Abstract
Obermoser et al. (2013) systematically analyzed and compared the blood transcriptomic response of the pneumococcal and influenza vaccines over multiple time-points spanning hours to tens of days. They then constructed web-based interactive figures to facilitate exploration of this large complex data set.
Advances in high-throughput multiplexed technologies for measuring states of biological systems from molecules to cells to whole organisms are rapidly turning biology into a “big data” science. Immunology is no exception. While the immune system and its response to perturbations such as infection and cancer are invariably complex involving intricate molecular and cellular orchestrations over space and time, the increasingly comprehensive view offered by global studies of immunity are complementing more traditional approaches to chip away at this complexity. Microarray profiling of peripheral blood before and after yellow fever and influenza vaccination, for instance, have revealed both expected (e.g., plasmablast) and new response signatures and have led to novel hypotheses on the genetic regulation of antibody and CD8+ T cell responses, which in turn could help facilitate rational design of new vaccines (Pulendran et al., 2010). The extent to which immune responses vary among different vaccines, however, remains largely unexplored at the genome-wide scale. In this issue of Immunity, Obermoser, Palucka, Chaussabel and colleagues systematically and comprehensively compare the blood transcriptomic responses of two widely-used vaccines: the pneumococcal vaccine composed of polysaccharide extracts and the influenza split vaccine that comprises inactivated viral components (Obermoser et al., 2013).
In addition to providing a comprehensive analysis of the shared and distinct transcriptome signatures induced by two important vaccines, this work contributes a rich data set for the investigation of vaccine responses in humans. However, exploration of such large data sets requires custom bioinformatics analysis, which is typically accessible only to individuals with computational and statistical expertise. Here the authors provide an innovative solution: for the first time in Immunity (and to the best of our knowledge, in any scientific journal), readers can further explore the data (not just the images) presented in all of the figures through an aesthetically appealing and functionally rich set of web tools called interactive figures (iFigures). These interactive data-browsing and analysis tools render such complex data sets immediately accessible to a diverse biological audience.
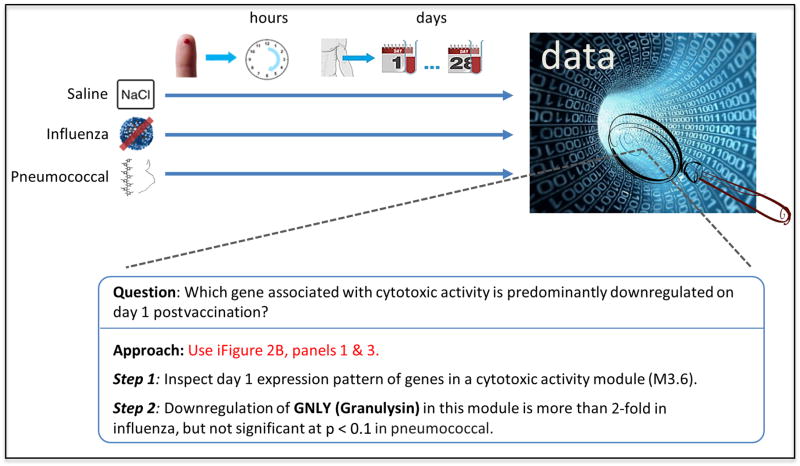
The design of this study is noteworthy (Figure 1). In addition to the typical time-points (days 0, 1, 3, and 7) profiled in previous studies of yellow fever and influenza vaccination, this work investigates longer term responses by sampling from days 10, 14, 21 and 28. To examine innate responses over a dense set of early time-points in the scale of hours after vaccination, the authors profile minute amounts of blood from finger-prick samples in a separate cohort of individuals. Also included is a parallel placebo arm with the same set of time-points where saline was administered in lieu of the vaccine. The lack of substantial response activity following saline injection provides reassurance that the data-generation platforms are robust; it also indicates that intramuscular injection elicits minimal transcriptomic activities in the blood, thus those observed in the main arms are vaccine-specific. Another nice feature is the inclusion of an extra pre-vaccination time-point (day -7) for obtaining more robust baseline estimates. The cohort sizes are relatively small, making robust inference of correlates of immunogenicity and control for confounding by outlier individuals difficult. However, the sample size is sufficient for deriving robust vaccine signatures, and the careful design and methodology of the study will serve as an invaluable reference for future blood transcriptomic studies in humans, such as ones involving pediatric subjects, dense time-points and minute blood volumes.
Figure 1.
The top panel depicts the study design: three parallel arms for saline, influenza or pneumococcal vaccine injections respectively; these arms were performed twice in different cohorts, once for profiling samples from early time-points in the scale of hours and the other for time-points in the scale of days following injection. This study generates large-scale data that can be further explored with interactive,, which accelerate finding answers to specific questions.
A major challenge in any study involving large-scale data is the transformation of such data sets into biological insights, with one of the main obstacles being the curse of the high dimensionality of such data sets. The authors address this in a couple of ways. First, they reduce the dimensionality of transcriptomic responses from tens of thousands of genes to 62 functionally annotated gene modules, which are sets of co-expressed genes derived from multiple independent blood transcriptomic datasets (Chaussabel et al., 2008). Similarly, they draw insights from a host of early responding genes by comparing them to a manageable set of innate transcriptomic signatures, which are obtained from the stimulation of in vitro blood cell culture with diverse stimuli, including cytokines, Toll-like receptor (TLR) ligands, live viruses and killed bacteria. Second, iFigures provide a point-and-click web interface to expand the static views of traditional figures and allow the reader to explore the data from different perspectives, thus offering a great level of flexibility and transparency. The reader can now test the robustness of the authors’ conclusions to different P-value or fold-change cutoffs, inspect the flow-cytometry gating details when examining figures depicting cell-population frequencies, select and further investigate the genes inside modules of interest (Figure 1) — these are but a few of the many exploratory possibilities. While releasing the raw data, as the authors are also doing with this work, is crucial to enable unrestricted exploratory analysis, model building, integration with data from other studies and further hypothesis generation, iFigures extend the reach of many of these exercises from computational experts to biologists with limited bioinformatics expertise.
The comparative exercise undertaken in the paper revealed that within hours both vaccines can induce meaningful transcriptomic responses in the blood, with interferon and inflammatory/TLR inducible genes predominant for the influenza and pneumococcal vaccines respectively. For both vaccines plasmablast responses were apparent by day 7, but the pneumococcal vaccine had a more sustained response that lasted until day 10 as seen in both microarray and flow cytometry data. These interesting observations raise further questions: e.g., how do differences in early responses affect the duration and level of the plasmablast response? Answers to this question can point to strategies for designing more potent vaccines. The current data also leave open the question of the mechanisms that drive the marked differences between the vaccines. Expression profiling of sorted cell subsets from the influenza vaccinees indicated that the interferon signature originated from neutrophils and monocytes, but which cell subset produced the interferon is less clear. A related question is where neutrophil stimulation took place—at the site of injection or further downstream? To begin to answer these questions, future studies could consider simultaneous profiling of blood and tissue biopsies from the injection site; utilizing mouse models where tissue samples can be obtained more readily could also prove valuable. In general, correlating responses in tissues to those in blood would be highly informative and can start to reveal molecular and cellular mechanisms that give rise to blood response signatures.
Data from this and other large-scale studies of human immunity together with ongoing efforts (e.g., those from the Human Immunology Project Consortium and the National Institutes of Health’s Center for Human Immunology (Leslie, 2010)) will begin to provide increasingly comprehensive characterizations of the human immune system, e.g., before and after vaccine and drug intervention in healthy and disease individuals. Proper integration of these dense data sets will start to provide a human “immune state” atlas—akin to the Connectivity Map for functional genomics and drug discovery (Lamb et al., 2006)—useful for probing and comparison of immune statuses under diverse settings and for reverse-engineering functional interactions among immune-system components based on correlations among the observed phenotypes across individuals (Marbach et al., 2012). In addition to proper design (e.g., sufficiently large cohort sizes) and execution at the individual study level, achieving this goal requires overcoming several obstacles, such as the standardization of experimental practices and technology platforms, identification of confounding covariates and developing approaches to normalize across studies, as well as agreeing to common data dissemination and replication standards. Some of these, such as data processing standards and sharing policies, can be partially borrowed from similar efforts in related fields (e.g., The Cancer Genome Atlas (Chin et al., 2011)); others need further research innovation as well as collaboration and dialogue across the research community. The biggest challenge and opportunity, however, lies in the development of novel concepts and approaches to turn such data compendia into predictive constructs for unseen perturbations and immunological insights. For example, predictive models have been built for disease prognosis (van ’t Veer et al., 2002) and are being explored at levels ranging from molecular interactions to whole cells (Bonneau et al., 2007; Karr et al., 2012). Extending these by incorporating key elements of immunity (e.g., cell-cell interactions; spatial and temporal considerations) and proper abstraction and modeling across different biological scales will help usher in an era of immunology where systematic collection of large-scale datasets such as the one presented in this study are combined with computational and mathematical modeling to systematically chip away at the complexity of the human immune system in health and disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nature medicine. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Jr, Assad-Garcia N, Glass JI, Covert MW. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Leslie M. Biomedical research. Immunology uncaged. Science. 2010;327:1573. doi: 10.1126/science.327.5973.1573. [DOI] [PubMed] [Google Scholar]
- Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Kellis M, Collins JJ, Stolovitzky G. Wisdom of crowds for robust gene network inference. Nature methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G, Presnell S, Domico K, Xu H, Wang Y, Anguiano E, Thompson-Snipes L, Ranganathan R, Zeitner B, Bjork A, et al. Human Immunology 2.0: Interactive exploration of responses to vaccines on a systems scale. Immunity. 2013 doi: 10.1016/j.immuni.2012.12.008. current issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]