Abstract
Differentiation of human mesenchymal stem cells (hMSCs) requires the rewiring of energy metabolism. Herein, we demonstrate that the ATPase inhibitory factor 1 (IF1) is expressed in hMSCs and in prostate and colon stem cells but is not expressed in the differentiated cells. IF1 inhibits oxidative phosphorylation and regulates the activity of aerobic glycolysis in hMSCs. Silencing of IF1 in hMSCs mimics the metabolic changes observed in osteocytes and accelerates cellular differentiation. Activation of IF1 degradation acts as the switch that regulates energy metabolism during differentiation. We conclude that IF1 is a stemness marker important for maintaining the quiescence state.
Keywords: ATPase inhibitory factor 1, cellular differentiation, H+-ATP synthase, mitochondria, protein degradation
INTRODUCTION
A master regulator of energy metabolism in aerobic differentiated cells is the mitochondrial H+-ATP synthase, the engine of oxidative phosphorylation (OXPHOS) that catalyses the synthesis of ATP using as driving force the proton gradient generated by the respiratory chain. The ATPase inhibitory factor 1 (IF1) [1] is the physiological inhibitor of the H+-ATP synthase that is highly overexpressed in the mitochondria of cancer cells [2]. Overexpression of IF1 results in the inhibition of the ATP synthetic activity of the enzyme, the switch to an increased aerobic glycolysis [2] and the ROS-mediated signalling of several features of the oncogenic phenotype [3]. These findings supported a role for IF1 as a master regulator of energy metabolism and retrograde nuclear signalling in cancer.
Adult human mesenchymal stem cells (hMSCs) are characterized by their multi-lineage differentiation potential (pluripotency) and their self-renewal capacity [4]. Recently, the study of energy metabolism of stem cells has received attention because of its implication in nuclear reprogramming. Undifferentiated pluripotent stem cells have a low activity of mitochondrial OXPHOS and preferentially use aerobic glycolysis as a major source of energy supply [4]. In contrast, differentiated cells depend more heavily on OXPHOS [5, 6]. Interestingly, stemness factor-mediated nuclear reprogramming of somatic cells induces the downregulation of OXPHOS concurrently with the activation of glycolysis [7]. However, the mechanism and signalling molecule that gears the stem cell decision to change its energy metabolism and hence to cause onset of proliferation or differentiation programs are unknown.
Here we demonstrate that the enhanced bioenergetic activity of mitochondria on osteogenic induction is not supported by proliferation of the organelles but by the bioenergetic differentiation of pre-existing mitochondria. We show that IF1 is expressed in hMSCs and in stem cell niches of CD44-positive cells of the human prostate and colon but is not expressed in differentiated osteocytes. Remarkably, IF1 promotes aerobic glycolysis in hMSCs and its expression is stringently regulated by degradation during osteogenic induction. Our findings support a functional role for IF1 in the regulation of stem cell fate decisions.
RESULTS
Onset of OXPHOS upon osteogenic differentiation
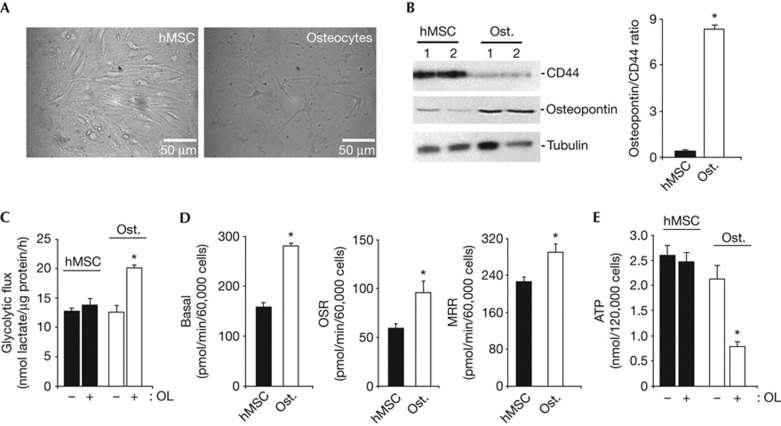
Osteogenesis from hMSCs promoted a large change in cellular morphology (Fig 1A) with concurrent upregulation of the osteogenic marker osteopontin (Fig 1B) and downregulation of the stem cell marker CD44 (Fig 1B). Hence, the osteopontin/CD44 ratio, an index of osteogenic differentiation, was significantly increased in osteocytes when compared to hMSCs (Fig 1B).
Figure 1.
Metabolic reprogramming of stem cells upon osteogenic differentiation. hMSCs were grown in the osteogenic induction media to promote differentiation. (A) Representative images of hMSCs and osteocytes. (B) Western blots of the expression of CD44, osteopontin and tubulin in two different preparations of hMSCs and Ost. The histograms show the calculated osteopontin/CD44 ratio. Bars are the mean±s.e.m. of four different samples. *P<0.05 when compared to hMSCs. (C) hMSCs and osteocytes were processed for the determination of the rates of aerobic glycolysis in the absence (−) or presence (+) of 6 μM OL. Bars are the mean±s.e.m. of 6–10 independent determinations. *P<0.05 when compared to OL-untreated cells. (D) The rates of basal respiration were determined in hMSCs and osteocytes. OSR and MRR were also measured after the addition of 6 μM OL and 0.75 mM DNP, respectively. Bars represent the mean±s.e.m. of three independent experiments. *P<0.05 when compared to hMSCs. (E) Cellular ATP concentrations were determined in the absence (−) or presence (+) of 6 μM OL. Bars are the mean±s.e.m. of three different experiments. *P<0.05 when compared to OL-untreated cells. DNP, 2,4-dinitrophenol; hMSCs, human mesenchymal stem cells; MRR, maximum respiratory rates; OL, oligomycin; OSR, oligomycin-sensitive respiration; Ost., osteocytes.
Basal rates of aerobic glycolysis were not significantly different between hMSCs and osteocytes (Fig 1C). Inhibition of OXPHOS with oligomycin (OL) did not affect the rates of aerobic glycolysis in hMSCs (Fig 1C) but promoted an increase in glycolysis in osteocytes (Fig 1C). Consistently, osteocytes showed a significant increase in basal, OL sensitive and maximum uncoupled respiratory rates (Fig 1D). Basal cellular ATP concentrations were not significantly different between hMSCs and osteocytes (Fig 1E). However, OL only promoted a significant decrease in ATP content in osteocytes (Fig 1E). The drop in ATP levels in response to OL treatment in osteocytes could not be attributed to an enhanced cell death (supplementary Fig S1 online), but could be considered as a result of a higher dependence of this cellular type on OXPHOS.
Osteogenesis triggers mitochondrial differentiation
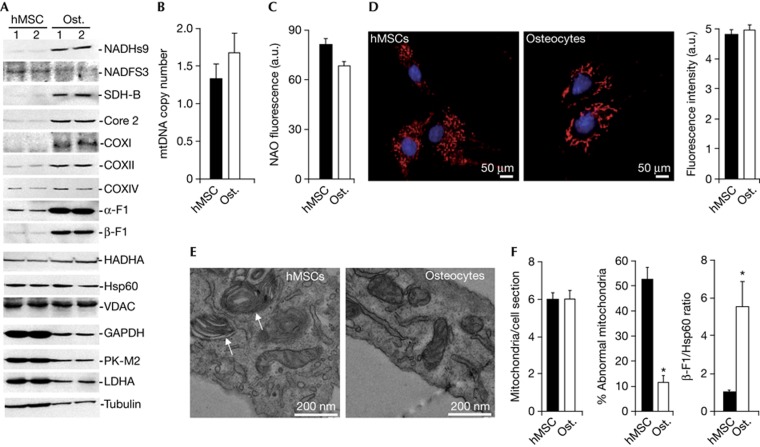
Cellular differentiation triggered a significant increase in the expression of nicotinamide adenine dinucleotide dehydrogenase subunit 9 from complex I, succinate dehydrogenase B from complex II, core 2 from complex III, cytochrome c oxidase subunits I and II from complex IV, and the α- and β-F1-ATPase subunits of the H+-ATP synthase from complex V of OXPHOS (Fig 2A; supplementary Fig S2A online). A concurrent decrease in the expression of the glycolytic proteins glyceraldehyde-3-phosphate dehydrogenase, pyruvate kinase isoform M2 (PK-M2) and lactate dehydrogenase A was observed (Fig 2A; supplementary Fig S2A online). Hence, the overall mitochondrial capacity of the cell (β-F1-ATPase/glyceraldehyde-3-phosphate dehydrogenase ratio) [8] (see supplementary Information online) was significantly augmented in osteocytes (supplementary Fig S2A online). Other mitochondrial proteins such as nicotinamide adenine dinucleotide Fe-S protein 3 from complex I, cytochrome oxidase subunit IV from complex IV, hydroxyacyl-CoA dehydrogenase of β-oxidation, porin (VDAC, voltage-dependent anion channel) and the structural mitochondrial protein Hsp60 revealed no relevant changes during osteogenic induction (Fig 2A; supplementary Fig S2A online). Analysis of OXPHOS complexes in blue native gels revealed no relevant differences in their assembly between hMSCs and osteocytes (supplementary Fig S2B online), supporting that their assembly is not limiting OXPHOS in hMSCs.
Figure 2.
Differentiation of mitochondria upon osteogenic induction. (A) Representative western blots of the expression of mitochondrial proteins (NADHs9, NDUFS3, SDH-B, Core 2, COXI, COXII, COXIV, α-F1-ATPase, β-F1-ATPase, HADHA, Hsp60 and VDAC), glycolytic enzymes (GAPDH, PK-M2 and LDHA) and tubulin in two different preparations of hMSCs and osteocytes. (B) hMSCs and osteocytes were processed for the determination of the relative mtDNA copy number (12S/β-F1-ATPase ratio). Bars represent the mean±s.e.m. of seven independent determinations. (C) The mitochondrial mass was determined by the content of cardiolipin as assessed by NAO fluorescence. Bars are the mean±s.e.m. of six independent determinations. *P<0.05 when compared to hMSCs. (D) Fluorescence microscopy of hMSC and osteocytes stained with MitoTracker Red and Hoechst. Images show a partially fragmented mitochondrial network in hMSCs when compared to the thread-like morphology of mitochondria observed in osteocytes. The histogram shows the quantification of the MitoTracker signal in hMSCs and osteocytes. The results shown are the mean±s.e.m. of 45–60 different cells. (E) Representative electron microscopy images of mitochondria in hMSCs and osteocytes. Onion-like mitochondria (white arrows) are observed in hMSCs. The histograms show the quantification of the number of mitochondria per cell section and the percentage of abnormal organelles in ultrathin sections of hMSCs and osteocytes, respectively. The results shown are the mean±s.e.m. of 45–50 different cells. *P<0.05 when compared to hMSCs. (F) The histogram shows the mean±s.e.m. of the calculated β-F1-ATPase/Hsp60 ratio in hMSCs and osteocytes in four different samples. *P<0.05 when compared to hMSCs. a.u., arbitrary units; COX, cytochrome oxidase; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; hMSCs, human mesenchymal stem cells; LDH-A, lactate dehydrogenase A; NADHs9, nicotinamide adenine dinucleotide subunit 9; OL, oligomycin; OSR, oligomycin-sensitive respiration; Ost., osteocytes; PK-M2, pyruvate kinase isoform M2.
In contrast with recent findings [6], we observed that osteogenic induction occurred in the absence of relevant changes in mitochondrial DNA (mtDNA) copy number (Fig 2B) as well as in mitochondrial mass as assessed by the cardiolipin content of the cell (Fig 2C). Analysis of mitochondrial morphology and abundance using MitoTracker Red revealed large differences in organelle shape (Fig 2D) and the absence of differences in mitochondrial content (the histogram in Fig 2D). Mitochondria in hMSCs were predominantly punctiform, whereas the thread-like morphology was more abundant in osteocytes (Fig 2D), in agreement with previous findings in pluripotent stem cells [9]. High-resolution electron microscopy further confirmed no significant differences in the number of mitochondria per cell section between osteocytes and hMSCs (Fig 2E). However, hMSCs contained a high percentage of mitochondria with abnormal structure when compared to mitochondria in osteocytes (Fig 2E). These results indicate that osteogenic induction promotes the bioenergetic differentiation of mitochondria rather than organelle proliferation, as supported by the increase in the β-F1-ATPase/Hsp60 ratio [8] (see supplementary Information online) observed in osteocytes when compared to hMSCs (Fig 2F).
We observed no relevant differences in the expression of β-F1-ATPase messenger RNA (mRNA) between hMSCs and osteocytes (supplementary Fig S3A online), suggesting that the expression of β-F1-ATPase (Fig 2A) is controlled at the level of mRNA translation during differentiation, which is in agreement with similar findings in development and in oncogenesis [10]. miR-127-5p targets the 3′ untranslated region of β-F1-ATPase mRNA and inhibits its translation [11]. However, both hMSCs and osteocytes do not express significant levels of miR-127-5p (supplementary Fig S3B online), indicating that it is not involved in repressing the bioenergetic differentiation of mitochondria in hMSCs.
IF1 regulates energy metabolism of stem cells
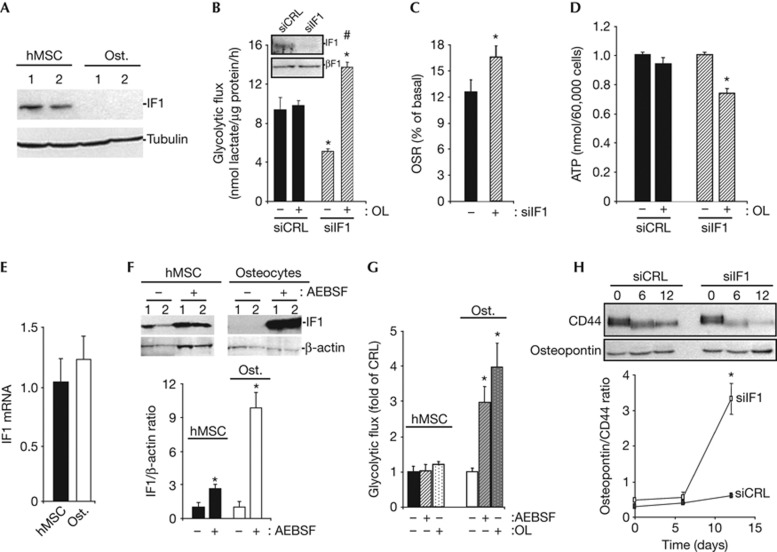
Western blot analysis revealed that osteocytes do not express IF1 when compared to hMSCs (Fig 3A). Short interfering RNA-mediated silencing of IF1 in hMSCs resulted in a significant reduction in the rates of aerobic glycolysis (Fig 3B). In this situation, OL treatment promoted a significant increase in lactate production (Fig 3B). Consistently, downregulation of IF1 in hMSCs promoted an increase in the activity of the mitochondrial H+-ATP synthase (Fig 3C). Although basal cellular ATP concentrations were not affected by IF1 downregulation (Fig 3D), a significant drop in cellular ATP content occurred in the presence of OL in IF1-silenced hMSCs (Fig 3D). These results suggest that IF1 is part of the switch that controls energy metabolism on osteogenic induction of hMSCs (Fig 1). The regulated expression of IF1 does not affect the proliferation rate of hMSCs (supplementary Fig S4 online), in contrast to data obtained in colon cancer cells [3] suggesting a cell-type specific variability in nuclear responses to IF1 signalling.
Figure 3.
IF1 degradation mediates metabolic reprogramming of stem cells during osteogenic differentiation. (A) Representative blots of IF1 and tubulin in two different preparations of hMSCs and osteocytes. Osteogenic induction is accompanied by repression of IF1 expression. (B–D,H) hMSCs were transfected with control (siCRL, closed bars) or siIF1 siRNA (siIF1, hatched bars) to regulate the expression of IF1. (B) Rates of aerobic glycolysis in the absence (−) or presence (+) of 6 μM OL. Bars are the mean±s.e.m. of six independent determinations. * and #P<0.05 when compared to siCRL or siIF1 untreated cells, respectively. (C) The rates of OSR were determined after addition of 6 μM OL. Data are shown as percentage of the basal respiration. Bars are the mean±s.e.m. of eight independent determinations. *P<0.05 when compared to siCRL. (D) Cellular ATP concentrations were determined in the absence (−) or presence (+) of 6 μM OL. Bars are the mean±s.e.m. of 12 independent determinations. *P<0.05 when compared to OL-untreated cells. (E) IF1 mRNA expression was assessed by RT–qPCR in hMSCs and osteocytes. Bars are the mean±s.e.m. of six independent determinations. (F) hMSCs and osteocytes were treated 24 h with (+) or without (−) 400 μM of the serine-protease inhibitor AEBSF to block the activity of mitochondrial proteases, and the expression of IF1 and β-actin (loading control) analysed by western blot in two different preparations. Plots are the mean±s.e.m. of the calculated IF1/β-actin ratio in four different experiments. *P<0.05 when compared to untreated (−) cells. (G) The rates of aerobic glycolysis in the absence (−) or presence (+) of 400 μM of AEBSF or 6 μM OL were determined in hMSCs and osteocytes. Bars are the mean±s.e.m. of six independent determinations. *P<0.05 when compared to untreated cells. (H) The expression of CD44 and osteopontin was analysed by western blot at the indicated times to assess the rate of differentiation after silencing of IF1. The plot represents the calculated osteopontin/CD44 ratio in siCRL and siIF1 cells. *P<0.05 when compared to siCRL. AEBSF, 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride; CRL, control; hMSCs, human mesenchymal stem cells; IF1, inhibitory factor 1; mRNA, messenger RNA; OL, oligomycin; OSR, oligomycin-sensitive respiration; Ost., osteocyte; RT-qPCR, real-time quantitative polymerase chain reaction; siCRL, short interfering control; siIF1, short interfering IF1.
Regulation of IF1 expression in stem cells
hMSCs and osteocytes revealed no relevant differences in the cellular content of IF1 mRNA (Fig 3E). Twenty-four-hour treatment of hMSCs and osteocytes with AEBSF (a serine-protease inhibitor) promoted a large accumulation of IF1 in both cellular types (Fig 3F). However, the accumulation of IF1 in AEBSF-treated osteocytes exceeded by three-fold the accumulation observed in hMSCs, suggesting that normal differentiated cells degrade IF1 at a much faster rate than hMSCs. A short interfering RNA-based screen aimed at the identification of the protease involved in the degradation of IF1 in hMSCs (supplementary Fig S5A online), and HCT116 colon cancer cells (supplementary Fig S5B online) failed to provide a candidate responsible for the degradation of IF1 despite evidence of partial silencing of the seven proteases tested (supplementary Fig S5 online). Remarkably, whereas treatment of hMSCs with AEBSF did not induce significant changes in the rates of aerobic glycolysis (Fig 3G); the same treatment in osteocytes triggered a large increase in the glycolytic flux (Fig 3G), comparable to that induced by treatment with OL (Fig 3G). Moreover, silencing of IF1 accelerated the rate of differentiation, as revealed by the osteopontin/CD44 ratio after 12 days of osteogenic induction (Fig 3H), suggesting a role for IF1 in signalling the repression of cellular differentiation.
IF1 is a marker of stem cells
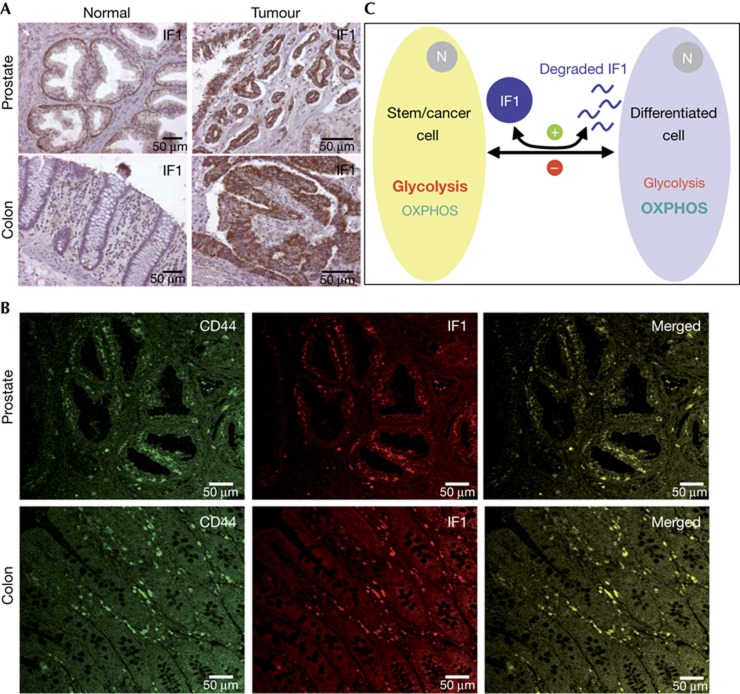
Immunohistochemistry of IF1 revealed that it is expressed in basal epithelial cells of the prostate (Fig 4A) and in the Lieberkühn crypts of the colon (Fig 4A). A high expression of IF1 was also observed in prostate and colon carcinomas (Fig 4A). Double-immunofluorescence microscopy analysis for CD44 and IF1 in human prostate and colon showed the co-localization of both markers in basal epithelial cells and Lieberkühn crypts, respectively (Fig 4B), which are the sites where stem cells are located in these tissues [12, 13].
Figure 4.
IF1 is a stem cell marker. (A) Immunohistochemistry of IF1 expression in human normal and tumour prostate and colon tissues. Images are shown at × 20–40 magnification. (B) Double-immunofluorescence microscopy of normal human prostate and colon stained with CD44 antibody (green) and with monoclonal IF1 antibody (red). The merged images revealed the co-localization of IF1 in stem cell niches of both tissues. Images are shown at × 40 magnification. (C) Scheme of the proposed mechanism by which IF1 (dark blue) regulates metabolic rewiring upon cellular differentiation/dedifferentiation. Stem and cancer cells (dark yellow) show a high expression of IF1, which promotes the inhibition of OXPHOS favouring aerobic glycolysis. Differentiation triggers an enhanced degradation of IF1 (+, green) and the activation of OXPHOS in the differentiated cell (light blue). The relevance of glycolysis as ATP supplier is diminished in this situation. Dedifferentiation of somatic cells into cancer cells results in the accumulation of IF1, the inhibition of OXPHOS and the activation of aerobic glycolysis. The accumulation of IF1 in cancer cells might be regulated by inhibiting the degradation of the protein (−, red). IF1, inhibitory factor 1; OXPHOS, oxidative phosphorylation.
DISCUSSION
Metabolic reprogramming during cellular differentiation [4–6] or in the dedifferentiation of somatic cells into iPSC [7] or cancer cells [3] is well established. Here we show that upon osteogenic differentiation of hMSCs, there is an upregulation of proteins of complex I, II, III, IV and V of OXPHOS. These changes result in an increase in the activity of the respiratory chain and of the H+-ATP synthase, and a higher dependence of osteocytes on OXPHOS to provide the ATP needed to sustain cellular specialization. Despite the enhancement of OXPHOS and the partial downregulation of the cellular content of glycolytic enzymes in osteocytes, the differentiated cells still show similar rates of glycolysis as hMSCs, indicating that the higher energy requirements of osteocytes cannot be covered by OXPHOS alone. Moreover, the maintenance of the glycolytic flux despite the diminished expression of glycolytic enzymes in osteocytes highlights the relevance of allosteric regulation of this metabolic pathway.
The mitochondrial phenotype of a given cell is the result of cell-type-specific programs that are regulated at both the transcriptional [14] and post-transcriptional [10] levels. Transcriptional programs usually result in the onset of mitochondrial proliferation [14], whereas post-transcriptional ones promote the rapid bioenergetic differentiation of the organelle in response to changing physiological cues (for a review, see Willers and Cuezva [10]). Our findings indicate that the enhanced activity of OXPHOS in osteocytes is not supported by an enhanced mitochondrial proliferation but by the bioenergetic differentiation of pre-existing stem cell mitochondria, in agreement with the findings upon pluripotent stem cell differentiation [9, 15]. Consistently, the bioenergetic accomplishment of mitochondria during osteogenesis also seems to be regulated at the post-transcriptional level [10].
Rewiring of energy metabolism to aerobic glycolysis has been shown to foster nuclear reprogramming, both during oncogenesis [16] and in dedifferentiation to iPSC [7]. However, the molecule that orchestrates the metabolic switch during stem cell differentiation remains elusive. Here, we show that IF1 regulates the energy metabolism of hMSCs by inhibiting the activity of OXPHOS and promoting aerobic glycolysis, reproducing the effect of OL in cellular metabolism [2]. Kinetic evidence indicates that IF1 inhibits the ATP hydrolase activity of the H+-ATP synthase [1]. However, it is likely that the binding of IF1 to the H+-ATP synthase also depends on the mass–action ratio and hence, when IF1 is expressed, as it is the situation in hMSCs, it can also inhibit the synthase activity of the enzyme. Indeed, silencing of IF1 in hMSCs resulted in a drop in glycolysis and a concurrent increase in ATP synthase activity. Conversely, its overexpression in different cell types has been shown to increase glycolysis and to inhibit the ATP synthase activity, mimicking the metabolic effects triggered by the ATP synthase inhibitor OL [2, 3]. Moreover, we document the expression of IF1 in hMSCs as well as in human prostate and colon stem cells, but not in osteocytes and other differentiated cells. Remarkably, disappearance of IF1 seems to be a prerequisite to facilitate the metabolic switch that accompanies cellular differentiation, emphasizing the prominent role that IF1 might play in the maintenance of stemness. However, other mechanisms might also operate, as it has been reported that human pluripotent stem cells have a low content of IF1 and their differentiation is triggered by repression of UCP2 [9].
Recent findings have stressed the relevance of post-translational modification of pluripotency-associated transcription factors in simultaneously maintaining pluripotency or inducing lineage-specific differentiation (for a review, see Cai et al. [17]). The results in this report add on the same idea by stressing the relevance of the degradation of IF1 in maintaining energy metabolism of the undifferentiated state of stem cells. The accumulation of IF1 in response to the inhibitor AEBSF indicates that the protein is degraded by a mitochondrial serine protease. The enhanced degradation of IF1 in osteocytes might thus result from the activation of any of these proteases as a surrogate process of the nuclear reprogramming that renders ongoing the differentiated state and/or because IF1 is experiencing a post-translational modification that makes it more susceptible to degradation. Our screening for the putative protease involved indicates that degradation of IF1 is more complex than originally anticipated and might involve an unexplored pathway for the degradation of mitochondrial proteins.
Cancer and stem cells show large phenotypic analogies regarding the molecular and functional activities of the proteins involved in glycolysis and in mitochondrial OXPHOS (Fig 4C). The expression of IF1 represents an additional phenotypic trait in common (Fig 4C). Deciphering the nuclear responses to the presence or absence of IF1 will certainly contribute to our understanding of stem cell fate decisions. Overall, we show that IF1 is a stem cell marker that regulates the energy metabolism of hMSCs. The regulated degradation of IF1 hinders self-renewal of stem cells to favour differentiation.
METHODS
Culture of hMSC. hMSCs were obtained from Lonza and cultured according to the manufacturer’s instructions; for more details see Supporting Information online.
Aerobic glycolysis and oxygen consumption rates. The rates of aerobic glycolysis were determined by the enzymatic determination of lactate in the medium in the absence or presence of 6 μM OL [18]. The oxygen consumption rates were determined in a XF24 Extracellular Flux Analyzer (Seahorse Biosciences) [2].
Determination of ATP. Cells were incubated for 1 h in the presence or absence of 6 μM OL and cellular ATP concentrations were determined using the ATP Bioluminiscence Assay Kit CLS II (Roche).
Western blot analysis. Cellular lysates were processed for blotting as indicated [2]. Details of the antibodies used are provided in Supporting Information online.
Determination of mtDNA copy number. After cellular DNA extraction, the quantification of mtDNA (mtDNA/nDNA) was performed by qPCR using the LightCycler 2.0 real-time PCR system as described [19].
Electron microscopy. For ultrastructural studies, hMSCs and osteocytes were fixed with 2% glutaraldehyde in 0.1 M Sörensen phosphate buffer, pH 7.4, and processed for electron microscopy as detailed elsewhere [18].
Fluorescence microscopy. Cells were incubated for 45 min with 500 nM Mitotracker Red FM (Invitrogen) followed by staining with Hoechst (1 mg/ml) for 10 min at 37 °C. Cellular fluorescence was analysed in an Axiovert 200 (Zeiss) microscope using a CCD camera. The red fluorescence intensity of the cells was calculated using ImageJ.
Statistical analysis. Statistical analyses were performed using a two-tailed Student’s t test. The results shown are the means±s.e.m. P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
C. Núñez de Arenas and M. Chamorro are acknowledged for expert technical assistance. I.M.R., J.G.B. and F.S. were supported by pre-doctoral fellowships from Junta Ampliación Estudios-Consejo Superior de Investigaciones Científicas, Formación Personal Investigador-Ministerio Educación y Ciencia and Formación Personal Investigador-Universidad Autónoma de Madrid Spain, respectively. This work was supported by grants from the Ministerio de Educación y Ciencia (BFU2010-18903), the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), ISCIII and Comunidad de Madrid (S2011/BMD-2402), Spain. The Centro de Biología Molecular Severo Ochoa recieves an institutional grant from the Fundación Ramón Areces.
Author contributions: M.S.-A. and J.M.C. designed the research; M.S.-A., J.G.-B., I.M.-R. and F.S. performed the research; M.S.-A. and J.M.C. analysed the data; M.S.-A. and J.M.C. wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Gledhill JR Montgomery MG Leslie AG & Walker JE (2007) How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc Natl Acad Sci USA 104: 15671–15676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cenizo L Formentini L Aldea M Ortega AD Garcia-Huerta P Sanchez-Arago M & Cuezva JM (2010) Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J Biol Chem 285: 25308–25313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formentini L Sánchez-Aragó M Sánchez-Cenizo L & Cuezva JM (2012) The mitochondrial ATPase Inhibitory Factor 1 (IF1) triggers a ROS-mediated retrograde pro-survival and proliferative response. Mol Cell 45: 731–742 [DOI] [PubMed] [Google Scholar]
- Rehman J (2010) Empowering self-renewal and differentiation: the role of mitochondria in stem cells. J Mol Med 88: 981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormos KV Anso E Hamanaka RB Eisenbart J Joseph J Kalyanaraman B & Chandel NS (2011) Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab 14: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT Shih YR Kuo TK Lee OK & Wei YH (2008) Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26: 960–968 [DOI] [PubMed] [Google Scholar]
- Folmes CD Nelson TJ Martinez-Fernandez A Arrell DK Lindor JZ Dzeja PP Ikeda Y Perez-Terzic C & Terzic A (2011) Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14: 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuezva JM et al. (2002) The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res 62: 6674–6681 [PubMed] [Google Scholar]
- Zhang J et al. (2011) UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J 30: 4860–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willers IM & Cuezva JM (2011) Post-transcriptional regulation of the mitochondrial H(+)-ATP synthase: a key regulator of the metabolic phenotype in cancer. Biochim Biophys Acta 1807: 543–551 [DOI] [PubMed] [Google Scholar]
- Willers IM Martínez-Reyes I Martínez-Diez M & Cuezva J (2012) miR-127-5p targets the 3'UTR of human β-F1-ATPase mRNA and inhibits its translation. Biochim Biophys Acta-Bioenergetics 1817: 838–848 [DOI] [PubMed] [Google Scholar]
- Lawson DA Xin L Lukacs RU Cheng D & Witte ON (2007) Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA 104: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen Hult LT Kleiveland CR Fosnes K Jacobsen M & Lea T (2011) EP receptor expression in human intestinal epithelium and localization relative to the stem cell zone of the crypts. PLoS One 6: e26816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638 [DOI] [PubMed] [Google Scholar]
- Birket MJ Orr AL Gerencser AA Madden DT Vitelli C Swistowski A Brand MD & Zeng X (2011) A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J Cell Sci 124: 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A Wang C & Schreiber SL (2005) Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci USA 102: 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N Li M Qu J Liu GH & Izpisua Belmonte JC (2012) Post-translational modulation of pluripotency. J Mol Cell Biol 4: 262–265 [DOI] [PubMed] [Google Scholar]
- Sanchez-Arago M Chamorro M & Cuezva JM (2010) Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis 31: 567–576 [DOI] [PubMed] [Google Scholar]
- Martinez-Reyes I Sanchez-Arago M & Cuezva JM (2012) AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. Biochem J 444: 249–259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.