Female-specific repression of msl2 mRNA in Drosophila provides a paradigm for control of gene expression by RNA-binding complexes. Sex-lethal (SXL) promotes Drosophila female sex determination by regulating msl2 splicing and translation. Using a two-step purification method, Graindorge et al. now identify Held-Out Wings (HOW) as a component of the msl2 5′ UTR-associated complex. Surprisingly, rather than affecting SXL-mediated regulation of splicing or translation, HOW facilitates nuclear retention of msl2 mRNA by SXL. Thus, this study uncovers a novel function for RNA-binding protein SXL in mRNA transport.
Keywords: Sex-lethal, Held-Out-Wings, nuclear mRNA retention, msl2
Abstract
Female-specific repression of male-specific-lethal-2 (msl2) mRNA in Drosophila melanogaster provides a paradigm for coordinated control of gene expression by RNA-binding complexes. Repression is orchestrated by Sex-lethal (SXL), which binds to the 5′ and 3′ untranslated regions (UTRs) of the mRNA and inhibits splicing in the nucleus and subsequent translation in the cytoplasm. Here we show that SXL ensures msl2 silencing by yet a third mechanism that involves inhibition of nucleocytoplasmic transport of msl2 mRNA. To identify SXL cofactors in msl2 regulation, we devised a two-step purification method termed GRAB (GST pull-down and RNA affinity binding) and identified Held-Out-Wings (HOW) as a component of the msl2 5′ UTR-associated complex. HOW directly interacts with SXL and binds to two sequence elements in the msl2 5′ UTR. Depletion of HOW reduces the capacity of SXL to repress the expression of msl2 reporters without affecting SXL-mediated regulation of splicing or translation. Instead, HOW is required for SXL to retain msl2 transcripts in the nucleus. Cooperation with SXL confers a sex-specific role to HOW. Our results uncover a novel function of SXL in nuclear mRNA retention and identify HOW as a mediator of this function.
Post-transcriptional control of gene expression is an important layer of regulation used in many biological situations to promote rapid and often reversible changes in the protein content of a cell. All steps of mRNA metabolism are subjected to regulation—from the selection of specific exons through alternative splicing to alternative polyadenylation, mRNA editing, export, localization, translation, and stability. Tight and coordinated regulation of these processes is mediated by the assembly of specific RNA-binding factors (proteins and/or noncoding RNAs [ncRNAs]) on the regulated transcripts, forming messenger ribonucleoprotein particles (mRNPs) whose composition changes during the life of the mRNA (for review, see Glisovic et al. 2008; Gebauer et al. 2012).
A paradigmatic example of coordinated regulation is provided by the sex-specific expression of male-specific-lethal-2 (msl2) in Drosophila melanogaster. MSL2 is the limiting subunit of the dosage compensation complex (DCC), which functions in males to hypertranscribe the single X chromosome in order to equalize its output to that of the two female X chromosomes. In females, msl2 expression needs to be repressed for viability (Kelley et al. 1995), and this is achieved by the binding of Sex-lethal (SXL), a female-specific protein, to uridine stretches in both the 5′ and 3′ untranslated regions (UTRs) of the msl2 transcript (for review, see Graindorge et al. 2011). To enforce efficient msl2 silencing, SXL targets multiple steps in the gene expression cascade. First, SXL acts at the splicing level by promoting retention of an intron in the 5′ UTR of msl2 pre-mRNA (Merendino et al. 1999; Forch et al. 2001). The retained intron contains SXL-binding sites that are required for subsequent steps of repression. After msl2 mRNA export into the cytoplasm, SXL coordinates its translational repression by targeting early steps of translation initiation (Bashaw and Baker 1997; Kelley et al. 1997; Gebauer et al. 1998). SXL bound to the 3′ UTR recruits Upstream of N-Ras (UNR) to inhibit ribosome recruitment (Abaza et al. 2006; Duncan et al. 2006, 2009). SXL bound to the 5′ UTR intron interferes with ribosomal scanning by a mechanism that involves ribosome recognition of an upstream AUG (Beckmann et al. 2005; Medenbach et al. 2011). Both mechanisms synergize to achieve full msl2 translational repression and are unlikely to involve simple steric hindrance because other RNA-binding proteins recognizing the same cis-regulatory elements cannot repress msl2 expression (Grskovic et al. 2003; Medenbach et al. 2011).
To gain insight into the mechanisms underlying msl2 regulation, we focused on the 5′ UTR, as this region is required for the control of splicing and translation. Using a two-step purification method termed GRAB (GST pull-down and RNA affinity binding), we identified the protein Held-Out-Wings (HOW) as a component of the msl2 5′ UTR mRNP. HOW interacts with SXL directly and binds to defined sequence elements in the 5′ UTR female-specific intron. HOW participates in msl2 5′ UTR-mediated regulation, but its depletion surprisingly does not affect splicing or translational control. Instead, HOW facilitates nuclear mRNA retention by SXL. These data uncover a novel function for SXL in nuclear mRNA retention and identify HOW as a cofactor in this function.
Results
Identification of candidate SXL cofactors for 5′ UTR-mediated regulation of msl2
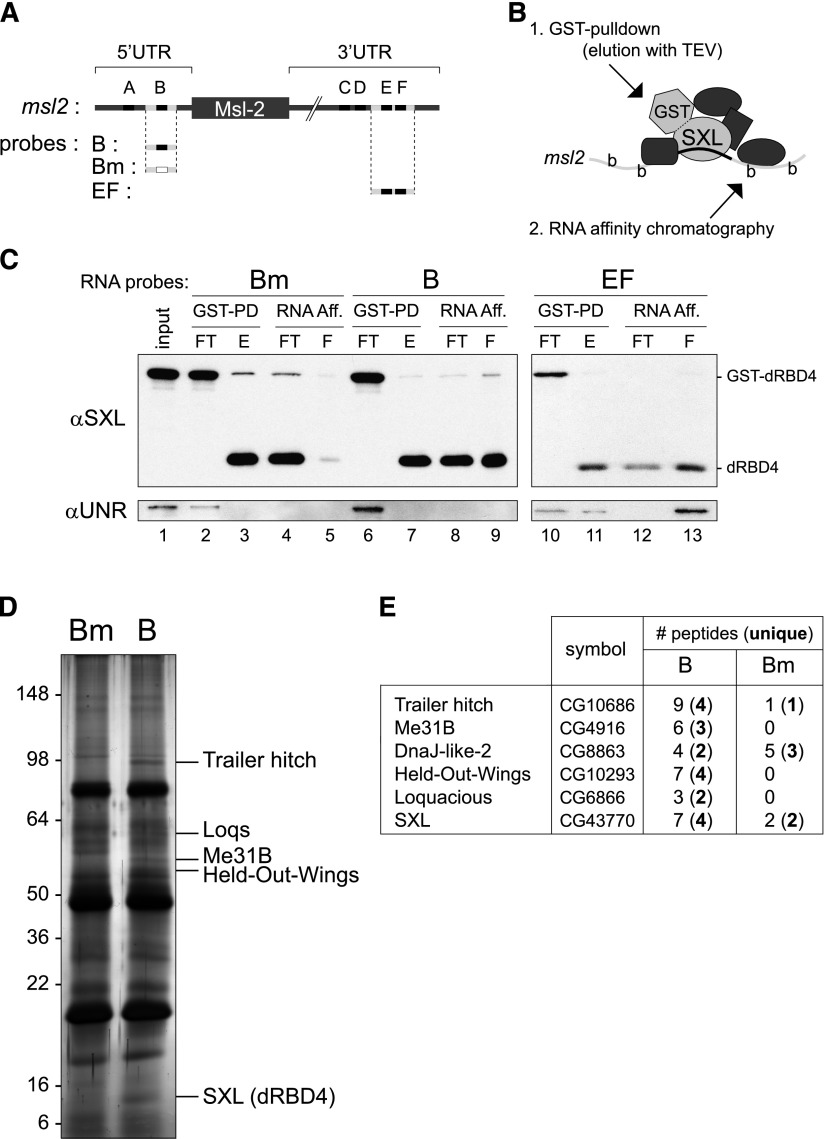
The female msl2 transcript contains long 5′ and 3′ UTRs (626 and 1047 nucleotides [nt], respectively). The 5′ UTR includes a sex-specific facultative intron with stretches of uridines located close to the splice sites that serve as SXL-binding sites, while the 3′ UTR contains a cluster of four SXL-binding sites near the 3′ end (Fig. 1A, sites A–F). Previous mutational studies have reduced the sequences required for translational repression to 70 nt in the 5′ UTR, including site B, and 46 nt in the 3′ UTR, including sites E and F (Fig. 1A, regions B and EF; Gebauer et al. 2003). Similarly, a fragment of SXL consisting of the RNA-binding domains and a 7-amino-acid extension is fully functional in translational repression (fragment dRBD4) (Grskovic et al. 2003). To identify SXL cofactors involved in 5′ UTR-mediated regulation, we aimed to isolate the SXL:msl2 RNP associated with the 5′ UTR. We optimized a two-step methodology, termed GRAB, designed to distinguish 5′ from 3′ UTR-associated complexes. Biotinylated RNA corresponding to region B (5′ UTR) was incubated with Drosophila embryo extracts in the presence of recombinant GST-dRBD4 in conditions competent for translational repression (see the Materials and Methods for details). Region EF was carried as a specificity control, as this region is known to bind SXL and the cofactor UNR (Abaza et al. 2006; Duncan et al. 2006). In addition, a fragment of RNA in which SXL-binding site B had been mutated was used as a negative control (Fig. 1A, Bm probe). After complex formation, the SXL:msl2 RNPs were selected as depicted in Figure 1B. First, GST pull-down was used to enrich for SXL-containing complexes, the GST moiety was separated by TEV cleavage, and the eluted complexes were subjected to a second round of purification using streptavidin magnetic beads. Proteins associated with the biotinylated RNAs were then eluted with Laemmli buffer. The GRAB procedure was monitored by Western blot against SXL and UNR (Fig. 1C). The results showed that the procedure is specific, as SXL was efficiently selected with the B and EF RNAs but not with the Bm RNA (Fig. 1C, cf. lanes 9,13 and 5). Furthermore, UNR was found associated with EF but not with B or Bm RNAs, indicating that GRAB enables differential isolation of cofactors associated with the 5′ or 3′ RNPs.
Figure 1.
GRAB was used to identify components of the silenced SXL:msl2 mRNP. (A) Schematic representation of msl2 mRNA and the RNA fragments used in the GRAB procedure. SXL-binding sites are shown as black boxes (A–F); Bm RNA contains a substitution of U16 by (CU)8, which abrogates SXL binding (white box). (B) Cartoon of the GRAB purification protocol. Recombinant GST-SXL (GST-dRBD4 derivative) and biotinylated RNA were incubated with Drosophila embryo extracts. The complex was first selected by GST pull-down and eluted with TEV protease, followed by RNA affinity chromatography using streptavidin beads. (C) The GRAB procedure was monitored by Western blot against SXL and UNR. (FT) Flow-through; (E) eluate; (F) final bound material. The following amounts were loaded for each fraction (percentage of reaction mix): input (0.05%), FTGST (0.05%), EGST (1.25%), FTRNA (1.25%), ERNA (5.8%). (D) Proteins selected by GRAB were separated by SDS-PAGE, silver-stained, and identified by mass spectrometry. (E) List of identified proteins. Proteins identified with less than two unique peptides were not considered for further studies.
To identify factors associated with the 5′ UTR:SXL RNP, PAGE gels were silver-stained and examined for proteins enriched in the B eluate versus the Bm eluate, which were excised from the gel and identified by mass spectrometry (Fig. 1D). Figure 1E lists the identified proteins together with the corresponding number of peptides. In addition to the bait protein dRBD4, Trailer-hitch (Tral), Me31B, HOW, and Loquacious (Loqs) were specifically associated with the 5′ UTR:SXL RNP. All of these are RNA-binding proteins implicated in mRNA metabolism, and some, such as Tral and the RNA helicase Me31B, have been found previously to work in association (Wilhelm et al. 2005). We selected Tral and HOW for further studies, as they were identified with the highest number of peptides.
HOW cooperates with SXL to repress msl2 expression via the 5′ UTR
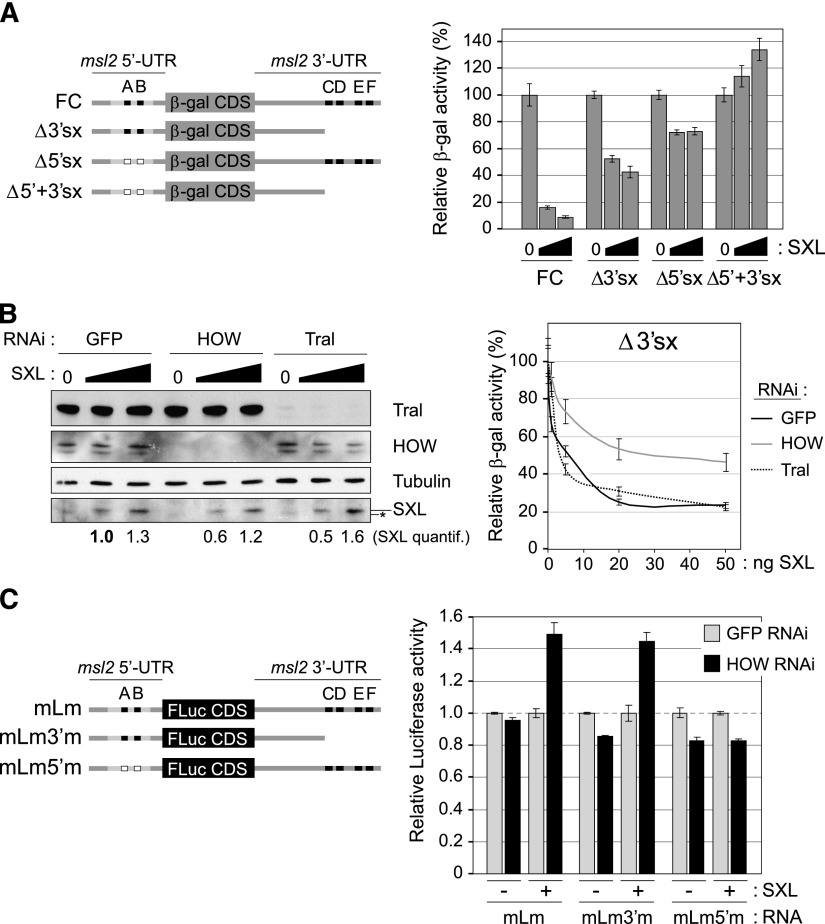
To test whether Tral and HOW were involved in msl2 regulation, we first recapitulated SXL-mediated msl2 repression in Drosophila SL2 cells. These cells lack endogenous SXL and are thus dependent on the addition of exogenous SXL for repression, enabling dose response analysis. To assess UTR-dependent msl2 regulation, we cotransfected SXL with msl2 reporters containing the β-galactosidase coding sequence (CDS) flanked by the msl2 UTRs in either their wild-type form or versions lacking the SXL-binding sites (Fig. 2A, left panel). This system recapitulates bona fide repression by SXL, as repression is efficient and specific (Fig. 2A, cf. FC and Δ5′+3′sx; Bashaw and Baker 1997; Duncan et al. 2006). Moreover, the system allows uncoupling of 5′ from 3′ UTR-mediated regulation by SXL (Fig. 2A, Δ3′sx and Δ5′sx).
Figure 2.
HOW cooperates with SXL to repress msl2 expression via the 5′ UTR. (A) Reconstitution of SXL-dependent msl2 repression in SL2 cells. (Left panel) msl2-βgal reporter constructs used in this study (Bashaw and Baker 1997). (Right panel) Relative levels of β-gal activity upon cotransfection of the reporters with increasing amounts of a SXL-encoding plasmid. Renilla luciferase was cotransfected as an internal control. The data were normalized for Renilla expression and plotted relative to the β-gal activity in the absence of SXL. (B) Depletion of HOW impairs SXL-mediated repression. (Left panel) Assessment of the depletion efficiencies by Western blot. A quantification of the SXL levels is shown at the bottom (normalized by Tubulin). The asterisk denotes a nonspecific band that migrates slightly faster than SXL. (Right panel) Repression of the Δ3′sx construct by SXL upon depletion of HOW and Tral; GFP depletion was used as a negative control. β-Gal activity was normalized by Renilla expression and corrected for the levels of the reporter RNA. A representative example of three independent experiments is shown. Error bars correspond to standard deviations of triplicate measurements. (C) The effect of HOW in msl2 silencing is mediated by the 5′ UTR. (Left panel) msl2-firefly reporter constructs used in this study. (Right panel) Reporter constructs were transfected as in A in either control cells (GFP RNAi) or HOW-depleted cells (HOW RNAi). Firefly luciferase activity was normalized by Renilla expression. The data were plotted relative to the activity obtained in control GFP RNAi cells in each condition. A representative example of four different experiments is shown. Error bars correspond to standard deviations of triplicate measurements. Considering the four experiments, P-values of 0.019 and 0.003 were obtained for the differences between GFP and HOW RNAi in the presence of SXL for the mLm and mLm3′m constructs, respectively.
We then used the Δ3′sx construct to assess 5′ UTR-dependent repression by SXL in the presence or absence of HOW and Tral. These genes were knocked down by RNAi, using RNAi against GFP as a negative control (Fig. 2B). The results showed that knockdown of HOW, but not Tral, impaired SXL-mediated repression. This effect was not due to a change in reporter RNA stability, according to the RNA normalization of the data (Fig. 2B) and quantification of transfected RNAs in independent experiments (Figs. 3, 4; Supplemental Fig. S1). Importantly, derepression of the reporter was not due to changes in SXL protein levels upon HOW depletion (Fig. 2B; data not shown). To test whether the effect of HOW was specific to the msl2 5′ UTR and was independent of the CDS or the 3′ UTR, we used firefly luciferase reporter constructs containing or lacking SXL-binding sites in either UTR. Loss of SXL repression was observed only when SXL-binding sites were present in the 5′ UTR (Fig. 2C), indicating that HOW cooperates with SXL to repress msl2 expression via the 5′ UTR.
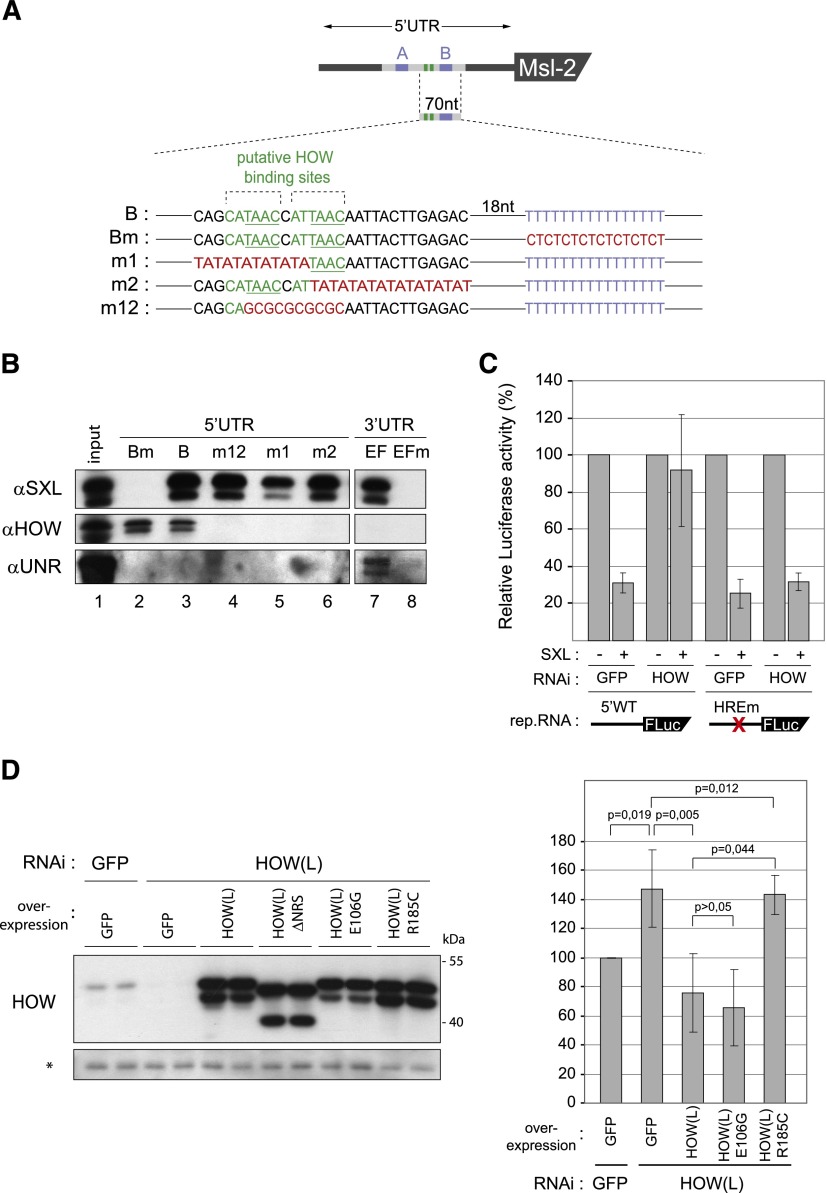
Figure 3.
Identification of the HREs in the 5′ UTR of msl2. (A) Schematic diagram of the RNAs used to map the HOW-binding sites. The female-specific alternative intron is depicted in gray. SXL-binding sites and putative HOW-binding sites are indicated in blue and green, respectively. The most invariant nucleotides of the HOW-binding site are underlined. Mutated sequences are detailed in red. (B) HOW binds two adjacent HREs in the msl2 5′ UTR. Biotinylated probes were incubated with Drosophila embryo extracts in the presence of recombinant His-SXL(ΔGN) and pulled down with streptavidin Dynabeads. SXL(ΔGN) is an extended version of dRBD4 containing the full C-terminal domain. The association of SXL, HOW, and UNR was detected by Western blot. A representative example of three independent experiments is shown. (C) HOW binding to the HREs mediates msl2 silencing. Constructs to assess 5′ UTR-mediated repression (mLm3′m) containing wild-type or mutated HREs were transfected in control (GFP) or HOW-depleted cells, together with 25 ng of a SXL-expressing plasmid (+) or empty vector (−). Firefly luciferase activity was normalized for cotransfected Renilla expression and corrected for the levels of the reporter RNA. The data were plotted relative to the firefly luciferase activity in the absence of SXL. Average values and standard deviations of four independent experiments are shown. The P-value for the loss of derepression after HOW depletion in the absence of the HREs was 0.043. (D) Overexpression of HOW(L), but not a mutant lacking RNA-binding activity, restores msl2 silencing in HOW-depleted cells. The following proteins were overexpressed in HOW-depleted cells: wild-type HOW(L), point mutants with impaired dimerization (E106G) or RNA-binding (R185C) activities, and GFP as control. (Left panel) The expression levels were assessed by Western blot using αHOW antibodies (the asterisk denotes an irrelevant band). mLm3′m-, Renilla-, and SXL-expressing plasmids were cotransfected into these cells as well as into control GFP RNAi cells. (Right panel) Firefly luciferase values were normalized as in C and were plotted relative to the activity obtained in control GFP RNAi cells. Average values and standard deviations of four independent experiments are shown.
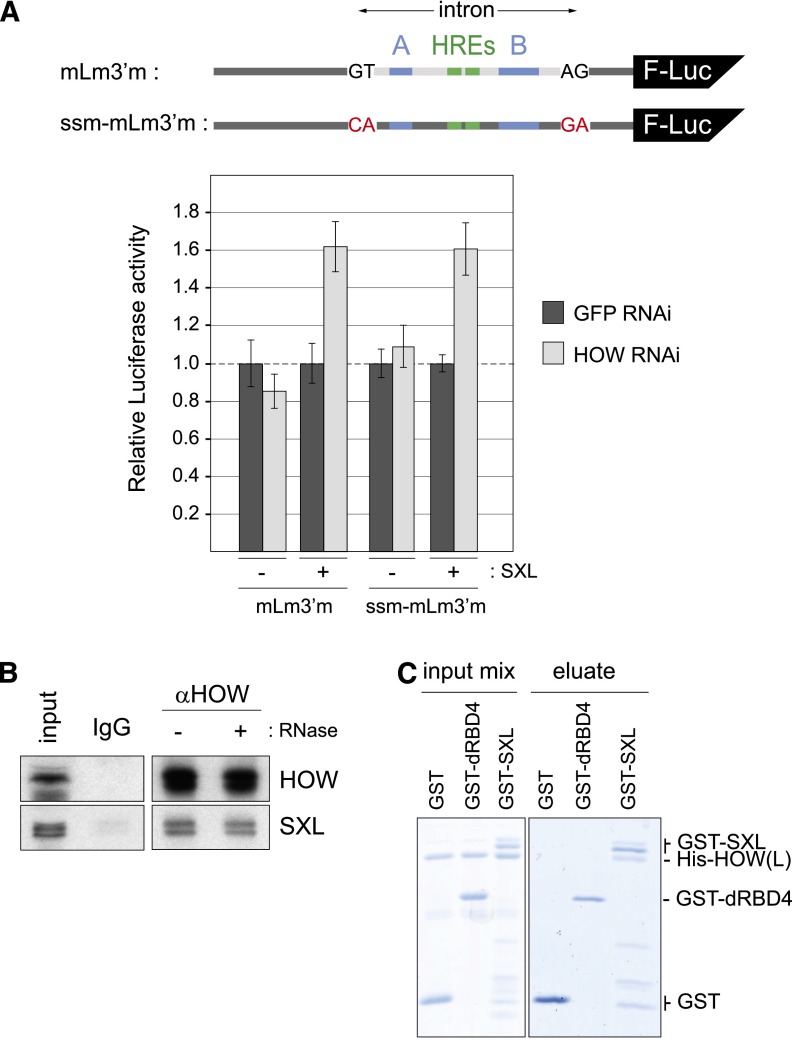
Figure 4.
SXL confers a sex-specific role for HOW in msl2 regulation. (A) Intron retention does not suffice to confer a role for HOW in msl2 silencing. A ssm construct containing the mutations indicated in red was cotransfected with Renilla luciferase as in Figure 2C and compared with a construct lacking these mutations. (Blue) SXL-binding sites; (green) HREs. A representative example of four different experiments is shown. Error bars correspond to standard deviations of triplicate measurements. Considering the four experiments, P-values of 0.023 and 0.006 were obtained for the differences between GFP and HOW RNAi in the presence of SXL for the mLm3′m and ssm-mLm3′m constructs, respectively. (B) SXL and HOW interact in the absence of RNA. HOW was immunoprecipitated from Drosophila embryo extracts, and the presence of SXL in the pellet was tested by Western blot. Immunoprecipitation with nonspecific IgG was used as a negative control. (C) SXL and HOW interact directly. Recombinant His-HOW(L) was incubated with GST, GST-dRBD4, or GST-SXL proteins. After GST pull-down, the precipitated proteins were separated by SDS-PAGE and stained with Colloidal Blue.
HOW belongs to the STAR (signal transduction and activation of RNA) protein family, whose members are involved in many aspects of RNA metabolism (Volk et al. 2008). Interestingly, the how gene encodes two protein isoforms generated by alternative splicing: HOW(L) and HOW(S). Both proteins contain a maxi-KH domain flanked by two Quaking domains but diverge at their C termini: HOW(L) carries an extension containing a nuclear retention signal (NRS) that is lacking in HOW(S) (Supplemental Fig S2A). HOW(M), a third isoform lacking the NRS, has been predicted from genomic data, although it was not detected in biological samples. As HOW is detected as a doublet in SL2 cells (Fig. 2B), we wished to determine whether these two bands corresponded to different HOW isoforms. We designed dsRNA fragments to specifically target the three isoforms of HOW by RNAi and observed that both bands disappeared by HOW(L) targeting while remaining unaffected when HOW(S) or HOW(M) were targeted (Supplemental Fig. S2B). We conclude that only HOW(L) is expressed at detectable levels in SL2 cells. In addition, assessment of HOW subcellular localization indicated that HOW was predominantly nuclear (Supplemental Fig. S2C; data not shown). These results suggest that nuclear HOW(L) is involved in msl2 regulation.
HOW mediates msl2 regulation by binding to specific elements in the 5′ UTR intron
Recent high-throughput studies have identified the binding sites of two HOW orthologs: Caenorhabditis elegans Gld-1 and mammalian QKI (Hafner et al. 2010; Jungkamp et al. 2011). The 5′ UTR of msl2 contains two such sites in tandem, located in the female-specific facultative intron upstream of SXL-binding site B (Fig. 3A). To test whether these sites were indeed required for HOW binding, we performed RNA pull-down experiments. The RNA probes were biotinylated and mixed with Drosophila embryo extracts supplemented with recombinant SXL, and the presence of endogenous HOW was tested by Western blot (Fig. 3B). We tested RNAs containing mutations in the putative HOW-binding sites (Fig. 3A, lines m1, m2, and m12) and the SXL-binding site (Fig. 3A, line Bm) and compared them with a wild-type 5′ UTR (Fig. 3A, line B). RNA corresponding to the 3′ EF region of msl2 was carried as a control (Fig. 3B, EF lane) as well as its mutated derivative lacking SXL-binding sites (Fig. 3B, EFm lane). As expected, SXL binding to the 5′ and 3′ UTR probes was lost after mutation of the SXL-binding sites (Fig. 3B, cf. lanes 2,3 and 7,8). Furthermore, as previously reported, UNR bound to the 3′ UTR in a SXL-dependent manner (Fig. 3B, lanes 7–8) and was not found to bind to the 5′ UTR (Fig. 3B, lane 3; Abaza et al. 2006). Thus, the patterns of SXL and UNR binding support the specificity of the experimental system. HOW was detected specifically at the 5′ UTR and did not bind to the 3′ UTR (Fig. 3B, lanes 3,7). Contrary to UNR, binding of HOW was independent of SXL, as elimination of the SXL-binding sites did not affect HOW binding (Fig. 3B, lanes 2,3). Binding of HOW, however, required the presence of both putative HOW-binding sites in the 5′ UTR (Fig. 3B, lanes 4–6). These results confirm the identity of the HOW-binding sites. For consistency with published reports, we call these elements HOW response elements (HREs).
We next wanted to test whether the HREs mediate msl2 repression by HOW. To this aim, a reporter construct with mutated HREs was cotransfected with SXL in order to monitor 5′ UTR-mediated repression. Transfections were performed in either control cells (GFP RNAi) or cells depleted of HOW (HOW RNAi) (Fig. 3C). Consistent with previous results, depletion of HOW reduced the capacity of SXL to repress the expression of the msl2 reporter containing the HREs (Fig. 3C, first through fourth bars). Although we initially expected that mutation of the HOW-binding sites would be equivalent to depletion of HOW, we did not observe any derepression of the HREm reporter (Fig. 3C, fifth and sixth bars). Significantly, however, mutation of the HREs restored SXL-mediated repression in HOW-depleted cells (Fig. 3C, seventh and eighth bars). This indicates that in the absence of the HREs, HOW is no longer required for SXL-mediated regulation, and therefore the effect of HOW is mediated by its binding to the HREs. The results further suggest that HOW competes with a factor that binds to the HREs to promote msl2 expression (see the Discussion).
Recent evidence indicates that phosphorylation of HOW facilitates its homodimerization and enhances its RNA-binding ability (Nir et al. 2012). The fact that two HREs are required for efficient binding of HOW to msl2 mRNA suggests that HOW could bind as a dimer. To test whether dimer formation is important for HOW-mediated control of msl2 and confirm that HOW binding to RNA is required for regulation, we tested the ability of wild-type HOW(L) or mutant derivatives unable to dimerize (E106G) (Nir et al. 2012) or bind to RNA (R185C) (Nabel-Rosen et al. 2005) to restore msl2 repression upon overexpression in HOW-depleted cells. While all HOW derivatives were expressed at similar levels (Fig. 3D, left panel), only wild-type HOW(L) and the E106G mutant were able to restore efficient repression (Fig. 3D, right panel). These results support our previous conclusion that RNA binding by HOW(L) is required for msl2 regulation and suggest that dimerization is dispensable for HOW activity on msl2 silencing.
SXL confers a sex-specific role for HOW in msl2 regulation
Depletion of HOW does not increase the expression of msl2 reporters in the absence of SXL (Fig. 2C), suggesting that HOW has no role in msl2 expression on its own, but rather that SXL conveys a sex-specific role for HOW in msl2 regulation. No differences could be observed in HOW expression in male and female flies or cells, indicating that sex specificity is not driven by HOW expression (Supplemental Fig. S3). Because the HREs are located in the female-specific intron, whose retention depends on SXL (Gebauer et al. 1998; Merendino et al. 1999), HOW function requires SXL-mediated intron retention. The requirement of SXL could be simply explained by the retention of this intron. Alternatively, SXL and HOW could further cooperate to regulate msl2 via specific protein–protein interactions. To address this question, we evaluated the repression of a reporter containing mutated splice sites so that intron retention was uncoupled from the presence of SXL (Fig. 4A, ssm [splice site mutant] construct). The results showed that the effect of HOW was still SXL-dependent in the ssm RNA, indicating that intron retention is not sufficient and that SXL and HOW act together to repress msl2 expression.
To test interactions between HOW and SXL, we performed coimmunoprecipitation experiments and found that endogenous SXL is associated with endogenous HOW in an RNA-independent fashion (Fig. 4B). Furthermore, GST pull-down analysis indicated that SXL and HOW interact directly (Fig. 4C). The RNA-binding domains of SXL, contained in the construct dRBD4, are not sufficient to mediate this interaction. As dRBD4 is fully functional in translational repression, these data suggest that HOW affects processes other than translation.
HOW cooperates with SXL to promote nuclear mRNA retention
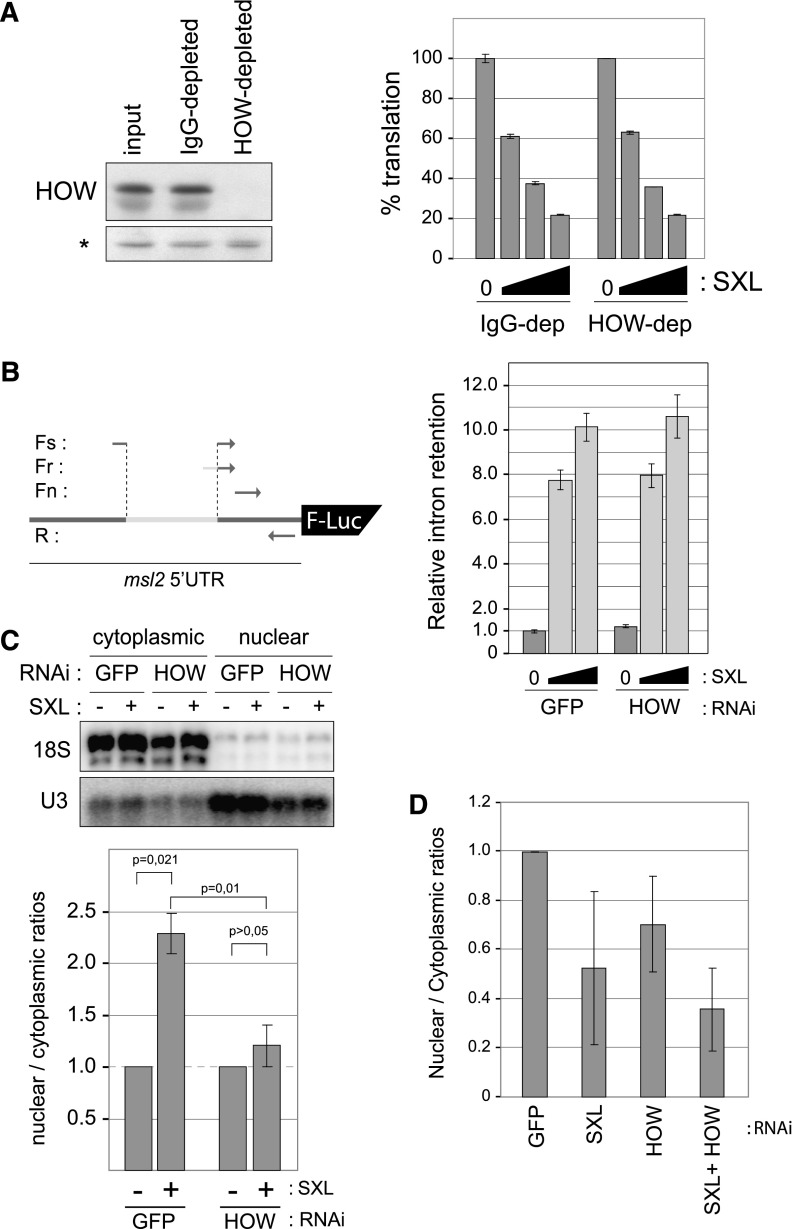
We next analyzed the molecular mechanism by which HOW and SXL cooperate to repress msl2 expression. Although HOW has been shown to regulate mRNA stability (Volk et al. 2008), our results ruled out an effect of HOW on mRNA steady-state levels (Fig. 2B; Supplemental Fig. S1). As mentioned above, indirect evidence suggested that HOW was not involved in translational repression. To test this directly, we depleted HOW from translation-competent embryo extracts and assessed the ability of SXL to repress the translation of msl2 reporters in these extracts (Gebauer and Hentze 2007). We used untreated and mock-depleted extracts as controls. The results showed no differences in SXL-mediated translational repression upon depletion of HOW from the extracts, arguing that HOW is not directly involved in msl2 translational repression in vitro (Fig. 5A).
Figure 5.
Effect of HOW on the metabolism of msl2 reporters. (A) Depletion of HOW does not affect the ability of SXL to repress translation. Embryo extracts were either untreated, mock-depleted (IgG), or HOW-depleted. (Left panel) The efficiency of depletion was assessed by Western blot. Detection of a nonrelevant protein (*) is shown as a control. (Right panel) The ability of dRBD4 to repress the translation of AUG-BLEF mRNA (Medenbach et al. 2011) was tested in these extracts. dRBD4 was used at 5×, 20×, and 50× molar excess over the mRNA. Renilla mRNA was cotranslated as an internal control. Firefly luciferase was corrected for Renilla expression, and the data were plotted as the percentage of translation in the absence of SXL. Similar results were obtained using full-length SXL or mLm3′m reporter RNA (not shown). Error bars represent the standard deviation of triplicate measurements. (B) HOW is dispensable for SXL-mediated intron retention. (Left panel) Strategy for the amplification of the msl2 splicing variants by qPCR. (Right panel) SL2 cells were depleted of HOW or mock-depleted (GFP). Cells were transfected with mLm3′m- and SXL-expressing plasmids, and the ratio of intron-containing versus spliced reporter mRNAs was calculated for each sample. The ratio obtained in the absence of SXL in the GFP RNAi condition was set to 1. A representative example of two independent experiments is shown. Error bars correspond to standard deviations of triplicate measurements. (C) HOW cooperates with SXL to promote nuclear mRNA retention. (Top panel) Assessment of correct nucleo-cytoplasmic separation by Northern blot. (Bottom panel) Cells were depleted of HOW or mock-depleted (GFP) and transfected with 25 or 50 ng of a SXL-expressing plasmid together with plasmids expressing mLm3′mut and Renilla reporters. RNA was then extracted from the nuclear and cytoplasmic fractions, and the amount of the firefly luciferase reporter was quantified by qPCR and normalized by cotransfected Renilla levels. The data were plotted as the ratio between the nuclear and cytoplasmic RNA levels, which was set to 1 in the absence of SXL. Average values and standard deviations of four experiments are shown. (D) Depletion of SXL and/or HOW from female Kc cells reduces nuclear accumulation of endogenous msl2 mRNA. The average of two independent experiments is shown.
HOW has been shown to modulate alternative splicing together with the splicing factor Crooked-Neck (Crn) (Rodrigues et al. 2012), raising the possibility that it may affect msl2 splicing in association with SXL. To test whether HOW cooperates with SXL to promote retention of the msl2 5′ UTR intron, we measured intron retention in cells depleted of HOW, carrying GFP depletion as a negative control. We employed quantitative PCR (qPCR) using the strategy depicted in the left panel of Figure 5B. The spliced variant of the mRNA was detected using a primer that spanned the exon–exon junction (Fig. 5B, Fs), while the nonspliced isoform was amplified with a primer that spanned the intron–exon boundary (Fig. 5B, Fr). Oligonucleotides mapping entirely to the constitutive exon were used for normalization (Fig. 5B, Fn). As expected, the presence of SXL increased intron retention (Fig. 5B, GFP). Retention did not decrease upon HOW depletion, indicating that HOW is dispensable for SXL-mediated intron retention (Fig. 5B, HOW).
Finally, we tested whether HOW promoted nuclear mRNA retention, sequestering msl2 transcripts from cytoplasmic translation. We examined the nucleo-cytoplasmic distribution of reporter mRNAs by qPCR upon SXL transfection and evaluated changes in this distribution upon HOW depletion. Correct nucleo-cytoplasmic separation was monitored by visualizing the distribution of the U3 snoRNA and 18S rRNA, which are located in the nucleus and cytoplasm, respectively (Fig. 5C, top panel). Strikingly, SXL promoted nuclear mRNA retention, an activity not previously reported for SXL (Fig. 5C, bottom panel, GFP). Importantly, depletion of HOW abrogated this retention (Fig. 5C, bottom panel, HOW). To confirm this novel activity of SXL, the subcellular localization of msl2 reporter transcripts was directly visualized by in situ hybridization (Supplemental Fig. S4). In the absence of SXL, reporter transcripts were observed in both the nucleus and the cytoplasm (Supplemental Fig. S4A–F). However, addition of SXL resulted in an almost strictly nuclear localization of msl2 transcripts (Supplemental Fig. S4G–L). To evaluate the role of SXL in nuclear mRNA retention in a more physiological setting, we tested the distribution of endogenous msl2 mRNA in female Kc cells, which express endogenous SXL. We found that nuclear localization of msl2 mRNA decreased upon depletion of either SXL, HOW, or both (Fig. 5D). Altogether, these data indicate that HOW cooperates with SXL to promote nuclear retention of msl2 mRNA.
HOW represses msl2 expression in Drosophila larvae
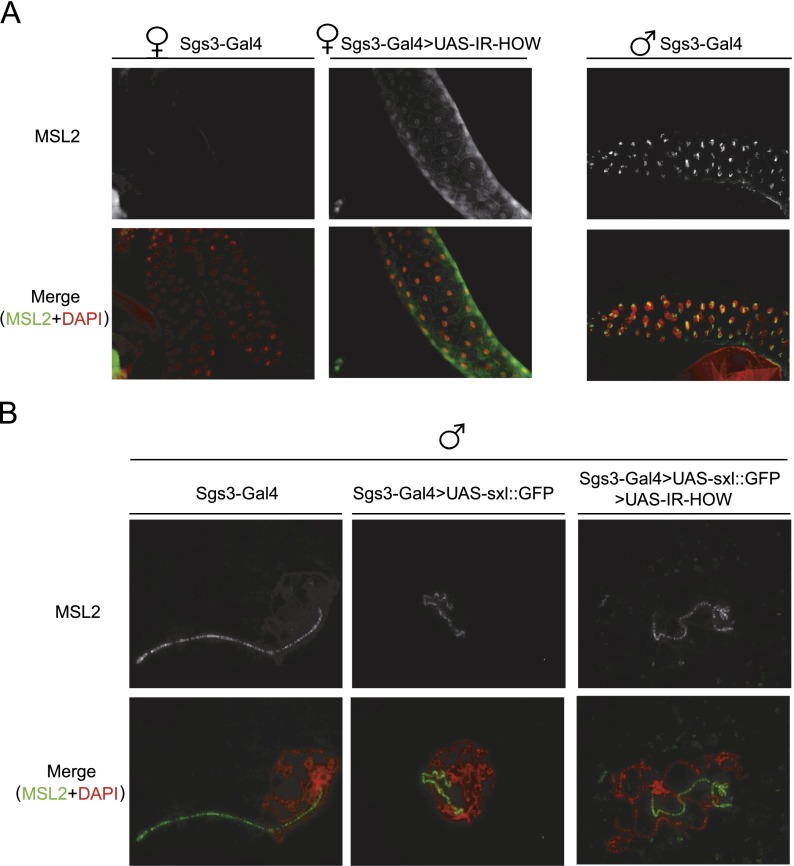
To investigate the influence of HOW in msl2 expression in the context of the whole animal, we followed two complementary approaches. First, we evaluated the expression of MSL2 in the salivary glands of female third instar larvae upon depletion of HOW. To deplete HOW, we crossed flies transformed with a transgene expressing a dsRNA against HOW (UAS-IR-HOW) with flies expressing the salivary gland-specific Sgs3-Gal4 driver. The results show that while no MSL2 staining is observed in parental females, a modest but reproducible MSL2 staining appears concentrated in the nucleus of salivary gland cells upon depletion of HOW (Fig. 6A, left and middle panels). Surprisingly, MSL2 staining is not concentrated in the X-chromosome territory, as observed for wild-type males (Fig. 6A, cf. middle and right panels). These results show that depletion of HOW derepresses msl2 expression in females. Second, we expressed SXL in the salivary glands of third instar larvae males and tested the effect of HOW depletion on the SXL-induced phenotype. Ectopic expression of SXL caused a robust phenotype consisting of a twisted and more fragile X chromosome with a distorted banding pattern (Fig. 6B, middle panel). The levels of MSL2 were difficult to assess given the altered structure of the X chromosome. However, we could detect a variable decrease of MSL2 staining on independent crosses (S. Patalano and F. Gebauer, unpubl.). Inappropriate DCC function has been previously shown to affect the structure of the X chromosome (Corona et al. 2002). Importantly, depletion of HOW almost completely suppressed this strong phenotype (Fig. 6B, right panel). These data indicate that, as observed in cell culture, HOW cooperates with SXL also in the context of the whole animal.
Figure 6.
HOW cooperates with SXL to repress msl2 expression in Drosophila larvae. (A) Depletion of HOW derepresses msl2 expression in females. Costaining of salivary glands from female SgS3-GAL4 or SgS3-GAL4>UAS-IR-HOW third instar larvae with αMSL2 antibodies (green) and DAPI (red). Control male SgS3-GAL4 salivary glands are shown. (B) Polytene chromosomes from males expressing a SXL-GFP fusion protein in the third instar larva salivary glands containing (Sgs3-Gal4>UAS-SXL∷GFP) or lacking (Sgs3-Gal4>UAS-SXL∷GFP;UAS-IR[HOW]) HOW were costained with αMSL2 (green) and DAPI (red). Control polytene chromosomes of SgS3-GAL4 male third instar larvae are shown on the left.
Discussion
SXL is the master sex determination switch in Drosophila and functions by modulating the splicing and translation of downstream targets (for review, see Salz and Erickson 2010). SXL-mediated regulation of the alternative splicing of its own transcript and transformer pre-mRNA initiates a cascade of events that leads to female-specific differentiation. Translational repression of notch and nanos mRNAs by SXL contributes to sexual dimorphism and female germ cell identity, respectively (Penn and Schedl 2007; Suissa et al. 2010; Chau et al. 2012). In addition, regulation of splicing and translation of msl2 mRNA leads to repression of X-chromosome dosage compensation in females. Here we show that SXL also functions by promoting nuclear mRNA retention. This process reinforces silencing of msl2 and requires the activity of the RNA-binding protein HOW.
HOW is a member of the STAR family of proteins, which includes well-studied homologs such as C. elegans GLD-1 and mammalian QKI (for review, see Volk et al. 2008). Members of this family contain an extended (macro-)KH RNA-binding domain and have roles in many aspects of RNA metabolism, including pre-mRNA splicing and mRNA subcellular distribution, stability, and translation. STAR proteins bind to similar AU-rich consensus hexamers (Israeli et al. 2007; Hafner et al. 2010; Jungkamp et al. 2011). Although a single hexameric consensus is sufficient for RNA recognition, tight binding requires two hexamers that can be recognized by STAR homodimers (Beuck et al. 2010; Carmel et al. 2010; Nir et al. 2012). Recent crystal structures of QKI and GLD-1 bound to RNA suggest that a distance of 10 nt between the two hexamers must be met for a STAR homodimer to recognize a single RNA molecule (Teplova et al. 2013). In the 5′ UTR of msl2, two hexamers separated by 1 nt are required for HOW binding (Fig. 3). This configuration of sites is unlikely to support dimer binding. Accordingly, a HOW mutant unable to dimerize can restore msl2 repression in HOW-depleted cells, while a mutant with impaired RNA-binding ability cannot, suggesting that RNA binding―but not HOW dimerization―is important for msl2 regulation (Fig. 3D).
Importantly, the effect of HOW on msl2 expression is sex-specific, as it is observed only in the context of SXL-mediated regulation (Figs. 2C, 4A). Thus, SXL confers a female-specific function to HOW. A SXL-dependent role on msl2 repression has been also observed for UNR (Abaza et al. 2006; Duncan et al. 2006). In the case of UNR, sex specificity was conferred through cooperative binding with SXL to the 3′ UTR of msl2. However, in the case of HOW, the mechanism is different because HOW binds efficiently to the 5′ UTR of msl2 in the absence of SXL (Fig. 3B). Our data suggest that SXL uses a two-step mechanism to convey a female-specific function to HOW. First, by promoting the retention of the 5′ UTR facultative intron, SXL provides the female transcripts with two HREs that are absent in male msl2 mRNAs. Second, SXL and HOW cooperate at an additional step, as intron retention per se does not suffice to explain the role of HOW in msl2 repression (Fig. 4A). Consistent with this, HOW and SXL interact directly (Fig. 4B,C), and HOW is required for SXL-mediated nuclear retention of msl2 mRNA (Fig. 5). Our data show that functional interaction between HOW and SXL also occurs in the context of the whole organism (Fig. 6).
The how gene produces two alternatively spliced variants: HOW(L) is primarily found in the nucleus, while HOW(S) localizes to both the cytoplasmic and nuclear compartments (Nabel-Rosen et al. 1999, 2002). These two isoforms have opposite effects in the regulation of some biological traits, and thus their balance must be tightly regulated. For example, HOW isoforms have contrary effects on tendon cell differentiation due to antagonistic modulation of the key transcription factor Stripe: While HOW(L) destabilizes stripe mRNA, HOW(S) promotes stabilization of this transcript (Nabel-Rosen et al. 2002). HOW(L) is the isoform responsible for regulation of msl2, as it is the only isoform expressed in SL2 cells (Supplemental Fig. S2), and its depletion from these cells leads to derepression of msl2 reporters (Figs. 2–4). mRNA destabilization is a major function of HOW(L). In addition to stripe, HOW(L) has been shown to destabilize string/cdc25, miple, dpp, and diap1 mRNAs (Nabel-Rosen et al. 2005; Israeli et al. 2007; Toledano-Katchalski et al. 2007; Reuveny et al. 2009). However, the effect of HOW on msl2 mRNA cannot be attributed to destabilization because RNA levels were not altered upon HOW depletion (Supplemental Fig. S1), and differences in the expression of msl2 reporters remained after correction for RNA abundance (Figs. 2–4).
In addition to mRNA stability, HOW has been shown to facilitate neurexin IV exon inclusion during glia cell maturation by interactions with the splicing factor Crn (Edenfeld et al. 2006; Rodrigues et al. 2012). Because HOW interacts with SXL (Fig. 4B) and the HREs overlap with the location of a putative branch point in the msl2 5′ UTR intron, we tested whether HOW works together with SXL to promote the retention of this intron. Careful quantification of the msl2 splicing isoforms, however, did not reveal any effect of HOW in SXL-mediated intron retention (Fig. 5B). Furthermore, mutation of the splice sites did not eliminate the effect of HOW on msl2 silencing (Fig. 4A). We also tested whether HOW cooperates with SXL to repress the translation of msl2 reporters in a cell-free system that faithfully recapitulates msl2 translational regulation (Gebauer et al. 1999). HOW(L) binds upstream of a regulatory AUG that has been shown to contribute to inhibition of ribosomal scanning by SXL and could, in principle, participate in this mechanism (Medenbach et al. 2011). Nevertheless, depletion of HOW did not affect the ability of SXL to repress translation regardless of whether the msl2 reporters contained or lacked this AUG (Fig. 5A; data not shown). This implies that HOW adds a new layer to msl2 silencing in addition to the previously described splicing and translation mechanisms. The overlapping effects of splicing, transport, and translation inhibition ensures msl2 repression, which is essential for the survival of females.
We found that HOW depletion decreased nuclear msl2 mRNA retention by SXL an average of 2.2-fold in six different experiments (Fig. 5C; data not shown). The levels of msl2 mRNA regulation by HOW observed throughout this study are similar to that reported for other HOW targets (e.g., Nabel-Rosen et al. 1999, 2002; Reuveny et al. 2009). Furthermore, depletion of SXL and HOW reduces the nuclear accumulation of endogenous msl2 in Kc cells (Fig. 5D). How do both proteins cooperate to retain msl2 mRNA in the nucleus? Mutation of the HREs did not affect SXL-mediated regulation but restored efficient repression by SXL in HOW-depleted extracts (Fig. 3C). These results are consistent with the hypothesis that HOW, together with SXL, prevents the binding of a positive regulator (e.g., a factor required for efficient export) that recognizes overlapping sequences on the mRNA. Nuclear retention could be important to sequester the mRNA from the translation machinery. Alternatively, HOW-mediated retention could allow for the proper maturation of the silenced mRNP and the acquisition of the factor combination required for optimal repression. HOW(L) also promotes nuclear retention of stripe mRNA, although in this case, the repressive mechanism is combined with mRNA degradation (Nabel-Rosen et al. 1999, 2002). A more similar mechanism has been described in mammals, where the nuclear HOW ortholog QKI-5 promotes nuclear retention of MBP mRNA without altering the levels of the transcript (Larocque et al. 2002). Thus, nuclear mRNA retention may be a conserved function of STAR proteins. Interestingly, tandems of GLD-1-binding sites have been identified in the 5′ UTR of C. elegans transcripts (Jungkamp et al. 2011), raising the possibility that regulation by STAR proteins binding to the 5′ UTR is more general and that mRNAs other than msl2 are regulated by a similar mechanism.
Materials and methods
Fly stocks
Flies transgenic for the UAS-IR[HOW] (transformant ID 100775) and SgS3-Gal4 constructs were ordered from the Vienna Drosophila RNAi Center and Bloomington Stock Center, respectively. Flies containing the SXL-GFP transgene were obtained by classical P-element transformation.
Stainings
Salivary glands from third instar larvae raised at 29°C were dissected in cold PBS. Salivary glands were fixed in PBS with 4% paraformaldehyde for 20 min at 4°C; washed three times with PBS for 10 min; blocked in 1× PBS, 0.05% Tween 20, 0.2% bovine serum albumin for 30 min; and stained with rabbit anti-MSL2 antibody overnight (1:500 dilution). After three washes of 10 min in 0.05% PBST, salivary glands were incubated with secondary anti-rabbit FITC antibodies (Jackson) for 2 h at room temperature in the dark, washed three times for 10 min in PBST, and transferred to PBS. For polytene chromosome stainings, salivary glands were fixed for 3 min in 50% acetic acid and 3.7% formaldehyde, squashed on glass slides, and stained as described (Zink and Paro 1995). Primary anti-MSL2 and secondary FITC-conjugated anti-rabbit antibodies were used at 1:500 dilution. For both salivary gland and polytene chromosome stainings, slides were mounted in ProLong Gold anti-fade reagent with DAPI, and imaging was carried out using a Leica DMI 4000 B microscope with a Roper Scientific Coolsnap HQ camera.
Plasmids
DNA templates used to generate the B, Bm, m1, m2, m12, and EF biotinylated probes were produced by ligation of double-stranded oligonucleotides containing the relevant sequences into the SacI and XbaI sites of pGEM-3Z. The β-gal reporters used in Figure 2 were previously described (Bashaw and Baker 1997). The msl2 firefly luciferase plasmids used in transfection experiments were obtained by inserting the corresponding msl2 and luciferase sequences into the EcoRI and BamHI sites of pAc5.1B-NheI (a generous gift from Britta Hartmann). Inserts were obtained by PCR amplification of the construct mLm (containing the firefly luciferase ORF flanked by the full-length 5′ and 3′ UTRs of msl2) or mutant mLm derivatives lacking the SXL-binding sites (Gebauer et al. 1999). The HRE and splice site mutations were introduced into pAc-mLm3′m by PCR-directed mutagenesis. In the HRE mutant, the HREs were substituted by the sequence CGCGGCGC. To obtain pAc-Renilla, the CDS of Renilla luciferase was amplified by PCR and cloned into the NheI and HindIII sites of pAc5.1B-NheI. PCR reactions were performed with Phusion proofreading DNA polymerase (Finnzymes). The pAc-HOW(L) construct was obtained from the insertion of the PCR-amplified HOW(L) ORF into the EcoRI and BstBI sites of pAc5.1B-NheI. The E106G and R185C HOW(L) mutants were generated by PCR-directed mutagenesis. All constructs were verified by sequencing. Primer sequences used for PCR are summarized in the Supplemental Material.
Recombinant proteins
SXL and its derivative, dRBD4 (containing amino acids 122–301 of SXL), were expressed in Escherichia coli as N-terminal GST-tagged fusions and purified as previously described (Grskovic et al. 2003). His-SXL(ΔGN) (amino acids 122–354 of SXL) and His-HOW(L) were prepared at the Center for Genomic Regulation (CRG) Protein Facility according to standard procedures. All proteins were dialyzed against buffer D (20 mM HEPES at pH 8.0, 20% glycerol, 1 mM DTT, 0.01% NP-40, 0.2 mM EDTA).
RNA synthesis
Biotinylated RNAs were synthesized using the MEGAshort script kit (Ambion), adding bio-14-CTP (Invitrogen) at an equimolar ratio with CTP in the reaction mix. This ratio was found not to interfere with SXL-mediated translational repression and therefore allowed silencing complex formation. dsRNA for depletion experiments was synthesized using the Megascript kit (Ambion) and was purified on G25 columns (GE Healthcare). Primer sequences used to generate the DNA templates for dsRNA synthesis are detailed in the Supplemental Material.
mRNAs used in in vitro translation reactions were synthesized as previously described (Gebauer et al. 1999). All mRNAs contained a 7mGpppG cap and a poly(A) tail of 73 residues.
Antibodies and immunoprecipitation
αSXL and αUNR rabbit polyclonal antibodies were previously described (Abaza et al. 2006). αTral was a generous gift from J.E. Wilhelm's and E. Izaurralde's laboratories. Affinity-purified αHOW antibody was kindly provided by M. Blanchette; rabbit polyclonal αHOW antibodies were also raised in our laboratory against the full-length protein. αTubulin (Sigma, T6199) and αHistone H1 (Millipore, 045457) were provided commercially. For immunoprecipitation, αHOW antibodies were covalently cross-linked to protein A-Dynabeads (Invitrogen). Four milligrams of Drosophila embryo extract was incubated with 120 μL of beads in a final volume of 600 μL containing 0.1 M sodium phosphate buffer (pH 8.0). Following three washes with cold PBS, samples were treated with RNase cocktail (Ambion) when indicated, and proteins were recovered with Laemmli buffer and resolved by SDS-PAGE.
GRAB
Sixty picomoles of biotinylated RNA was mixed with equimolar amounts of recombinant GST-dRBD4 in a 1-mL reaction containing 10 mg of Drosophila embryo extract, 100 U of RNAsin (Promega), 25 μg of tRNA, 1×x Complete protease inhibitor cocktail (Roche), and 1× translation condition buffer [1× TCB: 17 mM creatine phosphate, 80 ng/μL creatine kinase, 25 mM Hepes at pH 8.0, 0.6 mM Mg(OAc)2, 80 mM KOAc]. After 30 min of incubation at room temperature, the mix was supplemented with 100 μL of glutathione beads (GE Healthcare) previously equilibrated in TCB and incubated for 1 h at 4°C. Glutathione beads were then washed twice with 8 vol of cold PBS, and the complexes were eluted by incubation with 40 U of AcTEV protease (Invitrogen) in 200 μL of AcTEV buffer supplemented with 1 mM DTT for 1 h at 4°C. The eluate was then mixed with 60 μL of streptavidin Dynabead slurry (Invitrogen), which was preblocked for 10 min with 100 ng/μL tRNA in TCB. After 1 h of incubation at 4°C, the beads were washed twice with 800 μL of cold PBS, and the complexes were recovered with Laemmli buffer, separated by SDS-PAGE, and analyzed by immunoblotting or silver staining.
RNAi, transfections, and reporter activity assays
SL2 Drosophila cells were grown at 25°C in Schneider's medium supplemented with L-glutamine (Gibco), 1% (v/v) penicillin/streptomycin (Gibco), and 10% FBS (Invitrogen). For RNAi treatment, 1.0 × 106 cells were incubated with 15 μg of dsRNA in 1 mL of Schneider medium without serum and incubated for 30–60 min at 25°C. One volume of Schneider's medium containing 20% FBS was then added. For the add-back experiment of Figure 3D, dsRNA against the 3′ UTR of HOW(L) was used. After 3–5 d, cells were split to a density of 1.5 × 106 cells per milliliter. The following day, cells were cotransfected with msl2 reporter plasmids (100 ng of msl2-β-gal or 300 ng of the firefly luciferase reporters), Renilla luciferase control (20 ng), increasing amounts of pAc-SXL (or pAc empty vector), and, when appropriate, pAc-HOW(L) constructs (500 ng) using Effectene (Qiagen). Cells were harvested 2–3 d after transfection. B-Galactosidase and luciferase activities were measured with Galacto-Star (Tropix) or Dual Luciferase kit (Promega), respectively. β-Gal and firefly activities were corrected for cotransfected Renilla and, when indicated, for the levels of reporter RNA.
RNA extraction and quantification
Total, nuclear, and cytoplasmic RNA were extracted with Trizol (Invitrogen). To obtain total RNA, cell pellets were directly resuspended in Trizol and processed according to the manufacturer's instructions. To obtain nuclear and cytoplasmic RNA, cell pellets were washed with cold PBS and resuspended in hypotonic buffer (150 mM NaCl, 10 mM Tris at pH 7.5, 0.1% SDS, 1% DOC, 5 mM EDTA, 1% Triton X-100, protease inhibitors). After 5 min on ice, cells were broken with plastic pestles and centrifuged at 13,000 rpm for 5 min at 4°C. The supernatant was recovered as the cytoplasmic fraction. The pellet (nuclear fraction) was washed with PBS before adding Trizol.
For quantification, RNA was treated with Turbo DNase (Ambion) and reverse-transcribed using random primers and SuperScript II (Invitrogen). cDNAs were diluted threefold to 10-fold in water and used for qPCR amplification (primers are detailed in the Supplemental Material). Firefly luciferase RNA levels were normalized for cotransfected Renilla luciferase RNA levels.
To assess endogenous RNA levels in Kc cells, nuclear and cytoplasmic msl2 levels were normalized for U3 snoRNA and actin79B mRNA levels, respectively.
Fluorescence in situ hybridization
SL2 cells were fixed with 3.7% formaldehyde in PBS for 10 min, washed with PBS, and permeabilized with 70% ethanol for 2 h at 4°C. Cells were then washed with wash buffer (1× SSC, 10% formamide) and hybridized to antisense oligonucleotide probes (Stellaris probes, Biosearch Technologies, Inc.) in hybridization buffer (100 mg/mL dextran sulphate, 10% formamide, 2× SSC) overnight at 37°C. Following incubation for 30 min at 37°C in wash buffer and DAPI staining for 30 min, cells were washed with 2× SSC and mounted in GLOX buffer.
Biotin and GST pull-down
Biotin pull-down assays were performed in 500 μL of 1× TCB (see above) containing 100 pmol of biotinylated RNA, 100 pmol of His-SXL(ΔGN), 5 mg of Drosophila embryo extract, 50 U of RNAsin (Promega), 12.5 μg of tRNA, and 1× Complete protease inhibitor cocktail (Roche). After 30 min of incubation at room temperature, 100 μL of streptavidin Dynabead slurry (Invitrogen; preblocked for 10 min with 200 ng/μL tRNA in TCB) was added, and the mix was further incubated for 30 min. Beads were washed five times with 8 vol of PBS, and proteins were eluted by RNase treatment (2 μL of RNase cocktail [Ambion] in 30 μL of a buffer containing 10 mM Tris-HCl at pH 7.2, 1 mM MgCl2, 40 mM NaCl).
GST pull-down assays were performed in 40 μL of buffer D (see above) containing 40 pmol of either GST, GST-dRBD4, or GST-SXL and 40 pmol of His-HOW(L). After 30 min of incubation at room temperature, 10 μg of RNase A was added, and the incubation was extended for 30 min. The sample was diluted with 1 vol of capture buffer (CB; 20 mM HEPES at pH 8.0, 600 mM NaCl, 100 mM KOAc, 0.02% NP40, 0.2 mM EDTA) and bound to 20 μL of glutathione sepharose beads for 2 h at 4°C. Beads were then washed four times with 1 mL of CB, and bound proteins were recovered with Laemmli buffer and separated by SDS-PAGE.
Immunodepletions and in vitro translation assays
HOW was immunodepleted from embryo extracts using αHOW antibodies covalently coupled to magnetic protein A-Dynabeads (Biotech). One round of incubation for 30 min at room temperature was sufficient for efficient depletion. In vitro translation assays were performed as previously described (Gebauer et al. 1999).
Acknowledgments
We thank Juan Valcárcel, Jim Wilhelm, Marco Blanchette, Elisa Izaurralde, Christophe Antoniewski, and Talila Volk for kindly sharing their antibodies; Bruce Baker, Jan Medenbach, Britta Hartmann, and Talila Volk for reporter constructs; and Emilia Szostak for help with cloning. We thank Solenn Patalano for obtaining the SXL-GFP transgenic line. We also thank Juan Valcárcel, Lidia Pérez, Alena Shkumatava, Sylvain Meunier, and members of our laboratory for useful discussions and/or critical reading of this manuscript. We are grateful to the CRG Protein Service, the CRG/UPF Proteomics Facility, and the CRG Microscopy Unit for protein production and identification and help with confocal images, respectively. A.G. was supported by a fellowship from the Fondation pour la Recherche Médicale (FRM). C.C. was supported by the Agence Nationale de la Recherche (grant ANR BLAN 1210 01). This work was supported by grants BFU2009-08243 and Consolider CSD2009-00080 from the Spanish Ministry of Economy and Competitiveness.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.214999.113.
References
- Abaza I, Coll O, Patalano S, Gebauer F 2006. Drosophila UNR is required for translational repression of male-specific lethal 2 mRNA during regulation of X-chromosome dosage compensation. Genes Dev 20: 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ, Baker BS 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798 [DOI] [PubMed] [Google Scholar]
- Beckmann K, Grskovic M, Gebauer F, Hentze MW 2005. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell 122: 529–540 [DOI] [PubMed] [Google Scholar]
- Beuck C, Szymczyna BR, Kerkow DE, Carmel AB, Columbus L, Stanfield RL, Williamson JR 2010. Structure of the GLD-1 homodimerization domain: Insights into STAR protein-mediated translational regulation. Structure 18: 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel AB, Wu J, Lehmann-Blount KA, Williamson JR 2010. High-affinity consensus binding of target RNAs by the STAR/GSG proteins GLD-1, STAR-2 and Quaking. BMC Mol Biol 11: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK 2012. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci 109: 9465–9470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DFV, Clapier CR, Becker PB, Tamkun JW 2002. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, Gebauer F, Wilm M, Hentze MW 2006. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: Translational repression for dosage compensation. Genes Dev 20: 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KE, Strein C, Hentze MW 2009. The SXL–UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. Mol Cell 36: 571–582 [DOI] [PubMed] [Google Scholar]
- Edenfeld G, Volohonsky G, Krukkert K, Naffin E, Lammel U, Grimm A, Engelen D, Reuveny A, Volk T, Klambt C 2006. The splicing factor crooked neck associates with the RNA-binding protein HOW to control glial cell maturation in Drosophila. Neuron 52: 969–980 [DOI] [PubMed] [Google Scholar]
- Forch P, Merendino L, Martinez C, Valcarcel J 2001. Modulation of msl-2 5′ splice site recognition by Sex-lethal. RNA 7: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW 2007. Studying translational control in Drosophila cell-free systems. Methods Enzymol 429: 23–33 [DOI] [PubMed] [Google Scholar]
- Gebauer F, Merendino L, Hentze MW, Valcarcel J 1998. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA 4: 142–150 [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Corona DF, Preiss T, Becker PB, Hentze MW 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: Cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J 18: 6146–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Grskovic M, Hentze MW 2003. Drosophila sex-lethal inhibits the stable association of the 40S ribosomal subunit with msl-2 mRNA. Mol Cell 11: 1397–1404 [DOI] [PubMed] [Google Scholar]
- Gebauer F, Preiss T, Hentze MW 2012. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb Perspect Biol 4: a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge A, Militti C, Gebauer F 2011. Posttranscriptional control of X-chromosome dosage compensation. Wiley Interdiscip Rev RNA 2: 534–545 [DOI] [PubMed] [Google Scholar]
- Grskovic M, Hentze MW, Gebauer F 2003. A co-repressor assembly nucleated by Sex-lethal in the 3′UTR mediates translational control of Drosophila msl-2 mRNA. EMBO J 22: 5571–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli D, Nir R, Volk T 2007. Dissection of the target specificity of the RNA-binding protein HOW reveals dpp mRNA as a novel HOW target. Development 134: 2107–2114 [DOI] [PubMed] [Google Scholar]
- Jungkamp AC, Stoeckius M, Mecenas D, Grun D, Mastrobuoni G, Kempa S, Rajewsky N 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell 44: 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81: 867–877 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Wang J, Bell L, Kuroda MI 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199 [DOI] [PubMed] [Google Scholar]
- Larocque D, Pilotte J, Chen T, Cloutier F, Massie B, Pedraza L, Couture R, Lasko P, Almazan G, Richard S 2002. Nuclear retention of MBP mRNAs in the quaking viable mice. Neuron 36: 815–829 [DOI] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW 2011. Translational control via protein-regulated upstream open reading frames. Cell 145: 902–913 [DOI] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841 [DOI] [PubMed] [Google Scholar]
- Nabel-Rosen H, Dorevitch N, Reuveny A, Volk T 1999. The balance between two isoforms of the Drosophila RNA-binding protein how controls tendon cell differentiation. Mol Cell 4: 573–584 [DOI] [PubMed] [Google Scholar]
- Nabel-Rosen H, Volohonsky G, Reuveny A, Zaidel-Bar R, Volk T 2002. Two isoforms of the Drosophila RNA binding protein, how, act in opposing directions to regulate tendon cell differentiation. Dev Cell 2: 183–193 [DOI] [PubMed] [Google Scholar]
- Nabel-Rosen H, Toledano-Katchalski H, Volohonsky G, Volk T 2005. Cell divisions in the Drosophila embryonic mesoderm are repressed via posttranscriptional regulation of string/cdc25 by HOW. Curr Biol 15: 295–302 [DOI] [PubMed] [Google Scholar]
- Nir R, Grossman R, Paroush Z, Volk T 2012. Phosphorylation of the Drosophila melanogaster RNA-binding protein HOW by MAPK/ERK enhances its dimerization and activity. PLoS Genet 8: e1002632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn JK, Schedl P 2007. The master switch gene sex-lethal promotes female development by negatively regulating the N-signaling pathway. Dev Cell 12: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveny A, Elhanany H, Volk T 2009. Enhanced sensitivity of midline glial cells to apoptosis is achieved by HOW(L)-dependent repression of Diap1. Mech Dev 126: 30–41 [DOI] [PubMed] [Google Scholar]
- Rodrigues F, Thuma L, Klambt C 2012. The regulation of glial-specific splicing of Neurexin IV requires HOW and Cdk12 activity. Development 139: 1765–1776 [DOI] [PubMed] [Google Scholar]
- Salz HK, Erickson JW 2010. Sex determination in Drosophila: The view from the top. Fly (Austin) 4: 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa Y, Kalifa Y, Dinur T, Graham P, Deshpande G, Schedl P, Gerlitz O 2010. Hrp48 attenuates Sxl expression to allow for proper notch expression and signaling in wing development. Proc Natl Acad Sci 107: 6930–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Hafner M, Teplov D, Essig K, Tuschl T, Patel DJ 2013. Structure–function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Genes Dev 27: 928–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano-Katchalski H, Nir R, Volohonsky G, Volk T 2007. Post-transcriptional repression of the Drosophila midkine and pleiotrophin homolog miple by HOW is essential for correct mesoderm spreading. Development 134: 3473–3481 [DOI] [PubMed] [Google Scholar]
- Volk T, Israeli D, Nir R, Toledano-Katchalski H 2008. Tissue development and RNA control: ‘HOW’ is it coordinated? Trends Genet 24: 94–101 [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell 9: 675–685 [DOI] [PubMed] [Google Scholar]
- Zink D, Paro R 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J 14: 5660–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]