PBS with 0.1% Triton three times and permeabilized with 0.25% Triton in PBS for 1 h at room temperature. The embryos were incubated with primary antibody overnight at 4°C after blocking in PBS with 0.1% Triton and 10% FBS for 1 h at room temperature. The primary antibodies used in this study were α-pY705-STAT3 (1:100; Cell Signaling); α-STAT3 (79D7; Cell Signaling, 1:100); α-STAT3 (H-190; 1:100; Santa Cruz Biotechnology), α-OCT4 (C-10; 1:200; Santa Cruz Biotechnology); α-NANOG (1:100; Bethyl Laboratories); α-CDX2 (1:200; BioGenex), α-GATA4 (1:100; Santa Cruz Biotechnology), α-GATA6 (1:100; R&D Systems), and α-active CASPASE3 (1:100; Cell Signaling). The embryos were then stained with secondary antibodies conjugated with Alexa Fluor 488 and FITC for 1 h at room temperature. DAPI was used for nuclear staining. The embryos were analyzed by confocal microscopy using a 60× oil immersion lens. Images were taken every 2 μm through the embryo.
RNA extraction, reverse transcription, and qRT-PCR for ICM cells of early blastocysts
ICM cells were isolated from early blastocysts by immunosurgery (Solter and Knowles 1975). Total RNA of ICM cells was purified using a PicoPure RNA isolation kit (Arcturus Bioscience). The RNA was then used for cDNA synthesis with the SuperScript III kit (Invitrogen) according to the manufacturer's instructions. One microliter of cDNA was RT-preamplified with a specific set of primers using TaqMan PreAmp Master Mix kit (Life Technologies) and used for real-time PCR using an ABI PRISM7000 Real-time PCR machine.
In vitro culture of blastocysts
E3.5 embryos were flushed from the uteri of the females containing copulatory plugs. Blastocysts were independently cultured in 24-well plates in ES medium with LIF. After 5 d of culture, photographs of the cultured embryos were taken. The outgrowths were fixed and stained with antibodies to pSTAT3 and OCT4. Their genotypes were finally determined by PCR (Nichols et al. 1998).
Acknowledgments
We thank Anne Ferguson-Smith of Cambridge University for helpful discussions and reading of the manuscript. This work was supported by the Cancer Science Institute of Singapore; start-up funds from the DPRT office, MACC, and YLL School of Medicine of the National University of Singapore; and grants from BMRC (SICS-09/1/002), NMRC (IRG09may057), and Ministry of Education of Singapore (MOE2010-T2-1-084) (all to X.Y.F.). Work at Indiana University was supported by an NIH grant to X.Y.F. (R01 CA125568). A*STAR IMB supported the work of D.S., B.B.K., D.M.M. and C.L.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.221176.113.
References
- Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T 1994. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77: 63–71 [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeuf H, Hauss C, Graeve FD, Baran N, Kedinger C 1997. Leukemia inhibitory factor-dependent transcriptional activation in embryonic stem cells. J Cell Biol 138: 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr 1997. STATs and gene regulation. Science 277: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr, Kerr IM, Stark GR 1994. Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421 [DOI] [PubMed] [Google Scholar]
- De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB 2004. Maternal β-catenin and E-cadherin in mouse development. Development 131: 4435–4445 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. 2009. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol 11: 197–203 [DOI] [PubMed] [Google Scholar]
- Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE Jr 1990. ISGF3, the transcriptional activator induced by interferon α, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci 87: 8555–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XY, Schindler C, Improta T, Aebersold R, Darnell JE Jr 1992. The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci 89: 7840–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18: 675–685 [DOI] [PubMed] [Google Scholar]
- Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, et al. 2009. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell 5: 597–609 [DOI] [PubMed] [Google Scholar]
- Hawkins K, Mohamet L, Ritson S, Merry CL, Ward CM 2012. E-cadherin and, in its absence, N-cadherin promotes Nanog expression in mouse embryonic stem cells via STAT3 phosphorylation. Stem Cells 30: 1842–1851 [DOI] [PubMed] [Google Scholar]
- Hirano T, Ishihara K, Hibi M 2000. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19: 2548–2556 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MS, Kitchen JR, Wang X, Threat TA, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, Cui Y, et al. 2000. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development 127: 1737–1749 [DOI] [PubMed] [Google Scholar]
- Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, Zinkernagel R, Bluethmann H, Kohler G 1994. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature 368: 339–342 [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F 1999. The transcription factor GATA6 is essential for early extraembryonic development. Development 126: 723–732 [PubMed] [Google Scholar]
- Li M, Sendtner M, Smith A 1995. Essential function of LIF receptor in motor neurons. Nature 378: 724–727 [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Martin GR 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Kemler R 2010. Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol 344: 129–137 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Wiese S, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Yoshida K, Kishimoto T, Sendtner M, Taga T 1999. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci 19: 5429–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K 1998. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell 93: 397–409 [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A 2012. Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol 4: a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Chambers I, Smith A 1994. Derivation of germline competent embryonic stem cells with a combination of interleukin-6 and soluble interleukin-6 receptor. Exp Cell Res 215: 237–239 [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Nichols J, Chambers I, Taga T, Smith A 2001. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development 128: 2333–2339 [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123: 917–929 [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K 2009. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460: 118–122 [DOI] [PubMed] [Google Scholar]
- Raz R, Lee CK, Cannizzaro LA, d'Eustachio P, Levy DE 1999. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci 96: 2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, et al. 1998. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell 93: 373–383 [DOI] [PubMed] [Google Scholar]
- Rothstein JL, Johnson D, DeLoia JA, Skowronski J, Solter D, Knowles B 1992. Gene expression during preimplantation mouse development. Genes Dev 6: 1190–1201 [DOI] [PubMed] [Google Scholar]
- Schindler C, Darnell JE Jr 1995. Transcriptional responses to polypeptide ligands: The JAK–STAT pathway. Annu Rev Biochem 64: 621–651 [DOI] [PubMed] [Google Scholar]
- Schindler C, Shuai K, Prezioso VR, Darnell JE Jr 1992. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257: 809–813 [DOI] [PubMed] [Google Scholar]
- Scholer HR, Balling R, Hatzopoulos AK, Suzuki N, Gruss P 1989a. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J 8: 2551–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P 1989b. A family of octamer-specific proteins present during mouse embryogenesis: Evidence for germline-specific expression of an Oct factor. EMBO J 8: 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A 2009. Nanog is the gateway to the pluripotent ground state. Cell 138: 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336: 688–690 [DOI] [PubMed] [Google Scholar]
- Solter D, Knowles BB 1975. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci 72: 5099–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Wang R, Wang S, Lin J 2005. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci 102: 4700–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ 1992. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359: 76–79 [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132: 2093–2102 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S 1997. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci 94: 3801–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA 2010. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6: 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, et al. 1995. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development 121: 1283–1299 [DOI] [PubMed] [Google Scholar]
- Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, Yin Z, Kano A, Iwamoto Y, Li E, Craft JE, et al. 2003. STAT3 deletion during hematopoiesis causes Crohn's disease-like pathogenesis and lethality: A critical role of STAT3 in innate immunity. Proc Natl Acad Sci 100: 1879–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336: 684–687 [DOI] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A 2007. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134: 3827–3836 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Ueda J, Mizutani E, Saitou M, Wakayama T 2010. Survival and death of epiblast cells during embryonic stem cell derivation revealed by long-term live-cell imaging with an Oct4 reporter system. Dev Biol 346: 90–101 [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292 [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Chambers I, Nichols J, Smith A, Saito M, Yasukawa K, Shoyab M, Taga T, Kishimoto T 1994. Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech Dev 45: 163–171 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Taga T, Saito M, Suematsu S, Kumanogoh A, Tanaka T, Fujiwara H, Hirata M, Yamagami T, Nakahata T, et al. 1996. Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc Natl Acad Sci 93: 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE Jr 1994. Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264: 95–98 [DOI] [PubMed] [Google Scholar]