Abstract
Genetically engineered mouse models are invaluable to investigators in nearly all areas of biomedical research. The use of genetically engineered mice has allowed researchers to explore fundamental functions of genes in a mammal that shares substantial similarities with human physiology and pathology. Genetically engineered mice are often used as animal models of human diseases that are vital tools in investigating disease development and in developing and testing novel therapies. Gene targeting in embryonic stem cells allows endogenous genes to be specifically altered. As knowledge regarding precise genetic abnormalities underlying a variety of dermatological conditions continues to emerge, the ability to introduce corresponding alterations in endogenous gene loci in mice, often at a single base pair level, has become essential for detailed studies of these genetic diseases. In this review, we provide examples of mouse models harboring modified endogenous gene(s), generated using the technique commonly referred to as the “knock-in” approach, to exemplify the important and sometimes superior role of this methodology in dermatological research.
Keywords: knock-in, knockout, gene targeting, ES cell, targeted mutation, homologous recombination, mouse model, skin
Introduction
Mouse (Mus musculus) is a widely used mammalian model organism for functional genomic and reverse genetic studies to address questions relating to human health and welfare in nearly every biomedical field. In many respects, murine skin shares striking similarities with human skin and mouse models have been extremely useful in examining skin-related gene functions and in modeling skin diseases.
Mouse models harboring genetic abnormalities can be classified as either having a spontaneous mutation or as being genetically engineered by man. Spontaneous mutations affecting the skin typically result in overt skin and coat phenotypes and are often readily identified. Unfortunately, many human skin diseases do not naturally occur in mice. Therefore, most skin phenotypes must be induced by manipulation of environmental conditions or via modification of the mouse genome by, for example, gene targeting.
Gene targeting
A variety of techniques are available for creating genetically engineered mouse models (GEMMs), including the use of transgenes, targeted endogenous gene alterations, random gene inactivation, and random mutagenesis (Table 1). Targeted endogenous gene alterations allow for very specific gene modification, i.e., a pre-selected gene locus can be disrupted or altered at its natural location to generate targeted mutant mice. Targeted disruption or deletion of a gene (“knockout”) via homologous recombination in ES cells is a very powerful method for studying events associated with the absence of a gene product. The knockout technology has had a tremendous impact on gene function studies, drug discovery, and many other areas of biomedical research. Not surprisingly, shortly after the completion of the human and mouse genome projects, public and private entities began strategically generating knockout mouse models across the entire genome [1, 2]. The ultimate acknowledgement of the importance of gene targeting occurred when the 2007 Nobel Prize in Medicine and Physiology was awarded to three investigators (Capecchi, Smithies, and Evans) “for their discoveries of principles for introducing specific gene modifications in mice by the use of embryonic stem cells.”
Table 1.
Genetically Engineered Mouse Models
| Transgenic | Targeted mutation | Random gene inactivation | Random mutagenesis | ||
|---|---|---|---|---|---|
| Knockout | Knock-in | ||||
| Application | Expression of normal, modified, or exogenous gene(s) | Deletion or disruption of gene expression | Genomic modification by targeted point mutation, addition, or deletion | Random disruption of gene expression | Generation of random point mutations across the genome |
| Procedure | Microinjection of recombinant DNA into pronucleus of oocyte and implantation into oviducts of foster female | Homologous recombination in ES cells, injection of targeted ES cells into blastocytes, and implantation into oviducts of foster females | Gene trapping in ES cells | Chemicals or radiation Male mice are exposed to | |
| Characterization | Transgene(s) is inserted randomly in the mouse genome. Protein(s) encoded by the transgene(s) is expressed. | Targeted gene(s) is inactivated. Phenotype is due to the absence of targeted gene product. | Targeted gene(s) is modified. Phenotype is due to the expression of modified gene product. | Gene-trap vector randomly inserts into a gene locus and interrupts transcription at the site of integration. Phenotype is generally due to loss-of-function. | mutagen and their progeny are screened for phenotypes (forward genetics). Multiple randomly mutated genes need to be mapped. |
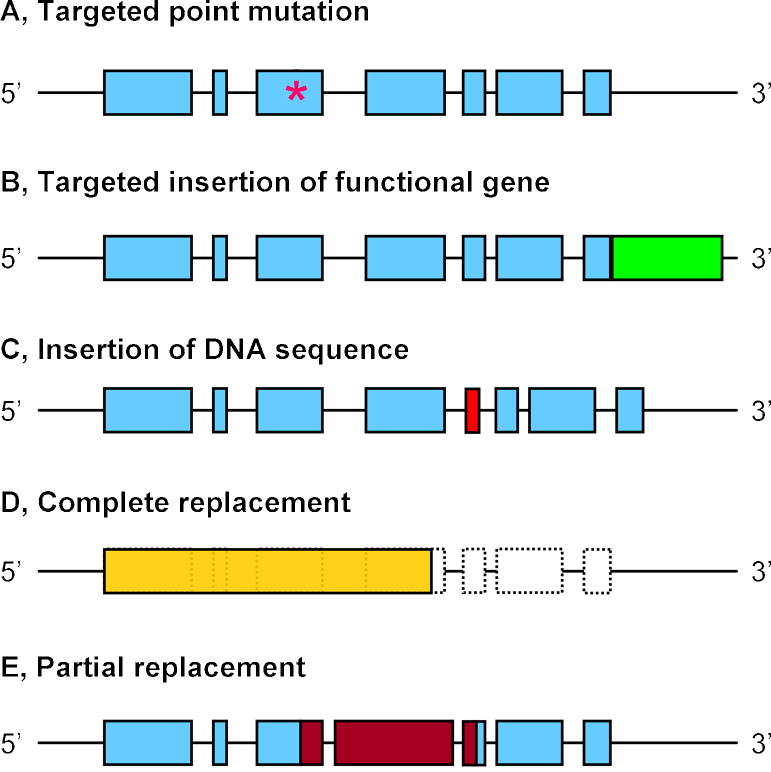
Another form of targeted gene alteration is commonly referred to as the “knock-in” approach. In the knock-in approach, target gene expression is normally not completely disrupted by the introduced modification. Instead, the modification creates a subtle point mutation or introduces additional genetic material. The knock-in approach has also been used to partially or completely replace an endogenous gene locus with a foreign gene, to generate human-mouse chimeric genes, or to replace an entire mouse gene with its human analog (Fig. 1). This technique has evolved to such a robust degree that researchers are now able to use the knock-in approach to correct genetic defects in mouse ES cells and use these corrected cells to generate phenotypically normal mice [3].
Fig. 1.
Various applications of genetically engineered knock-in mouse models. (A) Placing a subtle point mutation (asterisk) within a coding sequence is the most common use of the knock-in technology. (B) Insertion of a functional gene or sequence (e.g., lacZ, nuclear localization signal, etc.) expressed in tandem or as a fusion protein (green box) with the endogenous gene. (C) Targeted insertion of a DNA sequence (red box) that is not expressed but functions at the genomic or RNA level (e.g., loxP sites, siRNA). (D) Complete replacement of an endogenous gene locus (shaded box) with another related or unrelated functional gene (yellow box), such as a human homolog, cre recombinase, etc. (E) Partial replacement of a DNA sequence that has critical functions with a corresponding region of a related gene (red boxes).
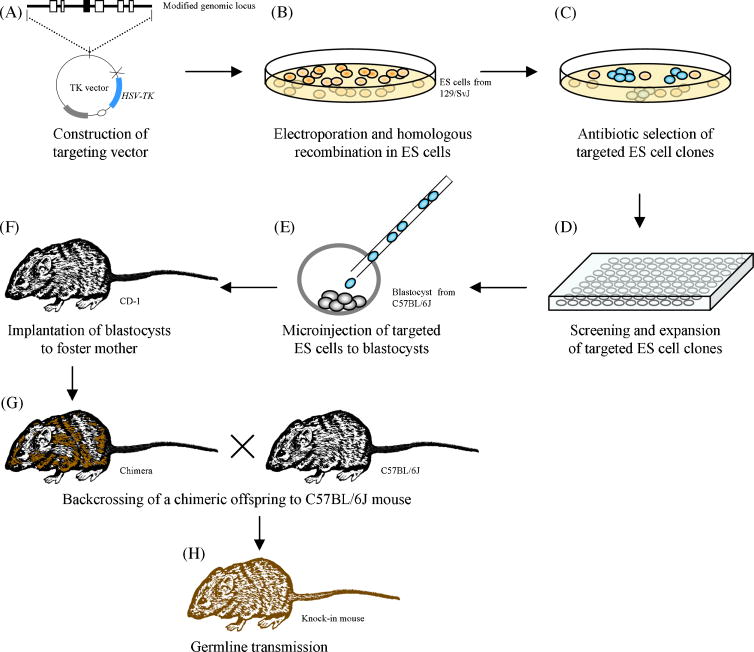
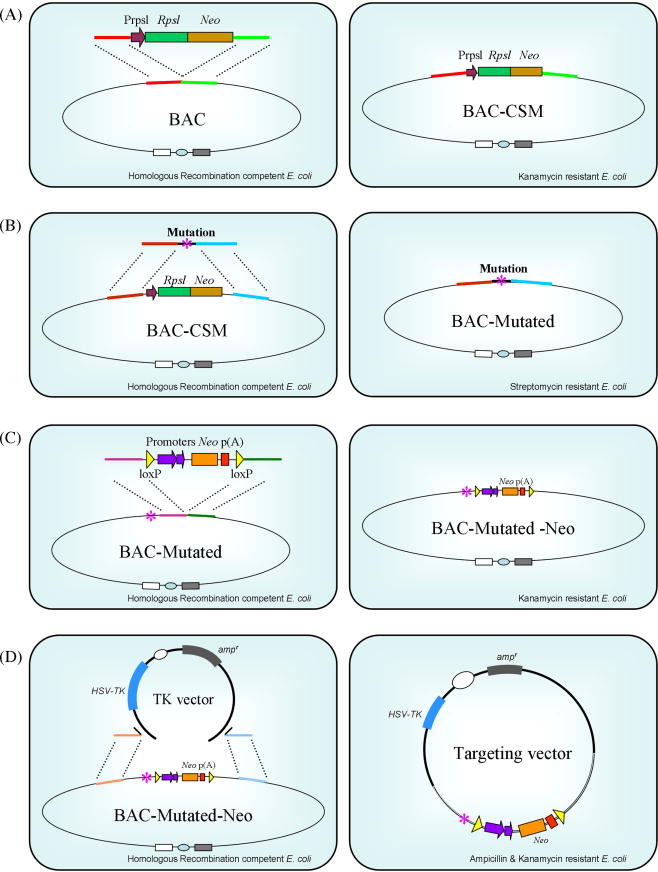
Key events associated with the generation of targeted mutant mice, including knock-in mice, via homologous recombination in ES cells are outlined in Figure 2. Efficient ES cell targeting is highly dependent on the targeting vector. Compared to the generation of knockout targeting vectors, the construction of a knock-in targeting vector is substantially restricted by the location of the intended modification within the endogenous gene. In many cases, it is practically impossible to introduce a subtle mutation or insertion using conventional restriction and ligation methods. Advancement of technologies such as cre-recombinase mediated site-specific recombination, the use of bacterial artificial chromosome (BAC) vectors and bacterial homologous recombination has allowed for greater flexibility and success in creating vectors for targeted gene modification [4]. Figure 3 illustrates the use of BAC vectors and bacterial homologous recombination in creating ES cell targeting vectors. A recently developed high-throughput VelociGene system which takes the advantage of high targeting efficiency of large BAC vectors that can carry virtually any genetic modification up to 70 kb and “loss-of-native-allele” assays, further streamlined ES cell gene targeting by circumventing the need of positive-negative selection and the use of isogenic DNA sequences [5].
Fig. 2.
Procedure of gene targeting through homologous recombination in mouse embryonic stem cells. (A) Construction of a targeting vector. The targeting vector contains the modified gene locus (filled box), and typically harbors a neo-cassette for positive selection (not shown) and the HSV-TK gene for negative selection of correctly targeted ES cells. (B) The targeting vector is linearized and introduced into ES cells (typically from 129/SvJ background) by electroporation. (C) ES cells that incorporated the targeting vector form colonies (blue) after selection with appropriate antibiotics. (D) Selected colonies are characterized by southern blotting to identify cells in which the desired gene targeting event occurred. (E) Correctly targeted ES cells are expanded and injected into C57BL/6J host embryos. (F) Blastocysts harboring a mixture of C57BL/6J embryonic cells and the targeted 129/SvJ ES cells are transferred to the oviducts of pseudo-pregnant CD-1 foster mothers. (G) Chimeric newborns are back-crossed with C57BL/6J mice. (H) Breeding of chimeric progeny carrying the targeted mutation to achieve germline transmission.
Fig. 3.
Bacterial homologous recombination in the construction of targeting vectors. (A) Left, a homologous recombination-competent bacterial strain harboring a BAC vector is shown. An RpsL-Neo cassette, as selection marker and counter selection marker (CSM) genes, is introduced via electroporation. The RpsL-Neo cassette is flanked with short DNA sequences (red and green) that are homologous to the target gene locus in the BAC vector. Right, selection for kanamycin resistant bacterial clones (conferred by Neo) identifies BAC vectors that have the RpsL-Neo cassette inserted into the designated gene locus through homologous recombination (BAC-CSM). (B) Left, a DNA fragment containing a subtle mutation (asterisk) and DNA sequences (brown and blue) that are homologous to the target gene locus in the BAC-CSM vector replaces the RpsL-neo cassette. Right, streptomycin selection for mutant BAC clones (resistance is conferred by the absence of RpsL) (BAC-Mutated). (C) Insertion of a neo-cassette as in (A) and (B). The neo-cassette is driven by dual prokaryotic and eukaryotic promoters and is flanked by loxP sites. This configuration enables the neo gene to be used to select for antibiotic resistance in E. coli (kanamycin) as well as in ES cells (geneticin). (D) Left, the modified gene locus in the BAC vector (BAC-Mutated-Neo) is subcloned into a gene targeting vector (TK Vector) through homologous recombination. The backbone of the targeting vector contains HSV-TK and ampicillin resistant (ampr) genes. Right, ampicillin and kanamycin selection identifies targeting vectors that contain both the plasmid backbone and the modified gene locus.
GEMMs in skin research and the significance of the targeted gene modification approach
Transgenic mouse models have been remarkable tools for gene function studies and modeling of a variety of skin conditions [6]. The knockout approach is also pervasive in skin research as illustrated by the fact that almost every gene known to be important in skin development, homeostasis, and disease has been examined using knockout mouse models. Despite the obvious strengths of transgenic and knockout mouse models, the forced over-expression of a transgene or complete ablation of an endogenous gene imposes certain limitations on gene function studies and human disease modeling. One overt limitation is the possibility of interference with the normal expression patterns of genes surrounding the often randomly inserted transgene or near the inactivated gene locus. Even though the evolution of inducible and conditional approaches have provided greater flexibility in utilizing these technologies in skin research, transgenic and knockout mice often cannot adequately recapitulate the genetic alterations observed in certain skin conditions.
To circumvent such limitations, the knock-in approach can be utilized to specifically modify an endogenous gene locus while maintaining its expression profile. A few examples of knock-in mouse models used in skin research are listed in Table 2. The combination of the knock-in strategy with inducible approaches also provides unique opportunities to mimic certain skin pathologies resulted from persistent or spontaneous mutant gene expression. The following examples demonstrate the critical and, sometimes, superior roles of knock-in mouse models as compared with transgenic and knockout mouse models in skin gene function studies, skin disease modeling, and skin cancer studies.
Table 2.
Examples of knock-in mouse models used in skin research
| Gene | Modification | Application | Reference | |
|---|---|---|---|---|
| Gene Function | Runx1 | Targeted replacement of Runx1 (exon 7–8) with lacZ | Expression pattern of Runx1 during skin embryogenesis | [7, 8] |
| Gata-3 | Targeted replacement of Gata- 3 (exon 1–2) with lacZ | Regulation of hair follicle morphogenesis | [9, 10] | |
| Ccd6 | Targeted insertion of EGFP to Ccr6 locus | Expression of Ccr6 in langerhans cells | [11] | |
| Apc | Targeted mutation of Apc (Apc 1638T) | Nipple-associated cutaneous cysts | [12] | |
| Col5α2 | Targeted substitution of Col5α2 (exon 6) with neo - cassette | Regulatory role of Col5α2 during matrix assembly | [13] | |
| Egfr | Targeted replacement of mouse Egfr with human EGFR cDNA | Hair follicle and hair cycle defects | [14] | |
| Dsc1 | Targeted deletion of exon 17 of Dsc1 (Dsc1 ΔE17) | Cytoplasmic domain of Dsc1 in epidermal development | [15] | |
| Krt75 | Targeted point mutation of Krt75 (Krt75 N159del) | Dominant effects of mutant Krt75 in hair and nail formation | [16] | |
| Disease Models | Krt10 | Targeted mutation of Krt10 (Krt10 R154C) | Inducible mouse model of EHK | [17] |
| Krt14 | Targeted point mutation of Krt14 (Krt14 R131C) | Inducible mouse model of EBS | [18] | |
| Skin Cancer | k-Ras | Targeted point mutation of k- ras (k-ras G12D) | Study of spontaneous oncogenic k -ras activation in cancer | [19, 20] |
| Krt10 - Krt14 | Targeted replacement of head and tail domains Krt14 with corresponding domains of Krt10 | Role of intermediate filaments in skin cancer susceptibility | [21] | |
| Kit | Targeted point mutation of Kit (Kit V558Del) | Role of gain-of-function mutation of Kit in skin cancer | [22] | |
| Cdk4 | Targeted point mutation of Cdk4 (Cdk4 R24C) | Cdk4 activation in skin carcinogenesis | [23] | |
| p53 | Targeted humanization of core domains (exon 4–9) of p53 | Recapitulation of frequent DNA damage to human p53 mutation | [24, 25] | |
| p53 | Targeted point mutation of p53 (p53 R171A, p53 R172H, p53 R270H) | Recapitulation of Li-Fraumeni Syndrome by expressing gain- of-function mutant p53 at its endogenous gene locus | [26, 27] |
1. GEMMs for studying keratin gene function and keratin disorders
All epithelial cells contain keratin intermediate filaments (IFs) which are polymers of keratin protein heterodimers. Keratin IFs provide epithelial cells with resilient strength to withstand mechanical damage. Many keratin genes exist and expression of these genes exhibits a certain degree of redundancy. Expression of a particular keratin may be restricted to a specific tissue type and/or to specific differentiation and homeostatic stages. In humans, defects in keratin genes result in a wide spectrum of skin diseases with tremendous variation in clinical presentation [28].
Many GEMMs have been utilized for studying keratin genes. For example, several mouse models have been generated to study the genes encoding the keratin 6 (Krt6) [29–35]. In one study, the transgenic approach was used to express mutant Krt6a and the resulting mice displayed dramatic skin and hair phenotypes [29]. This study demonstrated the dominant-negative effects of Krt6a mutations and provided evidence for the involvement of Krt6a in skin blistering and hair loss. However, the constitutive expression of mutant Krt6a resulted in devastating damage to the skin which prevented further investigation of these Krt6a mutations.
Another GEMM, in which the Krt6a gene was ablated, exhibited re-epithelialization defects in wound healing [30]. Interestingly, the lack of Krt6a did not prevent the normal development or general function of epithelial tissues and ectodermal appendages. This result suggested that a closely related Krt6b gene might compensate in the absence of Krt6a. When both Krt6a and Krt6b were deleted from the Krt6 locus, double null mice developed hyperplastic lesions in the oral cavity which were associated with mechanical stress from food intake [31, 33]. Surprisingly, aside from fatal starvation resulting from upper digestive tract constriction, the absence of Krt6a and Krt6b did not produce a defect in the skin or its appendages. While these knockout models were useful in furthering our understanding of keratin genes (e.g., data from these GEMMs suggested redundancy of keratin genes in epidermal keratinocytes and indicated that some of these genes may be dispensable), no correlation between Krt6 gene function and any human skin disease was identified.
Pachyonychia congenita (PC) is the only known disease associated with KRT6A and KRT6B. PC-1 and PC-2 are caused by dominant mis-sense mutations in KRT6A or KRT16 (PC-1) and KRT6B or KRT17 (PC-2), respectively. It is clinically characterized by distally progressive hypertrophic onychodystrophy and focal hyperkeratosis of the palms and soles [36]. A number of transgenic and knockout mouse models have been made with PC-causing genes but none fully reproduced the pachyonychia congenita phenotypes [37]. We recently generated a knock-in mouse model carrying a dominant mutation in mouse Krt75 (previously known as K6hf), which corresponds to a frequently observed point mutation in KRT6A in PC patients [16]. Mice carrying this dominant mutation developed congenital hair phenotypes and partially mimicked the hypertrophic nail dystrophy of PC [16]. While this mouse model does not recapitulate every feature of pachyonychia congenita, it clearly demonstrated the role of Krt75 in hair and nail formation, which might not have been revealed using transgenic or knockout approaches.
2. Mouse models of epidermolytic hyperkeratosis
Epidermolytic hyperkeratosis (EHK), also termed bullous congenital ichthyosiform erythroderma (BCIE), is an autosomal dominant form of ichthyosis caused by mutations in the KRT1 and KRT10 genes which are expressed in suprabasal keratinocytes of the epidermis [38, 39]. Clinical features of EHK include blistering, erythroderma, and peeling of skin at birth, followed by the development of acanthosis and hyperkeratosis, predominantly over areas prone to pressure and mechanical stress [40]. Infants with EHK can develop large areas of skin erosion resulting in an increased risk of neonatal mortality. Histologically, EHK blisters exhibit keratinocyte cytolysis within the suprabasal layers of the epidermis with damaged cells exhibiting peri-nuclear clumps of keratin IFs.
A number of GEMMs were developed for EHK [17, 41–44]. The first mouse model expressed a mutant Krt10 transgene [42] and was reported shortly before the genetic basis of EHK was published [38, 45, 46]. This model demonstrated that Krt10 mutation could result in EHK phenotypes and that the presence of mutant Krt10 molecules caused disruption of the keratin IF network. The utility of this transgenic model for studying EHK was limited in that the transgene was a truncated Krt10, which was in contrast to the commonly observed Krt10 point mutations in EHK patients, and that the transgene was only expressed at 2–10% of the level of endogenous Krt10.
Krt10 knockout mice were also generated and surprisingly, Krt10 null mice displayed a well-developed and functional epidermis that did not exhibit tissue fragility or epithelial cell IF aggregates [41]. This model provided insight into keratin gene redundancy and the ability of other keratin filaments to compensate in the absence of Krt10, but did not recapitulate EHK phenotypes.
In another GEMM, an HPRT mini gene was inserted between exon 2 and exon 8 of Krt10 to interrupt transcription of the Krt10 allele [44]. Transcriptional activity of the targeted Krt10 allele was substantially reduced; however, a truncated K10 polypeptide (K10T) was produced. The K10T molecule exerted a dominant interference effect, resulting in the presence of keratin aggregates, suprabasal cytolysis of keratinocytes, and severe hyperkeratosis in heterozygous mice. Homozygosity resulted in fatal barrier defects [41]. To a certain extent, this mouse model resembled a knock-in mouse model because the targeted allele produced a mutant gene product. The full spectrum of EHK phenotypes could be exhibited in homo- and heterozygous mutant mice, such that fragile skin with small blisters and large areas of skin loss were observed in the homozygous and new born mice, whereas hyperkeratosis developed in heterozygous adults. The segregation of phenotypes in these mutant mice may be explained by the fact that the mutation in K10T mice is different from the mutations discovered in humans with EHK, and that the amount of mutant Krt10 protein in the epidermis was drastically less than that of endogenous normal K10 protein [41]
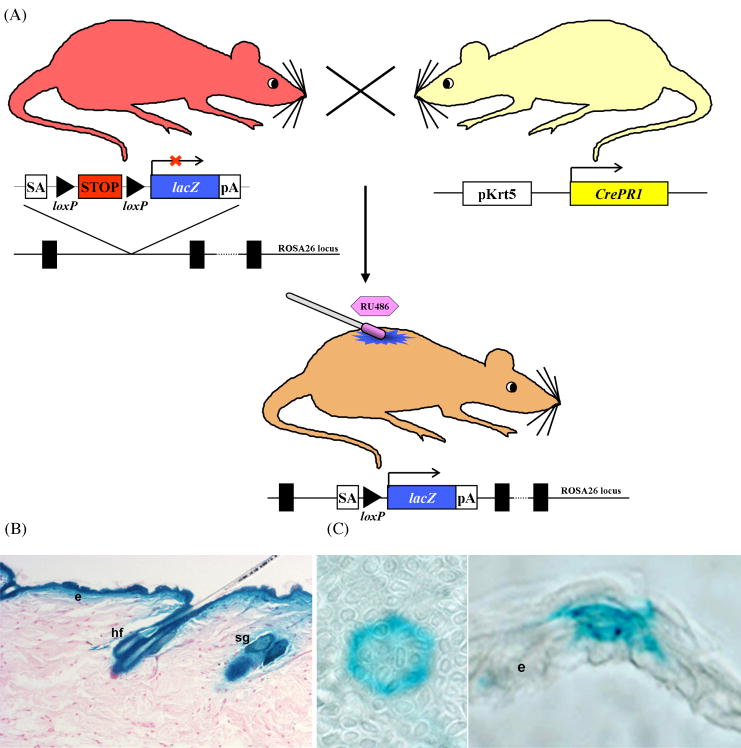
The most successful EHK mouse model developed thus far was created by modifying the endogenous Krt10 locus to introduce a frequently observed single-nucleotide mis-sense mutation [17]. This mouse model additionally utilized a cre-mediated inducible system (Fig. 4) to allow the activation of a conditional mutant Krt10 allele in a restricted area of the skin to circumvent neonatal fatalities commonly associated with congenital skin fragility syndromes. Topical application of an inducer, RU486, “activated” the mutant allele in the genome of epidermal keratinocytes, including epidermal stem cells resulting in continual production of progeny cells harboring the mutant allele, even after the inducer is no longer applied. These mice developed blisters that gradually resulted in hyperkeratotic lesions which persisted for life [17]. Moreover, this inducible knock-in model allowed for the mosaic form of EHK to be precisely reproduced, a result that had not been obtained using other GEMM techniques.
Fig. 4.
Cre recombinase-mediated tissue-specific inducible system. (A) The generation of an inducible lacZ mouse. A “target” mouse, where a splice-acceptor (SA) sequence, a STOP sequence (PGK-Neo-pA) flanked by loxP sites, and a lacZ gene were targeted into the ROSA26 locus, is shown at upper left. Transcription of the lacZ gene is blocked by the STOP sequence [61]. An “activator” mouse, expressing a fusion protein consisting of Cre-recombinase with a mutated ligand-binding domain of a progesterone receptor (CrePR1) under the control of skin-specific Krt5 promoter (pKrt5), is shown at upper right [62]. A bigenic inducible mouse obtained by crossing the above “target” (lacZ) and “activator” (Cre) mice is shown below. Topical application of an inducer (RU486) on the back skin of the bigenic mouse was able to activate Cre to excise the STOP signal placed between the two loxP sites, therefore, inducing expression of the lacZ gene in the skin. (B-C) X-gal staining of skin sections after induction of the conditional lacZ allele. Note the uniform expression of lacZ in the epidermis, hair follicles and sebaceous glands 6 months following activation of CrePR1 in utero in (B), confirming that K5CrePR1 is expressed in multipotent stem cells in B (Zhou et al., 2002). Topical application of RU486 to adult epidermis results in activation of K5Cre*PR1, and thus expression of lacZ in individual stem cells and their progeny is shown in (C) (Caulin et. al., 2007). Shown are both an en face (left) and cross (right) view of an epidermal proliferative unit expressing lacZ. Abbreviations: e, epidermis; hf, hair follicle; sg, sebaceous gland.
3. Oncogene activation and the development of nonmelanoma skin cancer
Tumor studies are best carried out in vivo since tumors are often highly heterogeneous and are dependent upon interactions with the stroma, the host immune system, and the tissue environment. During tumor development, cells typically undergo multiple genetic events, such as the activation of oncogenes or mutation of tumor suppressor genes. Mutations in ras genes are commonly found in human skin cancers [47, 48] and are frequently induced by the chemical tumor initiator, dimethylbenz[a]anthracene (DMBA), in mouse skin chemical carcinogenesis experiments [49–51]. The role of activated ras in skin tumor initiation has been well-established using a variety of GEMMs [52].
Mutations in the p53 gene, the most studied and highly characterized of the known tumor suppressor genes, are found in about 50% of all cancers [53]. Interestingly, p53-null mice rarely develop skin tumors prior to succumbing to internal tumor burden [54, 55]. However, more than 50% of human squamous cell carcinomas (SCCs) exhibit p53 mutations. For example, a mis-sense mutation resulting in the substitution of a histidine for arginine at amino acid 175 is frequently observed in human skin cancer [56, 57]. A transgenic mouse model in which an equivalent mutation in the murine p53 gene (p53R172H) was overexpressed in the epidermis provided early evidence of a gain-of-function property of p53 mutation in skin cancer [58]. In contrast to wildtype and p53-null mice, the p53R172H transgenic mice exhibited increased susceptibility to skin chemical carcinogenesis, tumor progression, and metastasis. However, the use of this model was limited in that the transgene was not expressed at levels comparable to endogenous p53, thus leaving room to question whether this mutant p53 molecule truly represented the effect of the same mutation in the endogenous p53 gene.
To effectively mimic the consequence of endogenous gene mutations during tumorigenesis, a knock-in approach is needed. Knock-in mouse models expressing mutant endogenous p53 genes that are analogous to those found in clinical tumor studies recapitulated distinctive features of Li-Fraumeni Syndrome [26, 27]. These models also provided evidence for gain-of-function properties of mutant p53 in cancer development. However, this study did not examine the potential gain-of-function property of mutant p53 in skin tumorigenesis since, much like the p53 knock-out mice, these mice succumbed to internal tumors without having developed skin tumors. To overcome this limitation, an inducible, tissue-specific knock-in mouse model was created in which mutant endogenous p53 (p53R172H) allele and endogenous K-ras (K-rasG12D) allele could be focally and conditionally activated in epidermal stem cells as illustrated in Figure 4 [20]. In the same study, conditional knock-in mutant K-ras mice were also mated with mice harboring a floxed p53 allele. This study was the first to directly compare, in a genetically equivalent manner, the role of p53 loss-of-function and gain-of-function in skin tumorigenesis with a common initiation event (activation of K-rasG12D). This study confirmed the gain-of-function capabilities of p53R172H. Cooperation between this mutant p53, but not loss of p53, and activated ras in skin tumor initiation was observed and a role for this p53 mutant in skin tumor progression and metastasis was confirmed [20]. This K-rasG12D/p53R172H model is the first skin cancer model capable of recapitulating multiple endogenous genetic events in a focal, inducible, and cell type-specific manner, and emphasizes the importance of the conditional, tissue-specific knock-in technique in skin cancer research.
Perspectives on future developments of mouse models for dermatological research using endogenous gene targeting
The utility of endogenous gene modifications in skin research reaches beyond merely studying skin gene functions and modeling of genodermatoses and cancer which have a clear genetic etiology. As an example, bullous pemphigoid (BP) is the most common antibody-mediated blistering autoimmune disease. It is caused by the cross-reaction of autoantibodies with the BP180 epitopes in the NC16A domain of COL17. However, the endogenous mouse Col17 does not cross-react with the disease causing IgG of humans. Therefore, it is not possible to establish in vivo models of BP by passive transfer of patient IgG into mice. In order to build a mouse model of this disease, Nishie and colleagues used strategic breeding of Col17 knockout mice with transgenic mice expressing human COL17 [59]. Transgenic mice expressing human COL17 rescued the lethal phenotype in Col17-deficient mice. Consequently, this strategy achieved complete humanization of the mouse BP antigen in the offspring of these mice, which permitted the cross-reaction of pathogenic human IgG with the human epitope and recapitulated the corresponding BP phenotypes [59]. This mouse model also became a successful in vivo model for testing potential recombinant peptide therapies for BP [59]. Utilizing a knock-in approach, a novel GEMM was recently established in which the endogenous Col17 locus was partially humanized by replacing the mouse BP180NC14A genomic locus with the corresponding immunogenic human BP180NC16A locus [60]. This strategy allowed the endogenously expressed mouse-human hybrid COL17 to cross-react with the pathogenic human autoantibody after passive transfer. Furthermore, this mouse model can be used directly in the mapping of the pathogenic epitope of BP180 in vivo [60].
The ability to modify the endogenous mouse genome within non-coding regions raises the possibility of building more complex genetic disease models, such as models for skin carcinoma that result from chromosomal translocations and inversions. In conjunction with other mutant mouse models and various inbred strains and the incorporation of a broad range of new technologies, such as reporter genes and in vivo imaging systems, protein tags, tissue-, organ-, or temporal-specific approaches, and RNA interference, the knock-in approach enables, but is not limited to, the engineering of mouse models that are capable of: studying modifier genes in epistasis and eventually recapitulating sophisticated polygenic or non-Mendelian skin conditions; accurately track endogenous gene expression patterns in the skin; overcome infertility or premature death caused by certain essential genetic modifications; properly recapitulate spontaneous genetic events in tumorigenesis; and to mimic the exact genetic and phenotypic changes of certain skin disease conditions, and be used unbiasedly in drug target validation, preclinical testing of novel therapeutic approaches, and biomarker discovery.
Acknowledgments
The authors sincerely thank Dr. Carlos Caulin at the M.D. Anderson Cancer Center and Drs. Neil Box and Peter Koch of the Department of Dermatology at the University of Colorado Denver for comments and critical reading of this manuscript. This work is supported in part by grants from the National Institute of Health (NIH) to DRR; research grants from the Foundation for Ichthyosis and Related Skin Types (FIRST) and International Pachyonychia Congenita Consortium (IPCC) to DRR and JC; and grants from the North American Hair Research Society (NAHRS) and the Council for Nail Disorders (CND) to JC.
Abbreviations
- BAC
bacterial artificial chromosome
- EHK
epidermolytic hyperkeratosis
- ES cell
embryonic stem cell
- GEM
genetically engineered mouse
- GEMM
genetically engineered mouse model
- IF
intermediate filament
- HSV-TK
herpes simplex virus thymidine kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Auwerx J, et al. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–7. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin CP, et al. The knockout mouse project. Nat Genet. 2004;36:921–4. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu LC, Sun CW, Ryan TM, Pawlik KM, Ren J, Townes TM. Correction of sickle cell disease by homologous recombination in embryonic stem cells. Blood. 2006;108:1183–8. doi: 10.1182/blood-2006-02-004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–88. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 5.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–9. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 6.Rothnagel JA, Greenhalgh DA, Wang XJ, Sellheyer K, Bickenbach JR, Dominey AM, Roop DR. Transgenic models of skin diseases. Arch Dermatol. 1993;129:1430–6. [PubMed] [Google Scholar]
- 7.North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–75. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 8.Lian JB, et al. Runx1/AML1 hematopoietic transcription factor contributes to skeletal development in vivo. J Cell Physiol. 2003;196:301–11. doi: 10.1002/jcp.10316. [DOI] [PubMed] [Google Scholar]
- 9.Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–8. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, Lim KC, Dai X, Alegre ML, Fuchs E. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–22. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucharzik T, Hudson JT, 3rd, Waikel RL, Martin WD, Williams IR. CCR6 expression distinguishes mouse myeloid and lymphoid dendritic cell subsets: demonstration using a CCR6 EGFP knock-in mouse. Eur J Immunol. 2002;32:104–12. doi: 10.1002/1521-4141(200201)32:1<104::AID-IMMU104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 12.Smits R, et al. Apc1638T: a mouse model delineating critical domains of the adenomatous polyposis coli protein involved in tumorigenesis and development. Genes Dev. 1999;13:1309–21. doi: 10.1101/gad.13.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrikopoulos K, Liu X, Keene DR, Jaenisch R, Ramirez F. Targeted mutation in the col5a2 gene reveals a regulatory role for type V collagen during matrix assembly. Nat Genet. 1995;9:31–6. doi: 10.1038/ng0195-31. [DOI] [PubMed] [Google Scholar]
- 14.Sibilia M, Wagner B, Hoebertz A, Elliott C, Marino S, Jochum W, Wagner EF. Mice humanised for the EGF receptor display hypomorphic phenotypes in skin, bone and heart. Development. 2003;130:4515–25. doi: 10.1242/dev.00664. [DOI] [PubMed] [Google Scholar]
- 15.Cheng X, Mihindukulasuriya K, Den Z, Kowalczyk AP, Calkins CC, Ishiko A, Shimizu A, Koch PJ. Assessment of splice variant-specific functions of desmocollin 1 in the skin. Mol Cell Biol. 2004;24:154–63. doi: 10.1128/MCB.24.1.154-163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Jaeger K, Den Z, Koch PJ, Sundberg JP, Roop DR. Mice expressing a mutant Krt75 (K6hf) allele develop hair and nail defects resembling pachyonychia congenita. J Invest Dermatol. 2008;128:270–9. doi: 10.1038/sj.jid.5701038. [DOI] [PubMed] [Google Scholar]
- 17.Arin MJ, Longley MA, Wang XJ, Roop DR. Focal activation of a mutant allele defines the role of stem cells in mosaic skin disorders. J Cell Biol. 2001;152:645–9. doi: 10.1083/jcb.152.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao T, Longley MA, Wang XJ, Roop DR. An inducible mouse model for epidermolysis bullosa simplex: implications for gene therapy. J Cell Biol. 2001;152:651–6. doi: 10.1083/jcb.152.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 20.Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, Lozano G, Roop DR. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest. 2007;117:1893–1901. doi: 10.1172/JCI31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Cheng X, Merched-Sauvage M, Caulin C, Roop DR, Koch PJ. An unexpected role for keratin 10 end domains in susceptibility to skin cancer. J Cell Sci. 2006;119:5067–76. doi: 10.1242/jcs.03298. [DOI] [PubMed] [Google Scholar]
- 22.Sommer G, et al. Gastrointestinal stromal tumors in a mouse model by targeted mutation of the Kit receptor tyrosine kinase. Proc Natl Acad Sci U S A. 2003;100:6706–11. doi: 10.1073/pnas.1037763100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rane SG, Cosenza SC, Mettus RV, Reddy EP. Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol Cell Biol. 2002;22:644–56. doi: 10.1128/MCB.22.2.644-656.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo JL, et al. UV-induced DNA damage and mutations in Hupki (human p53 knock-in) mice recapitulate p53 hotspot alterations in sun-exposed human skin. Cancer Res. 2001;61:8158–63. [PubMed] [Google Scholar]
- 25.Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001;20:320–8. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- 26.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Lane EB, McLean WH. Keratins and skin disorders. J Pathol. 2004;204:355–66. doi: 10.1002/path.1643. [DOI] [PubMed] [Google Scholar]
- 29.Wojcik SM, Imakado S, Seki T, Longley MA, Petherbridge L, Bundman DS, Bickenbach JR, Rothnagel JA, Roop DR. Expression of MK6a dominant-negative and C-terminal mutant transgenes in mice has distinct phenotypic consequences in the epidermis and hair follicle. Differentiation. 1999;65:97–112. doi: 10.1046/j.1432-0436.1999.6520097.x. [DOI] [PubMed] [Google Scholar]
- 30.Wojcik SM, Bundman DS, Roop DR. Delayed wound healing in keratin 6a knockout mice. Mol Cell Biol. 2000;20:5248–55. doi: 10.1128/mcb.20.14.5248-5255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wojcik SM, Longley MA, Roop DR. Discovery of a novel murine keratin 6 (K6) isoform explains the absence of hair and nail defects in mice deficient for K6a and K6b. J Cell Biol. 2001;154:619–30. doi: 10.1083/jcb.200102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong P, Domergue R, Coulombe PA. Overcoming functional redundancy to elicit pachyonychia congenita-like nail lesions in transgenic mice. Mol Cell Biol. 2005;25:197–205. doi: 10.1128/MCB.25.1.197-205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong P, Colucci-Guyon E, Takahashi K, Gu C, Babinet C, Coulombe PA. Introducing a null mutation in the mouse K6alpha and K6beta genes reveals their essential structural role in the oral mucosa. J Cell Biol. 2000;150:921–8. doi: 10.1083/jcb.150.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi K, Coulombe PA. Defining a region of the human keratin 6a gene that confers inducible expression in stratified epithelia of transgenic mice. J Biol Chem. 1997;272:11979–85. doi: 10.1074/jbc.272.18.11979. [DOI] [PubMed] [Google Scholar]
- 35.Rothnagel JA, Seki T, Ogo M, Longley MA, Wojcik SM, Bundman DS, Bickenbach JR, Roop DR. The mouse keratin 6 isoforms are differentially expressed in the hair follicle, footpad, tongue and activated epidermis. Differentiation. 1999;65:119–30. doi: 10.1046/j.1432-0436.1999.6520119.x. [DOI] [PubMed] [Google Scholar]
- 36.Leachman SA, et al. Clinical and Pathological Features of Pachyonychia Congenita. Journal of Investigative Dermatology Symposium Proceedings. 2005;10:3–17. doi: 10.1111/j.1087-0024.2005.10202.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Roop DR. Mouse Models in Preclinical Studies for Pachyonychia Congenita. Journal of Investigative Dermatology Symposium Proceedings. 2005;10:37–46. doi: 10.1111/j.1087-0024.2005.10206.x. [DOI] [PubMed] [Google Scholar]
- 38.Rothnagel JA, et al. Mutations in the rod domains of keratins 1 and 10 in epidermolytic hyperkeratosis. Science. 1992;257:1128–30. doi: 10.1126/science.257.5073.1128. [DOI] [PubMed] [Google Scholar]
- 39.Compton JG, et al. Linkage of epidermolytic hyperkeratosis to the type II keratin gene cluster on chromosome 12q. Nat Genet. 1992;1:301–5. doi: 10.1038/ng0792-301. [DOI] [PubMed] [Google Scholar]
- 40.DiGiovanna JJ. Ichthyosiform Dermatoses. In: Freedberg IM, Eisen AZ, Wolff K, Austen KF, Goldsmith LA, Katz SI, Fitzpatrick TB, editors. Dermatology in General Medicine. 5. Vol. 1. New York: McGraw Hill; 1999. [Google Scholar]
- 41.Reichelt J, Bussow H, Grund C, Magin TM. Formation of a normal epidermis supported by increased stability of keratins 5 and 14 in keratin 10 null mice. Mol Biol Cell. 2001;12:1557–68. doi: 10.1091/mbc.12.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuchs E, Esteves RA, Coulombe PA. Transgenic mice expressing a mutant keratin 10 gene reveal the likely genetic basis for epidermolytic hyperkeratosis. Proc Natl Acad Sci U S A. 1992;89:6906–10. doi: 10.1073/pnas.89.15.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bickenbach JR, Longley MA, Bundman DS, Dominey AM, Bowden PE, Rothnagel JA, Roop DR. A transgenic mouse model that recapitulates the clinical features of both neonatal and adult forms of the skin disease epidermolytic hyperkeratosis. Differentiation. 1996;61:129–39. doi: 10.1046/j.1432-0436.1996.6120129.x. [DOI] [PubMed] [Google Scholar]
- 44.Porter RM, Leitgeb S, Melton DW, Swensson O, Eady RA, Magin TM. Gene targeting at the mouse cytokeratin 10 locus: severe skin fragility and changes of cytokeratin expression in the epidermis. J Cell Biol. 1996;132:925–36. doi: 10.1083/jcb.132.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonifas JM, Bare JW, Chen MA, Lee MK, Slater CA, Goldsmith LA, Epstein EH., Jr Linkage of the epidermolytic hyperkeratosis phenotype and the region of the type II keratin gene cluster on chromosome 12. J Invest Dermatol. 1992;99:524–7. doi: 10.1111/1523-1747.ep12658061. [DOI] [PubMed] [Google Scholar]
- 46.Cheng J, Syder AJ, Yu QC, Letai A, Paller AS, Fuchs E. The genetic basis of epidermolytic hyperkeratosis: a disorder of differentiation-specific epidermal keratin genes. Cell. 1992;70:811–9. doi: 10.1016/0092-8674(92)90314-3. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 48.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 49.Balmain A, Ramsden M, Bowden GT, Smith J. Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature. 1984;307:658–60. doi: 10.1038/307658a0. [DOI] [PubMed] [Google Scholar]
- 50.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 51.Pazzaglia S, Mancuso M, Primerano B, Rebessi S, Biozzi G, Covelli V, Saran A. Analysis of c-Ha-ras gene mutations in skin tumors induced in carcinogenesis-susceptible and carcinogenesis-resistant mice by different two-stage protocols or tumor promoter alone. Mol Carcinog. 2001;30:111–8. doi: 10.1002/1098-2744(200102)30:2<111::aid-mc1019>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 52.Kemp CJ. Multistep skin cancer in mice as a model to study the evolution of cancer cells. Semin Cancer Biol. 2005;15:460–73. doi: 10.1016/j.semcancer.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Bartek J, et al. Aberrant expression of the p53 oncoprotein is a common feature of a wide spectrum of human malignancies. Oncogene. 1991;6:1699–703. [PubMed] [Google Scholar]
- 54.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 55.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 56.Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, Levine AJ. Gain of function mutations in p53. Nat Genet. 1993;4:42–6. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 57.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–78. [PubMed] [Google Scholar]
- 58.Wang XJ, Greenhalgh DA, Jiang A, He D, Zhong L, Medina D, Brinkley BR, Roop DR. Expression of a p53 mutant in the epidermis of transgenic mice accelerates chemical carcinogenesis. Oncogene. 1998;17:35–45. doi: 10.1038/sj.onc.1201890. [DOI] [PubMed] [Google Scholar]
- 59.Nishie W, et al. Humanization of autoantigen. Nat Med. 2007;13:378–83. doi: 10.1038/nm1496. [DOI] [PubMed] [Google Scholar]
- 60.Liu Z, et al. Identification of BP180 pathogenic epitope using humanized BP180NC16A mice. J Investig Dermatol. 2007;127:S12. [Google Scholar]
- 61.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 62.Zhou Z, Wang D, Wang XJ, Roop DR. In utero activation of K5. CrePR1 induces gene deletion. Genesis. 2002;32:191–2. doi: 10.1002/gene.10064. [DOI] [PubMed] [Google Scholar]