Summary
Embryonic stem cells (ESCs) are derived from mammalian embryos during the transition from totipotency, when individual blastomeres can make all lineages, to pluripotency, when they are competent to make only embryonic lineages. ESCs maintained with inhibitors of MEK and GSK3 (2i) are thought to represent an embryonically restricted ground state. However, we observed heterogeneous expression of the extraembryonic endoderm marker Hex in 2i-cultured embryos, suggesting that 2i blocked development prior to epiblast commitment. Similarly, 2i ESC cultures were heterogeneous and contained a Hex-positive fraction primed to differentiate into trophoblast and extraembryonic endoderm. Single Hex-positive ESCs coexpressed epiblast and extraembryonic genes and contributed to all lineages in chimeras. The cytokine LIF, necessary for ESC self-renewal, supported the expansion of this population but did not directly support Nanog-positive epiblast-like ESCs. Thus, 2i and LIF support a totipotent state comparable to early embryonic cells that coexpress embryonic and extraembryonic determinants.
Graphical Abstract
Highlights
-
•
Embryos cultured in 2i are blocked before extraembryonic lineage segregation
-
•
ESCs cultured in 2i express extraembryonic markers heterogeneously
-
•
Single 2i ESCs differentiate into both embryonic and extraembryonic lineages
-
•
LIF promotes proliferation of extraembryonically primed ESC populations
Embryonic stem cells (ESCs) are isolated from preimplantation embryos and can contribute to all tissues of the embryo, but not extraembryonic tissues (e.g., placenta). Therefore, they are considered pluripotent, not totipotent. Brickman and colleagues now show that single ESCs cultured in the presence of MEK and GSK3 inhibitors (2i) coexpress embryonic and extraembryonic markers and contribute to both embryonic and extraembryonic tissues (e.g., are totipotent). A role of the cytokine LIF in ESC self-renewal is to support this totipotent population.
Introduction
Embryonic stem cells (ESCs) are karyotypically normal cells derived from the inner cell mass (ICM) or epiblast of peri-implantation embryos. They are classically defined as pluripotent. In mouse, this is assessed by their ability to differentiate into all embryonic but not extraembryonic lineages when reintroduced into morulae or blastocysts. However, because this definition is based on retrospective function, the precise cellular phenotype of pluripotent cells is unknown, especially as there has been little characterization of the functional potential of single cells. Additionally, although ESCs have been shown to be pluripotent only in standard injections, they can generate extraembryonic primitive endoderm (PE) in vitro (Hayashi et al., 2010; Xu et al., 2002) and, in rare events, contribute to both embryonic and extraembryonic lineages in vivo (Beddington and Robertson, 1989; Canham et al., 2010; Lallemand and Brûlet, 1990; Macfarlan et al., 2012; Suemori et al., 1990). This suggests that ESC cultures contain a mixture of cells resembling precursors of embryonic epiblast and extraembryonic tissues but that epiblast-like precursors have a competitive advantage when reintroduced into chimeras.
ESCs can be maintained in the presence of LIF and either BMP4 or serum (Ying et al., 2003a). Under these conditions, numerous genes are expressed in a heterogeneous manner (Canham et al., 2010; Chambers et al., 2007; Hayashi et al., 2008; Kobayashi et al., 2009; Singh et al., 2007; Toyooka et al., 2008), implying that ESC cultures may harbor cells with distinct functional potentials. ESCs can also be cultured in minimal medium with MEK (mitogen-activated protein or extracellular signal-regulated kinase) and GSK3 (glycogen synthase kinase 3) inhibitors (2i) (Ying et al., 2008). These inhibitors shield ESCs from differentiation-inducing signals and are thought to generate a homogeneous early epiblast-like ground state for embryonic but not extraembryonic development (Nichols et al., 2009; Wray et al., 2010, 2011). Culture in 2i is often supplemented with LIF, which not only supports self-renewal of ESCs but also has a function in extraembryonic development, where it promotes trophoblast proliferation, differentiation, and invasion (Poehlmann et al., 2005; Prakash et al., 2011; Takahashi et al., 2003).
We previously described a sensitive reporter for the endoderm marker Hex utilizing a reiterated IRES element to translationally amplify expression of the fluorescent protein Venus, encoded downstream of Hex in the endogenous locus (Canham et al., 2010). Here, we utilize ESCs containing this reporter, and a transgenic reporter mouse derived from them, to explore the nature of the ground state and investigate the cell-intrinsic role of LIF in this defined context. We show that embryos and ESCs cultured in 2i are heterogeneous and contain a fraction of cells coexpressing markers of both embryonic and extraembryonic lineages. This population demonstrated an enhanced capacity to generate extraembryonic cell types, including trophoblast, in vitro, and single cells from this fraction were totipotent when assessed by morula aggregation in vivo. Thus, the combination of 2i and LIF promoted the expansion of individual totipotent cells reminiscent of the morula or early blastocyst stage, before lineage restrictions have occurred.
Results
Preimplantation Embryo Culture in 2i Captures an Early Blastocyst Stage of Development
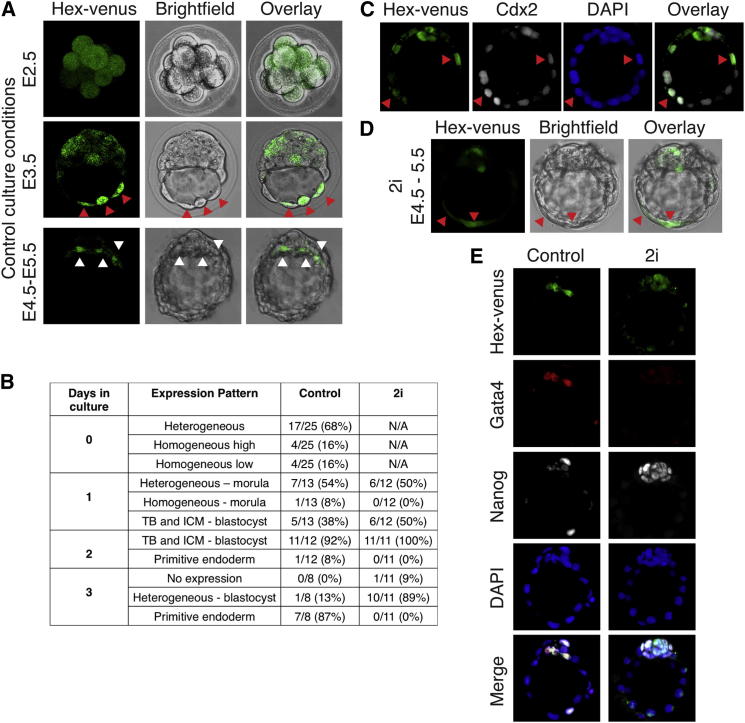
We generated a transgenic mouse line from our Hex-Venus (HV) reporter ESCs and assessed HV expression in preimplantation embryos. HV expression was observed at embryonic day (E) 2.5 (Figures 1A and 1B) and, in the majority of cases, was heterogeneous (Figures 1A and 1B). On average, 50% of blastomeres expressed HV at E2.5, although there was variation in the proportions between individual embryos. At E3.5, HV was expressed heterogeneously in both the ICM and trophoblast, being coexpressed with the trophoblast marker CDX2 in a number of cells (Figures 1A–1C). At E3.5, around one-third of the cells of the embryo expressed HV. By the late blastocyst stage (E4.5–E5.5), HV expression became restricted to the PE (Figures 1A, 1B, and 1E).
Figure 1.
Culture in 2i Does Not Eliminate Endoderm Marker Expression in the Early Embryo
(A) HV transgenic embryos were collected at E2.5 and cultured in vitro for different time periods before live confocal microscopy imaging. Representative images are shown for each stage. At E2.5, an extended focus image is shown. E3.5 and E4.5 images show confocal optical sections.
(B) Scoring of the HV expression pattern in embryos collected at E2.5 and cultured in either control or 2i medium. Day 0 = E2.5, day 1 = E3.5 (early blastocyst), day 2 = E4.5 (midblastocyst), and day 3 = E5.5 (late blastocyst, equivalent to E4.5 in vivo due to culture induced delay). Only HV-expressing transgenic embryos were scored. TB, trophoblast; ICM, inner cell mass.
(C) Confocal optical section of an E2.5 HV embryo cultured for 3 days immunostained for the trophoblast marker CDX2. Red arrowheads indicate trophoblast cells coexpressing HV and CDX2. Although in vivo PE is specified by E4.5, this segregation is delayed in vitro, and, hence, embryos were cultured until E5.5 to observe differentiated PE.
(D) HV embryos were flushed at E2.5 and cultured in 2i media for 3 days before live confocal microscopy imaging. Red arrowheads indicate HV-expressing trophoblast cells, whereas white arrowheads indicate HV+ PE cells.
(E) Confocal optical sections of immunostained transgenic HV embryos after flushing at E2.5 and 3 days in culture either in control or 2i medium.
Embryo culture in 2i has been reported to support the expansion of a homogeneous epiblast-like state at the expense of the expression of the late PE marker GATA4 (Nichols et al., 2009). We cultured transgenic HV embryos in 2i to determine whether the ICM differentiates into homogeneous epiblast. Embryos cultured in 2i, from E2.5 for 3 days, maintained heterogeneous HV expression (Figures 1B, 1D, and 1E) but did not segregate the PE and epiblast lineages, demonstrated by the absence of the late PE marker GATA4 (Figure 1E). Thus, 2i appears to block the progression of cultured embryos toward later blastocyst stages but does not eliminate the early endodermal precursor population.
Ground State In Vitro ESC Cultures Are Heterogeneous and Contain an Extraembryonically Primed Subpopulation
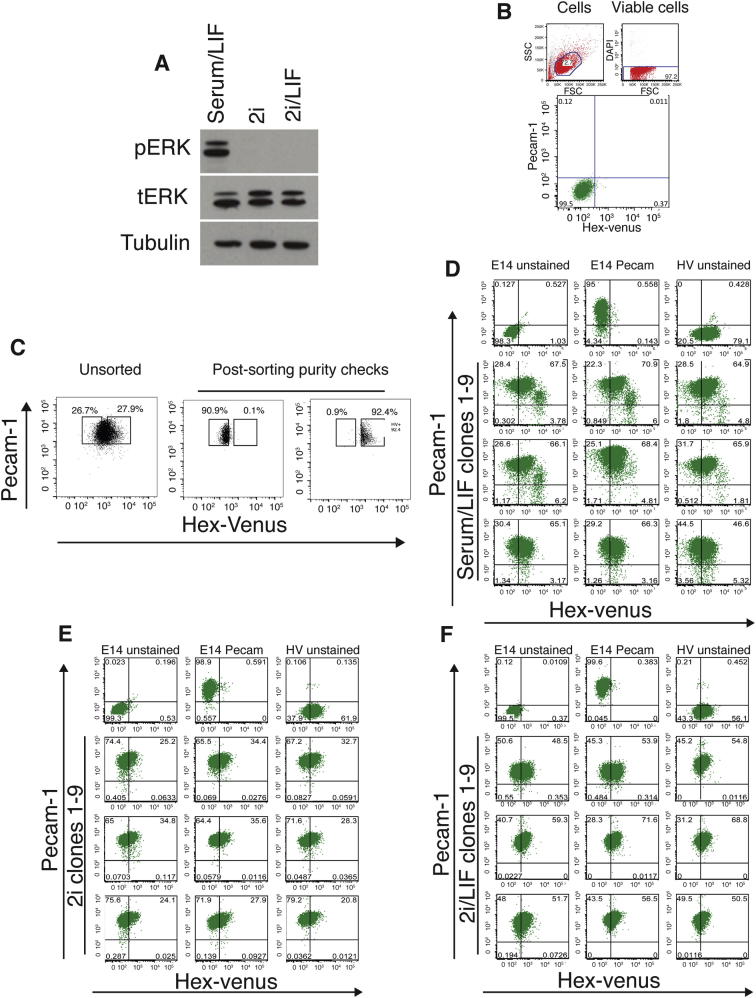
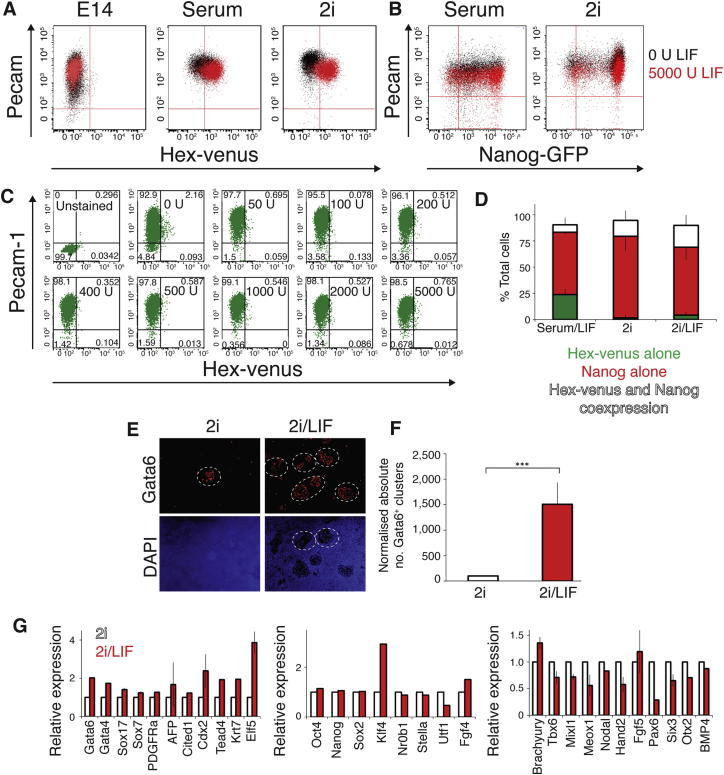
We then asked whether the in vitro ground state observed in ESCs is comparable to an early blastocyst stage in vivo, containing cells heterogeneously expressing low-level extraembryonic and pre-epiblast determinants (Chazaud et al., 2006). Previous observations suggested that ESCs in 2i maintained the expression of some endoderm markers (Canham et al., 2010; Marks et al., 2012). To assess the degree to which this represented heterogeneous gene expression, we clonally rederived HV ESC lines in 2i. We confirmed that ESCs in 2i demonstrated a block to extracellular signal-regulated kinase (ERK) signaling (Figure S1A) and found that at least nine independent clones showed heterogeneous expression of HV as judged by flow cytometry (gating methodology shown in Figures 2A and S1B–S1F). This was assessed for all three conditions (serum/LIF, 2i, and 2i/LIF). When single HV low (HV−) or HV high (HV+) cells were cultured in 2i or 2i/LIF, they regenerated heterogeneous cultures, although a proportion of cells maintained a bias toward regenerating mixed populations that contained a higher fraction of the seeding cell type (Figure 2B).
Figure S1.
HV Heterogeneity Can Be Observed after Clonal Rederivation of ESC Lines in 2i, Related to Figure 2
(A) Phospho ERK western blots of cells cultured in serum/LIF, 2i or 2i/LIF. Total ERK and Tubulin are shown as loading controls.
(B) Plots showing gating for all flow cytometry in this study. Gating for HV and PECAM-1 expression was based on a control E14 cell line without the HV reporter and unstained for PECAM-1. Cells were separated from debris by gating on forward (FSC) and side scatter (SSC). Dead cells, showing high DAPI levels, were eliminated.
(C) Purity checks after sorting by FACS of upper and lower 25% of HV expressing ESCs in 2i.
(D–F) Flow cytometry of individual clones in serum/LIF (D), in 2i (E) and 2i/LIF (F).
Figure 2.
ESCs Cultured in 2i Are Heterogeneous
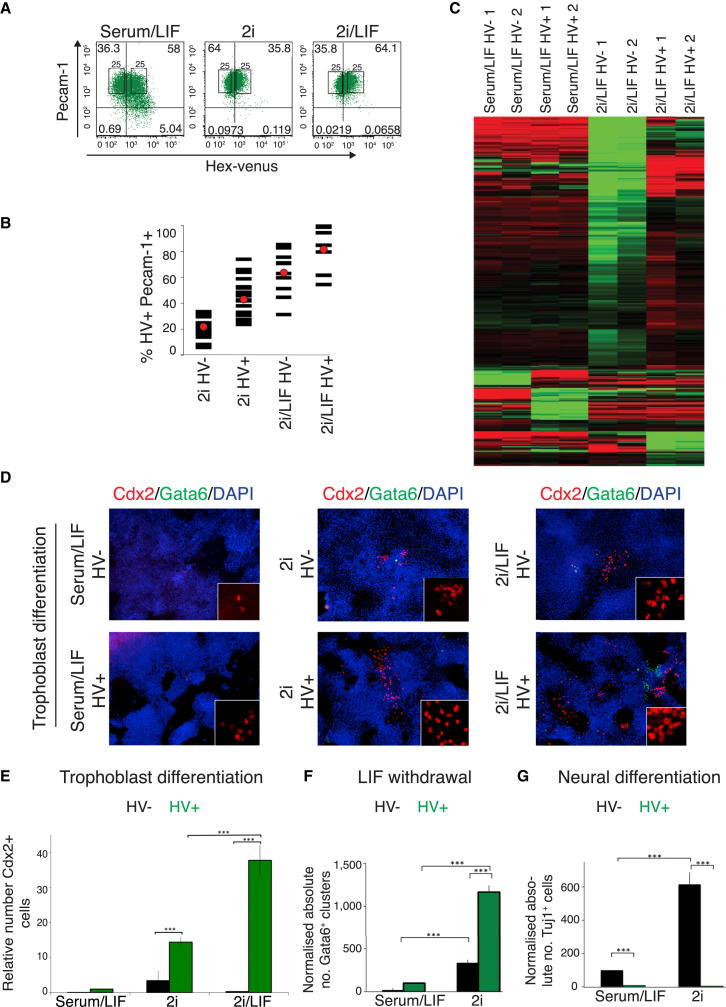
(A) Flow cytometry of HV and ESC marker PECAM-1 in cells cultured in serum/LIF, 2i, and 2i/LIF. Gates set using unstained E14 ESCs (Figure S1B). Black boxes indicate sorting gates for the upper and lower 25% of HV expression used to separate HV− and HV+ populations.
(B) Flow cytometry of clones (represented by individual bars) after expansion from single HV− or HV+ sorted cells from 2i or 2i/LIF. Red circles indicate mean.
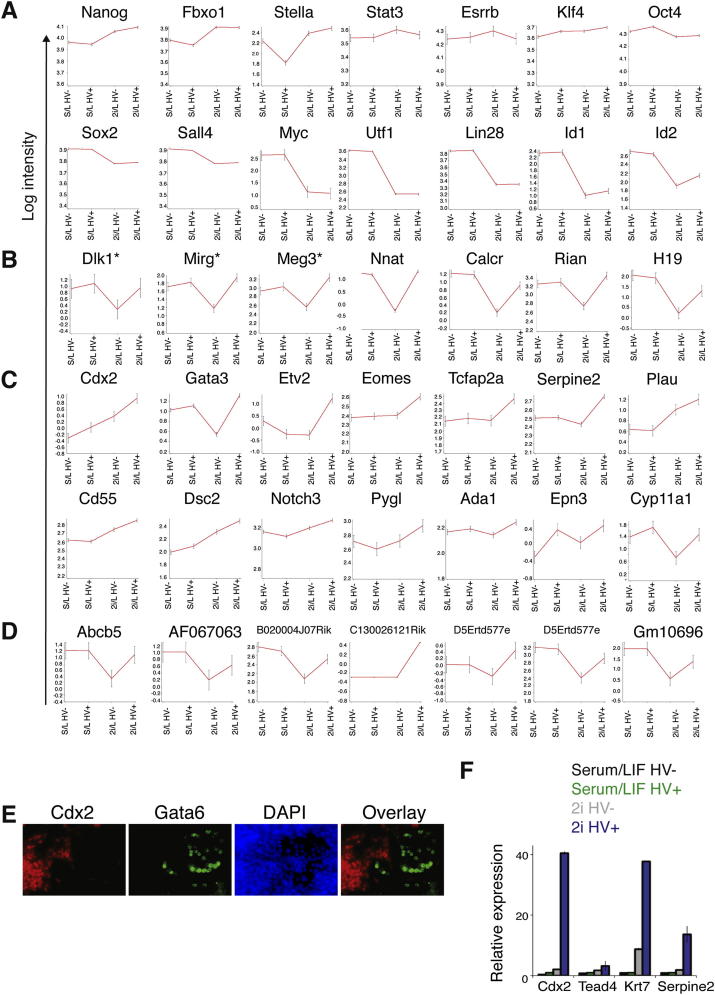
(C) Heat map of sorted HV− and HV+ populations from serum/LIF and 2i/LIF culture, based on gene expression data from RNA-seq. Heat map shows differentially expressed genes identified by pairwise comparison of all sorted fractions. Data were normalized by subtracting the average log expression from all samples. Genes are hierarchically clustered by average Euclidean distance. Two biological replicates are shown per sample. Red represents upregulation and green represents downregulation of expression.
(D) Immunostaining displaying representative images of CDX2-positive cells in trophoblast differentiation. High-magnification images are shown as insets.
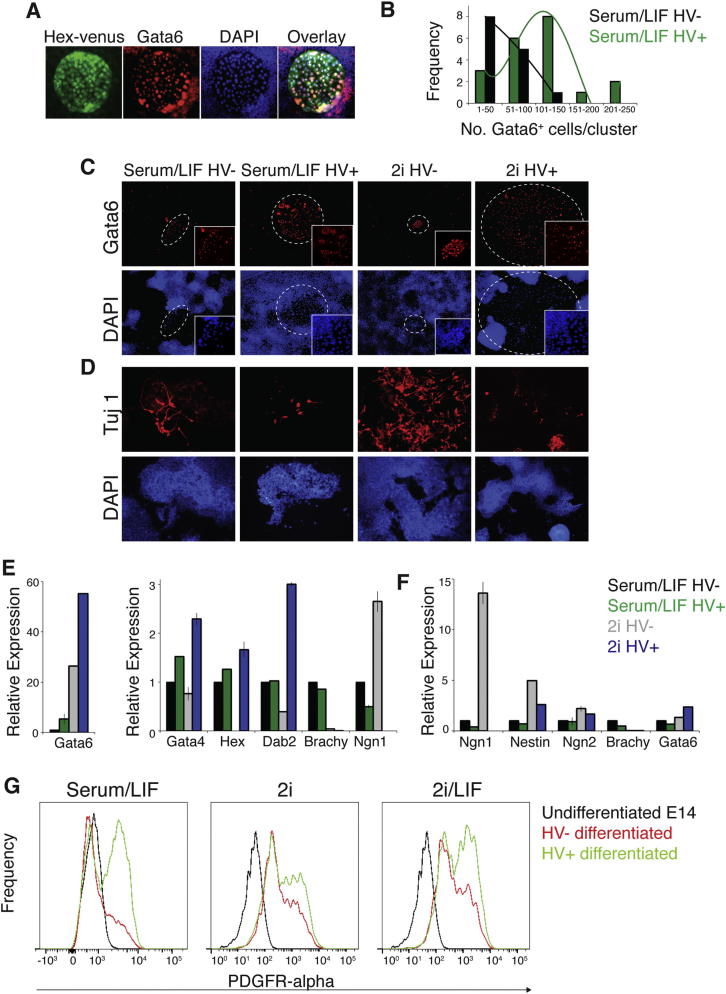
(E and F) Quantification of in vitro differentiation of sorted HV− and HV+ ESCs after differentiation in trophoblast stem cell conditions for 7 days (E), LIF withdrawal (F), or neural differentiation (G). (E) Trophoblast differentiation was quantified by counting CDX2-positive cells, which were negative for GATA6 and BRACHYURY (n = 3). Values are shown relative to HV+. (F) Endoderm differentiation was quantified by counting the number of GATA6-positive endodermal clusters (see also Figures S3A and S3C; n = 3). Values are shown relative to HV−.
(G) Neural differentiation was quantified by counting elongated TUJ1-positive neurons (n = 5). Values are shown relative to HV−. Error bars indicate mean ± SD of biological replicates.
See also Figures S1, S2, and S3.
We asked whether HV+ and HV− cells derived from serum/LIF or 2i/LIF culture conditions represented equivalent cell types by carrying out RNA sequencing (RNA-seq) genome-wide expression analysis on each population (Gene Expression Omnibus accession number GSE45182). Hierarchical clustering of gene expression based on an alternative pairwise comparison of the different populations (Figure 2C) suggested that HV+ and HV− states were more distinct in 2i/LIF than in serum/LIF (>2-fold change, false discovery rate [FDR] 0.05; Figure 2C; Tables S1 and S2).
While in this data set there was little change in early epiblast or pluripotency genes between the sorted 2i populations, we observed more than 20 imprinted genes that were enriched in the 2i/LIF HV+ compared to 2i/LIF HV− population; for example, the Dlk1-Dio3 cluster (Figures S2A and S2B) associated with efficient reprogramming (Liu et al., 2010). We also observed increased levels of trophoblast gene expression in the 2i/LIF HV+ population, including markers specifically expressed in trophoblast stem cells (Rugg-Gunn et al., 2012) (Figure S2C). In addition, endogenous retroviral (ERV) genes, enriched in an ESC population comparable to the two-cell-stage embryo (Macfarlan et al., 2012), such as Abcb5, AD067063, and Gm10696, were upregulated in the 2i/LIF HV+ population (Figure S2D). However, unlike the population of ERV-marked cells, 2i/LIF ESCs also continued to express pluripotency markers.
Figure S2.
Transcriptomics Suggest the Presence of a Population Primed toward Trophoblast as well as Endoderm, Related to Figure 2
(A–D) Common RNA-seq signatures in specific gene classes. The behavior of the individual genes belonging to the classes described in the text is shown. Plots compare mean log intensity values for individual genes among four sorted populations; serum/LIF HV-, serum/LIF HV+, 2i/LIF HV-, 2i/LIF HV+. Error bars represent s.d. between 2 biological replicates. (A) Markers of undifferentiated ESCs, (B) imprinted genes (∗ = member of Dlk1-Dio3 cluster), (C) trophoblast markers, (D) 2-cell stage embryo retroviral genes.
(E) High-magnification image of GATA6 and CDX2 immunostaining of ESCs, after 7 days differentiation in TSC conditions, demonstrating that there was no coexpression of these 2 markers.
(F) qRT-PCR of HV- and HV+ populations cultured in serum/LIF or 2i after LIF withdrawal for 4 days. Values were normalized to the housekeeping gene TBP and are shown relative to serum/LIF HV+. Error bars indicate mean ± SD.
This coexpression of pluripotency genes and trophoblast determinants is reminiscent of the stages of preimplantation development when blastomeres are competent to make all lineages. As ESCs are not thought to be able to generate trophoblast, we asked if 2i/LIF HV+ cells could differentiate into trophoblast in vitro. Figures 2D and 2E show that HV+ cells generated 40-fold more CDX2+ cells than HV− cells in trophoblast stem cell conditions (Quinn et al., 2006). CDX2+ cells appeared to be trophoblast-like, coexpressing neither the endoderm marker GATA6 nor the mesoderm marker BRACHYURY (Figure S2E; data not shown). We also observed that, upon differentiation by LIF withdrawal, only HV+ cells from 2i produced robust levels of trophoblast gene expression (Figure S2F). These observations revealed that HV+ ESCs in serum/LIF and 2i/LIF are fundamentally different from each other in both gene expression and functional capabilities, with cells from 2i/LIF demonstrating the additional capacity to generate trophoblast in vitro.
To determine whether 2i/LIF HV+ cells were restricted to the trophoblast lineage, we assessed their capacity to differentiate into endoderm and the epiblast-derived neural lineage. We observed a marked bias of the HV+ population to form endoderm, whereas the HV− population was biased toward a neural fate, even after prior culture in 2i (Figures 2F, 2G, and S3A–S3G; p < 0.001). Levels of differentiation were scored based on the number of GATA6+ cells (Figure S3B), GATA6+ colonies (Figure 2F), gene expression (Figures S3E and S3F), and flow cytometry to quantify the expression of an endodermal cell surface marker (Figure S3G). Absolute levels of differentiation were also higher in cells differentiated from 2i (Figures 2E–2G; p < 0.001).
Figure S3.
Quantification of Lineage Priming In Vitro, Related to Figure 2
(A) A typical GATA6+ endodermal colony, also expressing HV, as scored in differentiation assays.
(B) Quantification of number of GATA6+ cells per endodermal cluster.
(C and D) Immunostaining of ESCs differentiated by LIF withdrawal (C) or neural differentiation (D) after previous culture in serum/LIF or 2i and then sorting into HV- and HV+, PECAM-1+ ESCs. Cells were plated immediately into differentiation conditions after sorting. Endoderm levels were quantified by GATA6 immunostaining (n = 3) (values relative to serum/LIF HV+), (D) and neural differentiation by TUJ1 immunostaining (n = 5) (Values relative to serum/LIF HV-)
(E and F) qRT-PCR showing relative gene expression of sorted cells differentiated by LIF withdrawal (E) or neural differentiation (F). Data were normalized to the housekeeping gene TBP and is shown relative to serum/LIF HV sample. Values indicate mean ± SD of biological replicates.
(G) Flow cytometry of sorted HV- and HV+ populations following LIF withdrawal and staining for the endoderm marker PDGFR-alpha.
ESCs Cultured in 2i Can Contribute to Both Embryonic and Extraembryonic Lineages
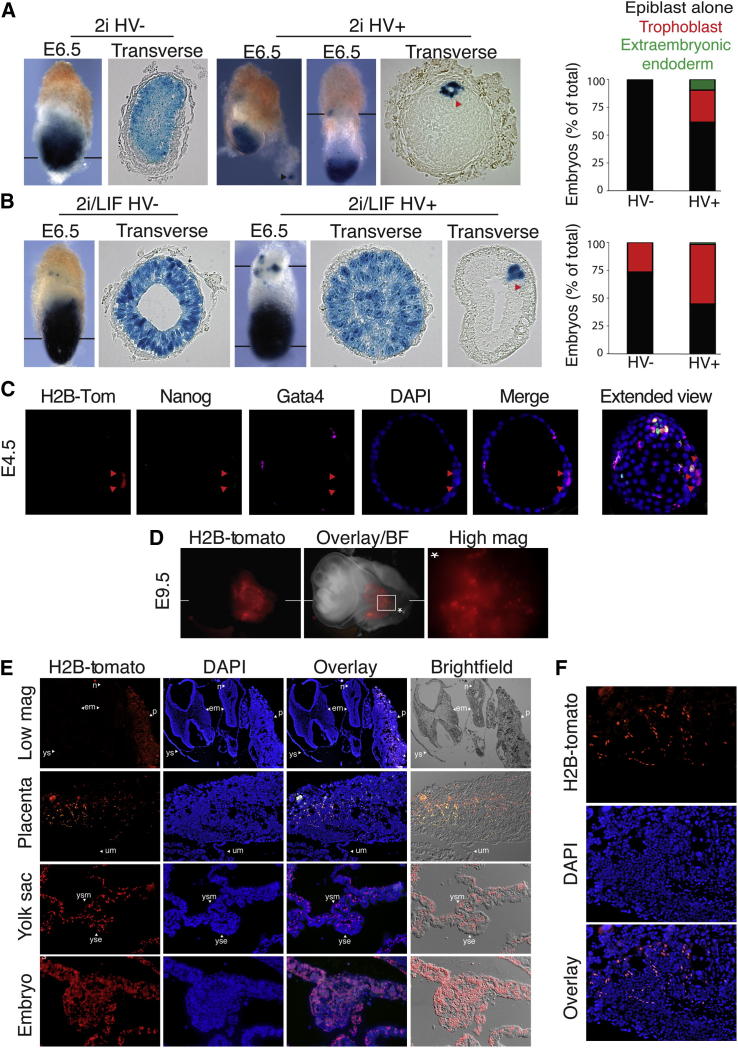
We tested the capacity of 2i-cultured HV− and HV+ ESCs to colonize embryonic and extraembryonic lineages in aggregation chimeras using HV ESCs constitutively expressing LACZ (Figures 3A and 3B). We previously found that HV− cells in serum/LIF contributed efficiently to the epiblast, whereas HV+ cells contributed to the extraembryonic endoderm and only weakly to epiblast (Canham et al., 2010). While HV− cells in 2i displayed high-level epiblast contribution (Figure 3A), HV+ cells from 2i contributed efficiently to the epiblast and to all extraembryonic lineages (i.e., exhibited totipotent properties) (Figures 3A, 3B, and S4A). In a large proportion of chimeras generated from the HV+ population, cells were detected in the trophoblast as well as visceral and parietal endoderm (n = 10/26; Figures 3A, 3B, and S4A), while the HV− population from 2i contributed only to epiblast (n = 18; Figure 3A). We assessed cells cultured in 2i as well as in 2i/LIF and found enhanced extraembryonic contribution from cells grown in 2i/LIF (n = 30/55) compared to those in 2i alone (n = 10/26). In 2i/LIF, a proportion of the HV− cells also contributed to extraembryonic lineages, although less efficiently than HV+ cells (n = 18/60; Figure 3B).
Figure 3.
Subpopulations of ESCs Cultured in 2i Demonstrate Extraembryonic Potential In Vivo
(A and B) HV cells, expressing a constitutive LACZ marker, cultured in 2i (A) or 2i/LIF (B), were sorted via FACS into HV− (A: n = 18; B: n = 60) and HV+ (A: n = 26; B: n = 55) PECAM-1+ cells and aggregated with wild-type F1 morulae. Embryos were collected at E6.5 and X-gal stained. Black bars indicate the plane of section. Arrowheads mark extraembryonic endoderm (black) and trophoblast (red).
(C–F) Sorted HV+ cells expressing a constitutive H2B-Tomato fusion protein, cultured in 2i/LIF, were used in morula aggregations. Embryos were cultured until E4.5 (C) or dissected at E9.5 (D). (C) Confocal optical section of an immunostained chimera after 3 days of culture (equivalent to E4.5 in vivo). Red arrowheads indicate H2B-Tomato cells within the trophoblast. Extended view throughout the embryo also shown for orientation. (D) E9.5 chimera. White line indicates plane of the low-magnification section in (E). White box indicates area of placenta shown in the high-magnification image marked with an asterisk (∗). (E) Wax sections of an E9.5 chimeric embryo showing HV H2B-Tomato cells in the placenta, yolk sac, and embryo. The uppermost panels show a low-magnification section of the whole embryo (shown in D) within the yolk sac and attached to the placenta. ys, yolk sac; yse, yolk sac endoderm; ysm, yolk sac mesoderm; em, embryo; n, notochord; p, placenta; um, umbilical cord. (F) High-magnification image of placental section.
See also Figure S4.
Figure S4.
HV+ ESCs Demonstrate Extraembryonic Potential in Morula Aggregations, Related to Figure 3
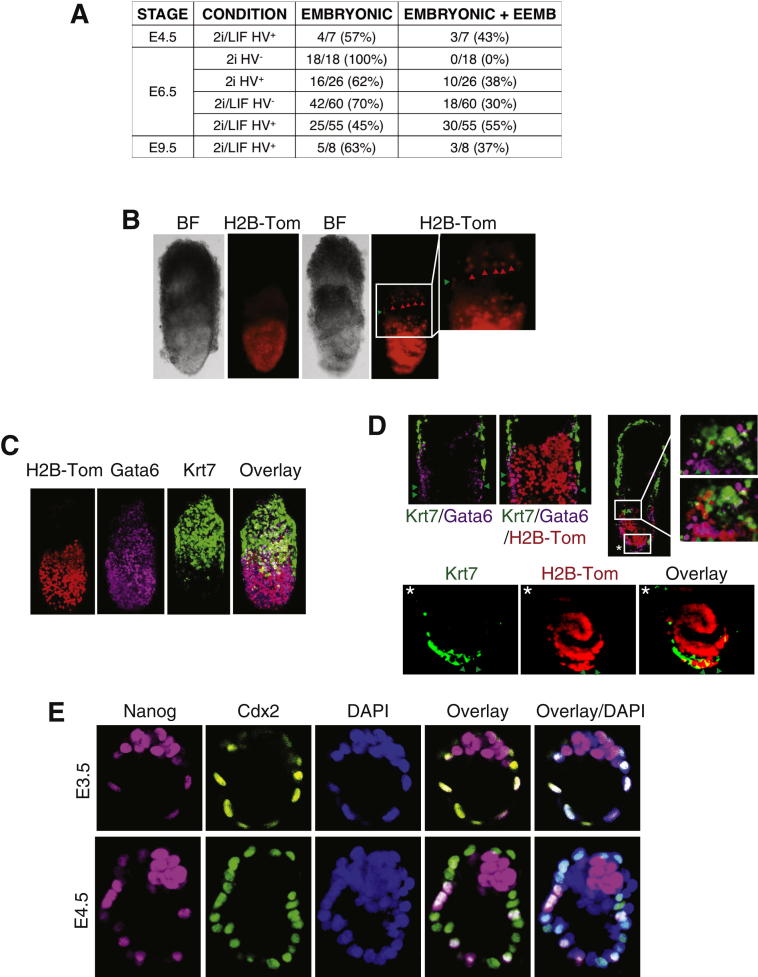
(A) Table containing a summary of all chimeras produced by morula aggregation in this study.
(B–D) HV ESCs were cultured in 2i/LIF. HV+ cells, expressing a constitutive H2B-Tomato fusion protein, were sorted and used in morula aggregations. Embryos were dissected at E6.5 (B-D) and imaged by confocal microscopy. (B) Example chimeras, one showing only epiblast contribution, the other both epiblast and extraembryonic contribution. Red arrowheads indicate H2B-Tomato cells in the trophoblast, green arrowheads indicate H2B-Tomato cells in the extraembryonic endoderm. (C) Whole-mount immunostaining of chimeras for an endoderm (GATA6) and trophoblast marker (KRT7). (D) Optical sections through whole-mount immunostained embryos. Upper panels show both visceral endoderm and trophoblast contribution at the epiblast/extraembryonic border. Lower panels indicate trophoblast giant cell contribution outside of the distal epiblast. White box marked with ∗ indicates area of high magnification in lower panel.
(E) Immunostaining of E3.5 and E4.5 wild-type blastocysts. Images show confocal optical sections.
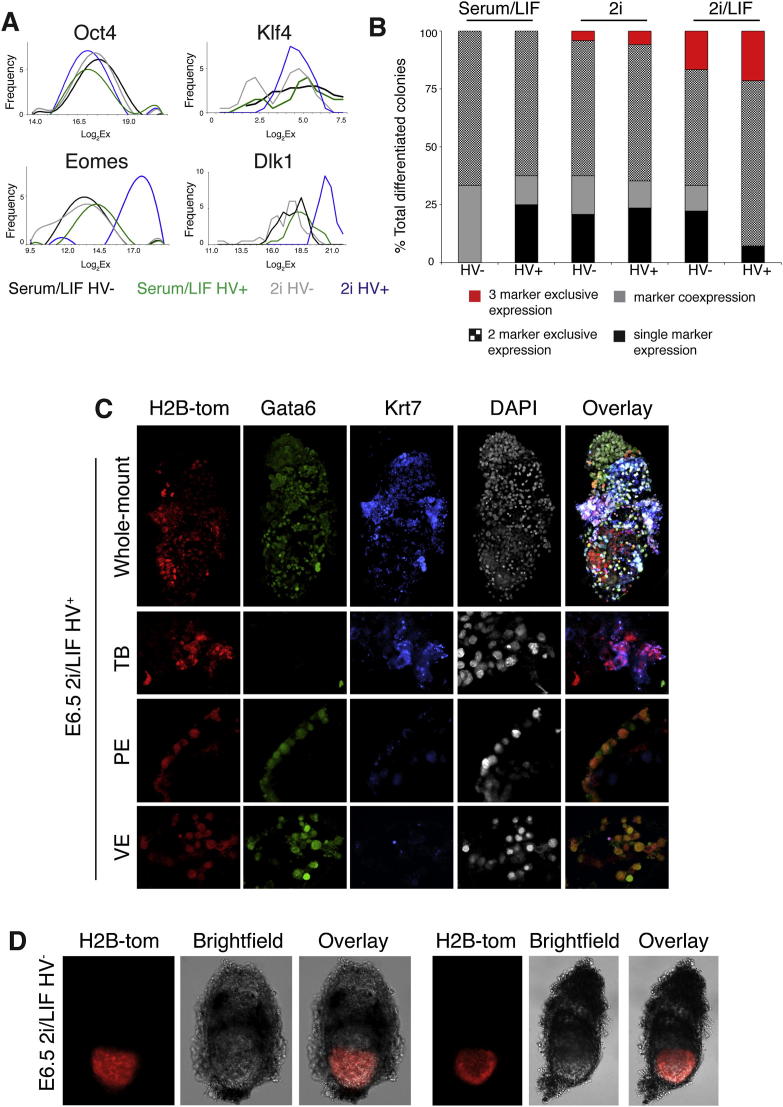
We additionally generated chimeras using HV cells expressing a constitutive H2B-Tomato fluorescent protein (Figures 3C–3F and S4B–S4D). We sorted HV+ cells from 2i/LIF conditions, aggregated these cells with wild-type morulae, and assessed chimera contribution by fluorescence and immunohistochemistry. Whole-mount immunostaining for the extraembryonic endoderm marker GATA6 and trophoblast marker KRT7 showed that H2B-Tomato ESCs in the extraembryonic region were found to express either GATA6 or KRT7 (Figures S4C and S4D). In late blastocysts, H2B-Tomato ESCs were found both in the epiblast and extraembryonic lineages (Figure 3C), and in E9.5 embryos, H2B-Tomato cells were integrated into the placenta and the yolk sac (Figures 3D–3F).
Single Cells Cultured in 21 Are Totipotent
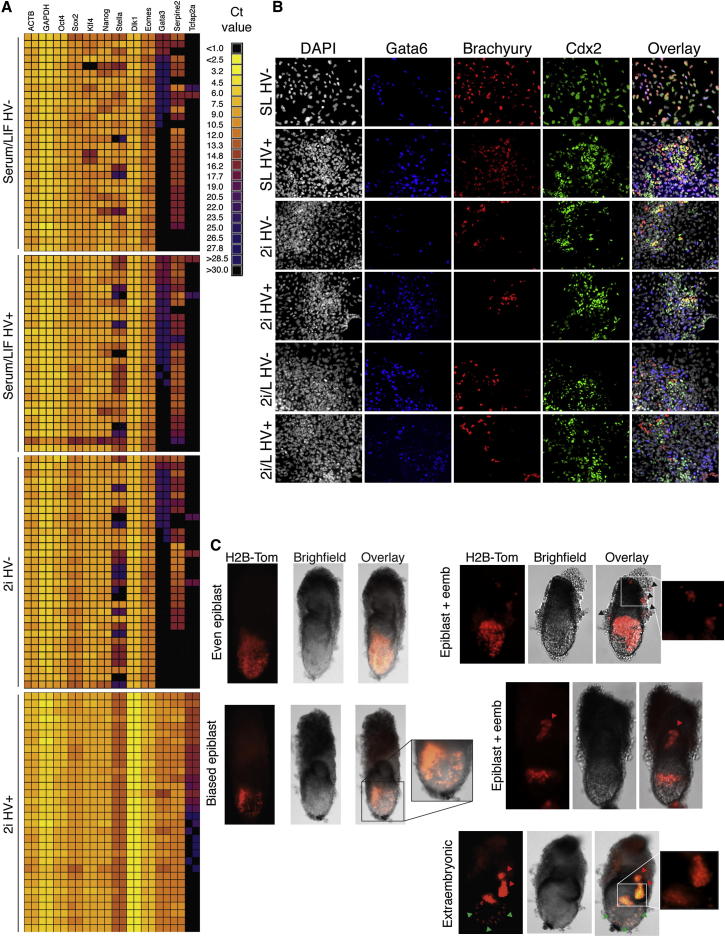
Although we observed contribution of 2i-cultured ESCs to both embryonic and extraembryonic lineages, the definition of totipotency is based on the capacity of a single cell to contribute to all lineages. To distinguish between the presence of individual totipotent cells or a population-based explanation for the totipotent activity of 2i ESC cultures, we assessed gene expression and functional properties of single ESCs. As totipotent cells in the early embryo coexpress both embryonic and extraembryonic determinants, we examined gene expression in single HV− and HV+ sorted ESCs from serum/LIF or 2i to ask if this is also the case in our ESC cultures. For this experiment, a BioMark HD System (Fluidigm) was used with custom designed DELTAgene Assay primer pairs (Fluidigm; sequences are provided in Extended Experimental Procedures). Consistent with our RNA-seq data, HV+ and HV− populations were more distinct in 2i than in serum conditions (Figure 4A). The extraembryonic markers Eomes, Gata3, Serpine2, and Tcfap2a showed considerable variability across all four populations, with low or no expression in HV− and HV+ cells from serum as well as HV− cells in 2i but dramatically enhanced expression in HV+ cells from 2i (Figure 4A). We also observed single-cell enrichment in the imprinted gene Dlk1, identified based on our global expression analysis, specifically in 2i HV+ cells (Figures 4A and S5A). Taken together, our single-cell data showed that individual cells in the 2i HV+ population simultaneously coexpressed epiblast and extraembryonic genes (Figures 4A and S5A). This reinforces the notion that HV+ cells in 2i are distinct from the HV+ population in serum and suggests that in 2i these single cells may possess totipotent properties.
Figure 4.
Single HV+ Cells in 2i Can Contribute to Both Epiblast and Extraembryonic Lineages
Single HV− or HV+ cells from serum/LIF or 2i were sorted by FACS and analyzed by qRT-PCR using the Biomark HD system (Fluidigm).
(A) Heat map for each condition and cell type showing the expression of genes with reliable primer melting curves. Adjacent squares show technical replicates for each cell.
(B) Single cells were sorted by flow cytometry into LIF withdrawal conditions. After 7 days, differentiated colonies were immunostained for GATA6, CDX2, and BRACHYURY.
(C) Single-sorted HV+ cells (sorted from the top 10% of HV expression), previously cultured in 2i/LIF, constitutively expressing H2B-Tomato were injected into morulae and dissected at E6.5 and chimeras (n = 23/64) were assessed for lineage contribution.
Examples of different contribution patterns of single cells are shown. Some cells showed even contribution across the whole epiblast while others showed a contribution that was biased toward a particular area of the epiblast. Red arrowheads indicate trophoblast contribution, green arrowheads indicate visceral endoderm contribution, and black arrowheads indicate parietal endoderm contribution. Epiblast contribution alone was observed in nine chimeras, extraembryonic contribution alone in one chimera, and 13 chimeras showed both epiblast and extraembryonic contribution.
See also Figure S5.
Figure S5.
Single HV+ Cells Are Totipotent in the Ground State, Related to Figure 4
Single cell qRT-PCR was carried out on a BioMark HD System (Fluidigm) using a 96 × 96 chip. DELTAgene Assay custom designed primers (Fluidigm) were used for amplification. Technical replicates were generated for each single cell.
(A) Histograms showing the distribution of expression of 4 genes in each population. Data shown unnormalized.
(B) Single cells were sorted by flow cytometry into LIF withdrawal conditions. After 7 days differentiated colonies were immunostained for GATA6, CDX2 and BRACHYURY. Quantification of marker expression patterns. 2 and 3 marker exclusive expression indicates colonies that contain independent cells positive for different lineage markers.
(C) Whole-mount immunostaining of an E6.5 chimeric embryo generated by injection of a single HV+ ES cell constitutively expressed H2B-Tomato, previously cultured in 2i/LIF, into a morula stage wild-type embryo. Upper whole-mount image shows an extended focus view of the entire embryo. Panels below show optical sections, generated by confocal microscopy, with ES cells contributing to trophoblast and visceral and parietal endoderm (VE and PE, respectively).
(D) E6.5 chimeras generated by injection of a single HV- cell that had been cultured in 2i/LIF. The majority of single cells contributed only to epiblast (11/13), with only 2 showing extraembryonic contribution.
Oct4 and Sox2 showed relatively uniform expression across all populations, while epiblast markers shown to be heterogeneously expressed, such as Nanog and Stella, exhibited more variable expression (Figure 4A). However, when 2i was compared to serum, we observed that Nanog expression became more homogenous. A total of 3 out of 30 ESCs in the serum/LIF HV+ population showed very low or no Nanog expression (data not shown). This was not seen in any other population analyzed. However, these Nanog-negative cells, although expressing genes including Oct4, showed lower levels of housekeeping genes and hence were excluded from further analysis.
Based on the coexpression of epiblast and extraembryonic genes in single cells, we asked whether individual cells grown in 2i could differentiate into both embryonic and extraembryonic cells in vitro. Single cells that had previously been cultured in serum/LIF, 2i, or 2i/LIF were sorted into medium without LIF and differentiated under clonal conditions. Multilineage differentiation was quantified based on the expression pattern of specific lineage markers (GATA6, CDX2, and BRACHYURY) within differentiated colonies (Figures 4B and S5B). Coexpression of these markers in the same cell is likely to represent mesendoderm, while the presence of all three in separate cells would suggest the presence of PE, trophoblast, and epiblast/mesoderm. Cells that had previously been cultured in serum/LIF generated colonies either exclusively expressing one or two markers or coexpressing multiple markers within the same cells. Conversely, cells cultured in 2i or 2i/LIF showed a decrease in single-marker expression and coexpression in favor of mutually exclusive expression of all three lineage markers within the same colony (Figures 4B and S5B). Thus, individual 2i-cultured and, in particular, HV+ cells have the capacity to generate all three lineages: epiblast, trophoblast, and PE. Additionally, cells cultured in 2i before differentiation showed an increase in the expression of CDX2 alone, indicative of trophoblast differentiation, compared to cells previously cultured in serum (data not shown). While this in vitro assay gives a crude estimate of the potency of individual ESCs, it is difficult to assign totipotency based solely on marker expression in the absence of embryonic context.
We therefore also assessed totipotency in vivo by asking whether single ESCs could contribute to all lineages when reintroduced into embryos. Single-cell morula injections of 2i/LIF HV+ cells were carried out and resulting chimeric E6.5 embryos were scored for contribution to different lineages (Figure 4C). More than half of the chimeras showed evidence of extraembryonic contribution (n = 23). One embryo showed contribution only to extraembryonic tissue, both endoderm and trophoblast, while 13 showed contribution both to extraembryonic tissue and epiblast (Figures 4C and S5C), indicating that this fraction contained a significant proportion of cells with extraembryonic potential. Of these 13, two showed contribution to the endoderm, eight showed trophoblast contribution, and three showed contribution to both the endoderm and trophoblast. Single HV− cells cultured in 2i/LIF were also injected as a control and, as observed for the multiple cell injections, only a small fraction of cells were able to contribute to the extraembryonic region (Figure S5D; n = 13).
LIF Supports Extraembryonic Priming
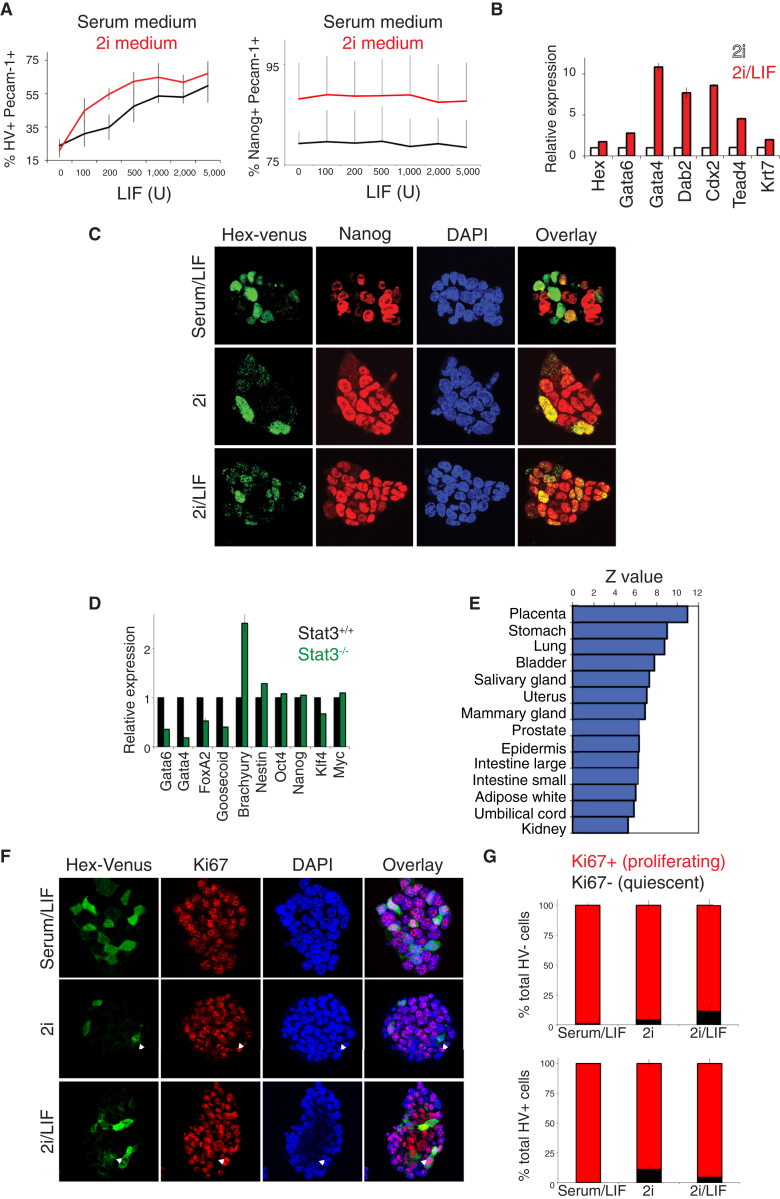
In our assessment of clonal ESC potential, we observed a significant enhancement of the potential of cells grown in LIF to generate the extraembryonic lineages (Figures 3A, 3B, and 4B). When either 2i or serum-containing medium was supplemented with increasing doses of LIF, the expression of the HV transgene and other extraembryonic markers increased (Figures 2A, 5A–5D, and S6A–S6C). However, a similar dose-response experiment using a Nanog-GFP ESC reporter line (Chambers et al., 2007) showed no change in GFP levels, even when LIF was present at a 5-fold excess of its normal saturating dose (Figures 5A and S6B). In both 2i and 2i/LIF, the majority of HV+ cells expressed NANOG, with this coexpressing cell population representing approximately 15%–20% of the culture (Figures 5C and S6D). In serum/LIF, NANOG and HV expression were predominantly mutually exclusive (Figure 5C).
Figure 5.
LIF Supports an Extraembryonically Primed Population of Cells
(A) Flow cytometry of HV or TNGB Nanog-GFP cells cultured in serum or 2i medium, with increasing LIF concentrations (n = 3). Raw data and controls are in Figures S6A–S6C. Error bars indicate mean ± SD of two biological replicates.
(B) qRT-PCR showing relative gene expression of extraembryonic markers in ESCs cultured in 2i or 2i/LIF. Data were normalized to the housekeeping gene TBP and are shown relative to 2i samples. Error bars indicate mean ± SD of biological replicates.
(C) Confocal optical section showing immunostaining of HV cells for NANOG after culture of cells in serum/LIF, 2i, or 2i/LIF.
(D) qRT-PCR showing relative gene expression in Stat3+/+ or −/− ESCs. Data are shown relative to wild-type expression levels. All qRT-PCR transcript levels in this figure were normalized to TBP and are shown as mean ± SD.
(E) Statistical significance of correlation of gene expression differences between ESCs cultured in the presence and absence of LIF and tissue-specific gene expression in the GNF Ver.3 database. Correlation was estimated from data on 5,085 genes, which represent the overlap between 10,000 most informative genes in our RNA-seq data and 10,000 most informative genes in the GNF database. Organs/tissues sorted by decreasing correlation.
(F) Confocal optical section of HV cells immunostained for the proliferation marker KI67. White arrowheads indicate KI67-negative cells.
(G) Quantification of KI67 immunostaining in HV+ and HV− ESCs. The absolute number of proliferating cells (expressing KI67, red bar) and quiescent cells (not expressing KI67, black bar) were counted. Error bars indicate SD of the mean from five colonies in each condition, analyzed by confocal microscopy.
See also Figure S5.
Figure S6.
LIF Increases HV Expression within an Undifferentiated Population, Related to Figure 5
(A and B) Flow cytometry of HV cells (A) and Nanog-GFP TNG cells (B) in the presence and absence of LIF. Control E14 cells are also shown. Cells were cultured at high density to prevent differentiation.
(C) While LIF stimulates the HV reporter (Figure S1) it does not enhance levels of autofluorescence in wild-type E14 ESCs.
(D) Quantification of colocalization of HV and NANOG in serum/LIF, 2i and 2i/LIF. Data, based on confocal optical sections of 5 individual colonies of cells per condition, shown as mean ± SD.
(E) Differentiation by LIF withdrawal from unsorted cultures in 2i or 2i/LIF. Cells were immunostained for GATA6 (n = 3).
(F) Quantification of GATA6 immunostaining. Values relative to 2i. Error bars indicate mean ± SD of biological replicates (∗∗∗p < 0.001). White dotted lines highlight GATA6+ endoderm clusters. See also Figure S3 and methods section.
(G) Summary of RNA-seq expression data for endoderm and trophoblast (left), canonical ESC (middle) and mesoderm and neurectoderm markers (right) for unsorted samples cultured in 2i or 2i/LIF. Error bars indicate mean ± SD of two biological replicates. Values are relative to 2i sample.
As the ability of LIF to mediate self-renewal is thought to be dependent on STAT3, we examined gene expression in Stat3−/− ESCs (Ying et al., 2008) compared to wild-type cells derived at the same time. Stat3−/− ESCs exhibited decreased levels of extraembryonic gene expression (endodermal and trophoblast) (Figure 5D), but, other than the STAT3 target gene Klf4, pluripotency markers were not affected.
Additionally, preculture of ESCs in 2i/LIF rather than 2i alone before differentiation by LIF withdrawal caused a significant increase in the levels of endoderm generated (Figures S6E and S6F).
We assessed the degree to which LIF induced extraembryonic gene expression in ESC culture by whole genome expression analysis, using RNA-seq, on unsorted ESCs cultured in 2i or 2i/LIF. Extraembryonic endoderm and trophoblast genes were both upregulated upon the addition of LIF, while neuroectoderm and mesoderm markers were reduced (Figure S6G). Moreover, genes upregulated by LIF in 2i demonstrated the strongest correlation with placental gene expression (Genomics Institute of the Novartis Research Foundation [GNF] Gene Expression Database; Figure 5E). In 2i, LIF increased the proportion of proliferating HV+ cells while decreasing the proportion of HV− proliferating cells (Figures 5F and 5G), suggesting that LIF promoted the expansion of the HV+ totipotent population.
Discussion
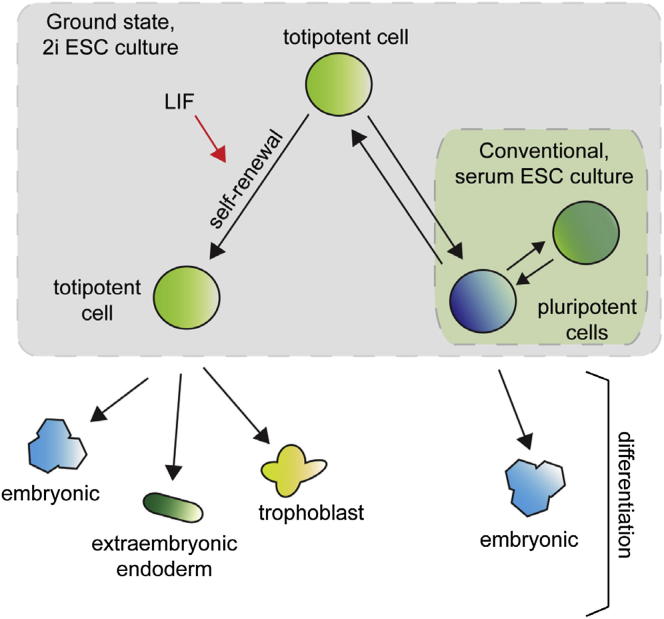
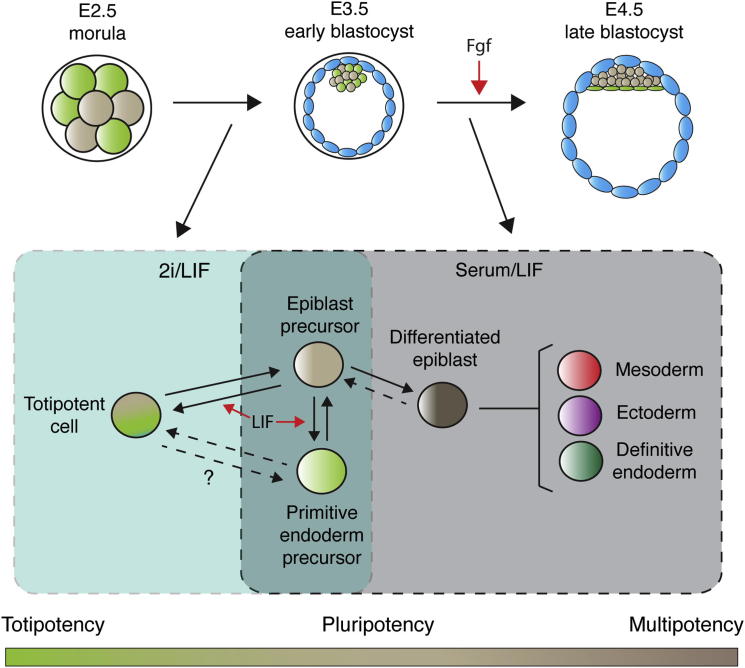
In this paper, we demonstrated that ESCs and embryos grown in 2i are heterogeneous with respect to extraembryonic gene expression. We identified a LIF-promoted population containing single cells that could give rise to trophoblast and PE as well as epiblast, a behavior characteristic of totipotent cells. This suggests that HV+ cells cultured in 2i may reflect an earlier developmental stage than widely believed to exist in ESC culture and that the role of LIF in supporting self-renewal may be a consequence of its capacity to support this population. A model illustrating these findings is shown in Figure 6.
Figure 6.
Model Comparing Standard Serum/LIF ESC Culture to 2i ESC Culture
The model relates serum/LIF and 2i ESC cultures to embryonic development. 2i/LIF cultures contain totipotent single cells that are comparable to early preimplantation stages when embryonic cells are still totipotent. As Fgf is needed for later lineage segregation, inhibiting Fgf signaling in 2i seems to block embryos and ESCs at a point prior to this commitment event. Serum/LIF ESC cultures are reminiscent of later embryonic stages when cells are pluripotent and contain more restricted cell types. Although partially differentiated mesoderm and ectoderm appear to be lost in 2i/LIF, endoderm is maintained. Based on our data, totipotent ESCs are able to dynamically interconvert to a pluripotent epiblast precursor state and may also exist in equilibrium with a similar primitive endoderm precursor, although we have not been able to distinguish this population in this study. LIF supports the expansion of the totipotent ESC state and primitive endoderm precursors in serum.
Culture in 2i is purported to maintain a “naïve” pluripotent state by shielding ESCs from differentiation-promoting signals (Nichols et al., 2009; Wray et al., 2011), but despite more homogeneous Nanog expression, there is little evidence that cells in 2i represent a single cell type. Moreover, 2i culture is regularly supplemented with LIF (Nichols et al., 2009; Ying et al., 2008) and, while LIF is known to upregulate pluripotency markers (Hall et al., 2009), its role in vivo is to support extraembryonic development (Stewart et al., 1992; Takahashi et al., 2008). In embryoid body differentiation, LIF selectively blocks primitive ectoderm differentiation while permitting PE differentiation (Shen and Leder, 1992). Additionally, the downstream effector of LIF, STAT3, binds directly to extraembryonic gene promoters such as Gata6, Gata3, and Eomes (Kidder et al., 2008). Thus, the role of LIF in supporting ESC culture may be mediated via a totipotent, extraembryonically primed cell type capable of effectively expanding and dynamically generating the heterogeneous distribution of cells normally observed in ESC culture.
Preimplantation embryos cultured in 2i retained the early heterogeneous expression of Hex, and ground state ESCs contained a population capable of generating PE and trophoblast. This suggests that 2i blocks the commitment of totipotent cells to embryonic or extraembryonic lineages. Cells and embryos maintain the coexpression of epiblast and extraembryonic markers that occurs in early preimplantation development and has been observed for the embryonic markers NANOG and OCT4 and the extraembryonic marker CDX2 (Dietrich and Hiiragi, 2007). The 2i MEK inhibitor PD0325901 blocks fibroblast growth factor signaling, which is important for the segregation of epiblast and PE (Hamazaki et al., 2006; Yamanaka et al., 2010; Chazaud et al., 2006; Nichols et al., 2009; Yamanaka et al., 2010). It is also important for the support and expansion of trophoblast (Quinn et al., 2006); thus, blocking this signal could halt embryonic-extraembryonic lineage segregation. Similar observations have been made for another PE reporter, PDGFR-alpha-GFP, where embryos cultured in 2i continued to express this marker heterogeneously within the ICM, although this expression was attributed to the persistence of the stable GFP (Yamanaka et al., 2010). We found that this low-level expression of extraembryonic markers correlated with totipotent function in ESCs, reminiscent of the activity of primed blastomeres expressing low-levels of PDGFR-alpha, shown to have greater lineage flexibility than their epiblast-primed counterparts (Grabarek et al., 2012).
Although it has been shown previously that ESCs can contribute to extraembryonic lineages (Beddington and Robertson, 1989; Canham et al., 2010; Lallemand and Brûlet, 1990; Macfarlan et al., 2012; Suemori et al., 1990), these events occurred at a low frequency. Thus, totipotent cells could exist in most ESC cultures, but because they are relatively rare, they are frequently not acknowledged in literature. Although relatively rare in standard culture conditions, our data suggest that these remarkable individual ground state ESCs have the capacity to generate the majority of the epiblast. While the extent of extraembryonic contribution that we observed was never as great as that which we observed in the epiblast, the contribution to trophoblast was at least comparable to that observed in chimeras generated from trophoblast stem (TS) cells (Quinn et al., 2006) and the extent of visceral endoderm contribution to that observed in chimeras generated from undifferentiated extraembryonic endoderm cells (Kunath et al., 2005).
It has been noted that some cells within standard ESC cultures express embryonic two-cell-stage transcripts and can contribute to both embryonic and extraembryonic lineages (Macfarlan et al., 2012). These cells lack expression of epiblast markers such as OCT4, NANOG, and SOX2, although these genes are expressed from early in mammalian development and throughout the period that mammalian embryos are considered totipotent. Rossant et al. demonstrated that, up until the early blastocyst stage, embryonic cells posses a similar level of flexibility to that demonstrated here for HV+ ground state ESCs. ICM cells contribute to the trophoblast in morula aggregations and isolated early ICMs implant into the uterus (Rossant and Lis, 1979). Additionally, the majority of outer trophoblast progenitor cells can contribute to the ICM and epiblast in blastocyst injections (Rossant and Vijh, 1980). We also observed that, at this early stage of blastocyst formation, a subset of trophoblast cells expressed high levels of NANOG in conjunction with CDX2 (Figure S4E). Similarly, NANOG, OCT4, and CDX2 have been observed to be coexpressed throughout early preimplantation development in both ICM and trophoblast cells (Dietrich and Hiiragi, 2007). Thus, at these stages, the ICM and trophoblast appear to retain plasticity, which is lost as embryos progress to the late blastocyst (Handyside, 1978; Hogan and Tilly, 1978; Rossant and Lis, 1979; Spindle, 1978), where certain ICM cells only exhibit pluripotency (Gardner and Rossant, 1979). Our findings suggest that 2i/LIF promotes the expansion of a totipotent population of cells, reminiscent of these early developmental stages in vivo.
Experimental Procedures
Cell Culture and Differentiation Assays
ESCs were cultured in serum/LIF (Canham et al., 2010), 2i (Ying et al., 2008), or 2i/LIF. Stat3−/− ESCs were maintained in 2i/LIF. H2B-Tomato cell lines were generated by introducing a H2B-Tomato vector under the control of a CAG promoter and upstream of IRES Puro cassette (a kind gift from H. Lickert) into the HV cell line.
LIF withdrawal and neural differentiation in monolayer culture were carried out as described previously (Fujikura et al., 2002; Ying et al., 2003b). For LIF withdrawal, 3 × 103 cells were plated per well of a six-well plate. Cells were cultured in standard serum-containing medium without LIF for 5 days. For neural differentiation, 104 ESCs were plated per well of a six-well plate in N2B27 medium for 9 days. For trophoblast stem cell differentiation assays, 104 cells were plated per well of a six-well plate. Cells were plated into trophoblast stem cell medium (70% mouse embryonic fibroblast-conditioned medium [R&D] and 30% TS cell medium [RPMI, GIBCO; glutamine and sodium pyruvate, GIBCO; 0.1 mM β-mercaptoethanol, Sigma; 20% fetal calf serum]). A total of 25 ng/ml Fgf4 (Peprotech) was added to medium along with 1 μg/ml heparin sulfate (Sigma). Cells were cultured for 7 days before analysis of differentiation levels by immunostaining.
Flow Cytometry
Cells were collected using Accutase dissociation buffer (A6964, Sigma) and antibody staining for PECAM-1 was carried out (Canham et al., 2010). Cells were sorted with a BD FACS Aria II cell sorter SORP or a BD FACS Aria III. DAPI-low, PECAM-1+ cells were sorted from the upper and lower 10% or 25% of HV expression. In flow cytometry purity checks, these sorted populations showed a good separation and demonstrated gene expression and functional differences upon further analysis (Figure S1B). To assess population interconversion, single cells were sorted twice, taking the top and bottom 1% of HV-expressing cells to ensure clear separation. Flow cytometry and PDGFR-alpha antibody staining after LIF withdrawal was carried out as previously reported (Rugg-Gunn et al., 2012). Analysis was performed using FlowJo software (Tree Star). Gating methodology is shown in Figure S1B and an example purity check is shown in Figure S1C.
Immunostaining and Quantification
Immunostaining was carried out as previously reported (Canham et al., 2010) utilizing antibody concentrations described in Table S1. Endoderm levels were quantified by counting defined clusters (Figure 3F; example shown in Figure S3A) or absolute numbers of GATA6+ cells (Figure S3B) and neural differentiation by counting absolute numbers of TUJ1+ elongated neurons. Absolute numbers of CDX2+ cells were counted. Two wells of a 12-well plate were analyzed per biological replicate and three biological replicates completed for each experiment. p values were calculated using one-way ANOVA tests. Images were acquired at 10× or 20× magnification. Colocalization studies with NANOG and KI67 were analyzed by confocal microscopy using an antibody against GFP to detect HV expression.
qRT-PCR
RNA was isolated from cells (RNeasy, 74104; QIAGEN). Complementary DNA (cDNA) was synthesized from 1 μg RNA using Superscript III according to the manufacturer’s guidelines (18080, Invitrogen). Quantitative RT-PCR (qRT-PCR) was carried out on a Lightcycler480 (Roche) using either SYBR green or UPL technologies (Roche) utilizing primer sequences specified in Table S2. Primers were used at a concentration of 1 μM. The housekeeping gene TBP was used for normalization of Ct values detected for each sample.
RNA-Seq
Approximately 10 μg of total RNA underwent two rounds of mRNA enrichment with Dynalbeads Oligo(dT)25 (61005, Life Technologies). Solid whole transcriptome libraries were made according to the Solid Total RNA-seq protocol with the exception of RNA fragmentation (4445374, Life Technologies). RNA was fragmented by chemical hydrolysis; heating to 95°C, 10 min in 1× RNase III buffer (AM2290, Life Technologies), and snap cooled on ice. ATP (0.83 mM, 11140965001, Roche) and 10 U of T4 PNK (M0201L, NEB) were added and incubated at 37°C for 30 min. RNA was purified using Purelink RNA Micro Kit (12183-016, Life Technologies). Equimolar pools of RNA-seq libraries were made following quantitative PCR quantification using a Kapa Library Quantification kit (KK4823, Kapa Biosystems). Emulsion PCR and templated bead enrichment was carried out with Solid EZ bead system according to the manufacturer’s guidelines. Enriched beads were sequenced on an ABi SOLiD 4 analyzer according to the manufacturer’s instructions to generate 50 bp reads in color space.
Ethics Statement
All animal work was carried in accordance with UK and European legislation and in particular according to the regulations described in the Animals (Scientific Procedures) Act of 1986 (UK). All work in this manuscript was authorized by and carried out under Project License 60/3715 issued by the UK Home Office. Genetic modification for the generation of mouse and mouse ESC lines were approved by the ethics committees of the University of Edinburgh and the University of Copenhagen.
HV Transgenic Mouse Line Generation
The HV mouse line was generated by blastocyst injection of E14 Ju09 HV ESCs into wild-type F1 blastocysts. E14 Ju09 HV cells were generated by homologous recombination using the HV construct described previously (Canham et al., 2010) into E14 Ju09 ESCs. Mice were backcrossed onto the C57BL/6 background and maintained as heterozygotes. PCR genotyping was carried out on tail biopsies using the following primers: 5′-CGGAGGCGAATCTGAAGCCAGC-3′ (forward), 5′-GCATACAGCGGGACTCCCACG-3′ (reverse).
Early Embryo Imaging and Culture
Wild-type C57BL/6 or transgenic HV mouse lines were used for all experiments. Mice were checked for copulation plugs each morning and embryos were considered E0.5 on the day of plug detection. Embryos were flushed either from oviducts at E2.5 or from the uterus at E3.5 in PB1 medium. Embryos were cultured in control KSOM medium (Millipore) or KSOM medium containing 1 μM PD032 and 3 μM CHIR99021 (2i) (Nichols et al., 2009) and imaged by confocal microscopy (Leica, TCS SP2, or TCS SP8) at 20X magnification. Embryos were scored (Figure 1D) as showing homogeneously high or low HV expression relative to one another or as demonstrating heterogeneous HV expression when a mix of high and low HV-expressing cells was present.
Chimera Generation
Chimeric mouse generation was performed by morula aggregation or morula injection of single cells. HV cells constitutively expressing LacZ-IRES-Puro from a CAG promoter or an H2B-Tomato fusion protein were sorted by flow cytometry into HV+ or HV− populations. Clusters of eight to ten cells were aggregated with wild-type F1 morulae or single cells were injected. For single cell injections, morulae were incubated in PB1 medium without calcium and magnesium for 15 min at room temperature to facilitate decompaction for ease of injection. Resultant embryos were cultured for 3 days in vitro (equivalent to E4.5 stage in vivo) or transferred to pseudopregnant female mice and harvested at E6.5 or E9.5 and subjected to X-gal staining (Canham et al., 2010) or immunostaining and fluorescence imaging. X-gal-stained embryos were cryosectioned and fluorescent embryos were wax sectioned.
Whole-Mount Immunostaining
E6.5 embryos were dissected and fixed in 4% PFA for 1–2 hr at 4°C. Embryos were washed twice with PBS for 10 min followed by two 1 hr washes in PBST (PBS with 5% serum and 0.1% Triton X-100) at 4°C. Primary antibodies were diluted to 1:100 in PBST and incubated overnight at 4°C. Embryos were washed twice in PBST for 15 min, at room temperature, followed by five 1 hr washes. They were incubated in the secondary antibody for 2 hr at room temperature followed by two 15 min washes and a further five 1 hr washes.
Single-Cell qRT-PCR
Single cells were sorted into 96-well PCR plates containing 5 μl CellsDirect reaction mix, 0.2 μl SuperScriptIII/Platinum Taq mix (CellsDirect One-Step qRT-PCR kit, Invitrogen), 2.8 μl DNA suspension buffer (TEKnova), and 1 μl 500 nM primer mix containing a mix of 48 DELTAgene Assays (Fluidigm) (sequences in Table S3). Controls of 100 and 1,000 cells were included. RT reaction conditions were 50°C, 15 min; 95°C, 2 min; and 22× (95°C, 15 s; 60°C, 4 min). An exonuclease step was performed to remove unincorporated primers at 37°C, 30 min and 80°C, 15 min. Amplification products were then diluted 5-fold in TE buffer. Amplified cDNA was mixed with SsoFast EvaGreen SuperMix with Low ROX (Bio-Rad). The same DELTAgene assays were used in qRT-PCR. Samples and assays were mixed with appropriate loading reagents and loaded onto a 96.96 gene expression Dynamic Array (Fluidigm). Samples were loaded in technical replicates. Arrays were read using a BioMark HD genetic analysis system (Fluidigm). Downstream analysis was completed in Microsoft Excel. Cells that expressed no or low levels of ACTB and GAPDH housekeeping genes, or with a Ct over 30, were excluded from further analysis. DELTAgene assays were custom designed by Fluidigm to cross introns and avoid amplifying genomic DNA. Assays showing poor melting curves were also excluded from analysis. Data were analyzed without normalization or also normalized to the median expression of all genes across the array. No significant difference was observed in the data generated from either analysis method.
Acknowledgments
We thank Lynsey Robertson, Sally Inverarity, Javier Martin Gonzalez, Kasper Bonderup, Ron Wilkie, Simon Monard, Gelo de la Cruz, and Valerie Wilson for technical assistance; Austin Smith, Heiko Lickert, and Ian Chambers for reagents; and Sophie Astrof, Sally Lowell, and the entire Brickman lab for critical discussion of this manuscript. This work was supported by grants from the Medical Research Council (MRC) (G0701428) and the Novo Nordisk Foundation (to J.M.B.), the Intramural Research Program of the National Institutes of Health, and the National Institute on Aging (Z01AG AG000656 and Z01AG000662 to M.S.H.K.). S.M.M. is supported by an MRC studentship, and J.M.B. is an MRC senior nonclinical fellow. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. S.M.M. conceived of and executed experiments and wrote the paper with J.M.B.; M.A.C. generated the HV mice and ESCs; J.N. designed and executed the experiments and helped with embryo culture; A.A.S. analyzed RNA-seq data sets; R.P.M. executed experiments; M.S.H.K. supervised data analysis; and J.M.B. designed and supervised experiments and wrote the paper with S.M.M.
Published: June 6, 2013
Footnotes
Supplemental Information includes Extended Experimental Procedures, six figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.04.034.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
The Gene Expression Omnibus accession number for the RNA-seq data reported in this paper is GSE45182.
Supplemental Information
Pair-wise comparisons (FDR < 0.05, >2-fold expression difference) were performed between HV− and HV+ populations to reveal significant changes in gene expression; 47 genes upregulated and 31 downregulated.
Pair-wise comparisons (FDR < 0.05, >2-fold expression difference) were performed between HV- and HV+ populations to reveal significant changes in gene expression; 60 genes upregulated and 272 downregulated.
References
- Beddington R.S., Robertson E.J. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- Canham M.A., Sharov A.A., Ko M.S., Brickman J.M. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Dietrich J.E., Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Fujikura J., Yamato E., Yonemura S., Hosoda K., Masui S., Nakao K., Miyazaki Ji J., Niwa H. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002;16:784–789. doi: 10.1101/gad.968802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.L., Rossant J. Investigation of the fate of 4-5 day post-coitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol. 1979;52:141–152. [PubMed] [Google Scholar]
- Grabarek J.B., Zyzyńska K., Saiz N., Piliszek A., Frankenberg S., Nichols J., Hadjantonakis A.K., Plusa B. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012;139:129–139. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Hamazaki T., Kehoe S.M., Nakano T., Terada N. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol. Cell. Biol. 2006;26:7539–7549. doi: 10.1128/MCB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handyside A.H. Time of commitment of inside cells isolated from preimplantation mouse embryos. J. Embryol. Exp. Morphol. 1978;45:37–53. [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Furue M.K., Tanaka S., Hirose M., Wakisaka N., Danno H., Ohnuma K., Oeda S., Aihara Y., Shiota K. BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell. Dev. Biol. Anim. 2010;46:416–430. doi: 10.1007/s11626-009-9266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B., Tilly R. In vitro development of inner cell masses isolated immunosurgically from mouse blastocysts. I. Inner cell masses from 3.5-day p.c. blastocysts incubated for 24 h before immunosurgery. J. Embryol. Exp. Morphol. 1978;45:93–105. [PubMed] [Google Scholar]
- Kidder B.L., Yang J., Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T., Arnaud D., Uy G.D., Okamoto I., Chureau C., Yamanaka Y., Heard E., Gardner R.L., Avner P., Rossant J. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132:1649–1661. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- Lallemand Y., Brûlet P. An in situ assessment of the routes and extents of colonisation of the mouse embryo by embryonic stem cells and their descendants. Development. 1990;110:1241–1248. doi: 10.1242/dev.110.4.1241. [DOI] [PubMed] [Google Scholar]
- Liu L., Luo G.Z., Yang W., Zhao X., Zheng Q., Lv Z., Li W., Wu H.J., Wang L., Wang X.J., Zhou Q. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J. Biol. Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlmann T.G., Fitzgerald J.S., Meissner A., Wengenmayer T., Schleussner E., Friedrich K., Markert U.R. Trophoblast invasion: tuning through LIF, signalling via Stat3. Placenta. 2005;26(Suppl A):S37–S41. doi: 10.1016/j.placenta.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Prakash G.J., Suman P., Morales Prieto D.M., Markert U.R., Gupta S.K. Leukaemia inhibitory factor mediated proliferation of HTR-8/SVneo trophoblast cells is dependent on activation of extracellular signal-regulated kinase 1/2. Reprod. Fertil. Dev. 2011;23:714–724. doi: 10.1071/RD10315. [DOI] [PubMed] [Google Scholar]
- Quinn J., Kunath T., Rossant J. Mouse trophoblast stem cells. Methods Mol. Med. 2006;121:125–148. doi: 10.1385/1-59259-983-4:123. [DOI] [PubMed] [Google Scholar]
- Rossant J., Lis W.T. Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev. Biol. 1979;70:255–261. doi: 10.1016/0012-1606(79)90022-8. [DOI] [PubMed] [Google Scholar]
- Rossant J., Vijh K.M. Ability of outside cells from preimplantation mouse embryos to form inner cell mass derivatives. Dev. Biol. 1980;76:475–482. doi: 10.1016/0012-1606(80)90395-4. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Cox B.J., Lanner F., Sharma P., Ignatchenko V., McDonald A.C., Garner J., Gramolini A.O., Rossant J., Kislinger T. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev. Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.M., Leder P. Leukemia inhibitory factor is expressed by the preimplantation uterus and selectively blocks primitive ectoderm formation in vitro. Proc. Natl. Acad. Sci. USA. 1992;89:8240–8244. doi: 10.1073/pnas.89.17.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.M., Hamazaki T., Hankowski K.E., Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Spindle A.I. Trophoblast regeneration by inner cell masses isolated from cultured mouse embryos. J. Exp. Zool. 1978;203:483–489. doi: 10.1002/jez.1402030315. [DOI] [PubMed] [Google Scholar]
- Stewart C.L., Kaspar P., Brunet L.J., Bhatt H., Gadi I., Köntgen F., Abbondanzo S.J. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Suemori H., Kadodawa Y., Goto K., Araki I., Kondoh H., Nakatsuji N. A mouse embryonic stem cell line showing pluripotency of differentiation in early embryos and ubiquitous beta-galactosidase expression. Cell Differ. Dev. 1990;29:181–186. doi: 10.1016/0922-3371(90)90120-l. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Carpino N., Cross J.C., Torres M., Parganas E., Ihle J.N. SOCS3: an essential regulator of LIF receptor signaling in trophoblast giant cell differentiation. EMBO J. 2003;22:372–384. doi: 10.1093/emboj/cdg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Takahashi M., Carpino N., Jou S.T., Chao J.R., Tanaka S., Shigeyoshi Y., Parganas E., Ihle J.N. Leukemia inhibitory factor regulates trophoblast giant cell differentiation via Janus kinase 1-signal transducer and activator of transcription 3-suppressor of cytokine signaling 3 pathway. Mol. Endocrinol. 2008;22:1673–1681. doi: 10.1210/me.2008-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Smith A.G. The ground state of pluripotency. Biochem. Soc. Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.H., Chen X., Li D.S., Li R., Addicks G.C., Glennon C., Zwaka T.P., Thomson J.A. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pair-wise comparisons (FDR < 0.05, >2-fold expression difference) were performed between HV− and HV+ populations to reveal significant changes in gene expression; 47 genes upregulated and 31 downregulated.
Pair-wise comparisons (FDR < 0.05, >2-fold expression difference) were performed between HV- and HV+ populations to reveal significant changes in gene expression; 60 genes upregulated and 272 downregulated.