Abstract
Satellite repeats in heterochromatin are transcribed into noncoding RNAs that have been linked to gene silencing and maintenance of chromosomal integrity. Using digital gene expression analysis, we showed that these transcripts are greatly overexpressed in mouse and human epithelial cancers. In 8 of 10 mouse pancreatic ductal adenocarcinomas (PDACs), pericentromeric satellites accounted for a mean 12% (range 1 to 50%) of all cellular transcripts, a mean 40-fold increase over that in normal tissue. In 15 of 15 human PDACs, alpha satellite transcripts were most abundant and HSATII transcripts were highly specific for cancer. Similar patterns were observed in cancers of the lung, kidney, ovary, colon, and prostate. Derepression of satellite transcripts correlated with overexpression of the long interspersed nuclear element 1 (LINE-1) retrotransposon and with aberrant expression of neuroendocrine-associated genes proximal to LINE-1 insertions. The overexpression of satellite transcripts in cancer may reflect global alterations in heterochromatin silencing and could potentially be useful as a biomarker for cancer detection.
Genome-wide sequencing approaches have revealed an increasing set of transcribed noncoding sequences (ncRNA), including “pervasive transcription” by heterochromatic regions of the genome linked to transcriptional silencing and chromosomal integrity (1, 2). In the mouse, heterochromatin is composed of centric (minor) and pericentric (major) satellite repeats that are required for formation of the mitotic spindle complex and faithful chromosome segregation (3), whereas human satellite repeats have been divided into multiple classes with similar functions (4). Accumulation of satellite transcripts in mouse and human cell lines results from DNA demethylation, heat shock, or the induction of apoptosis, and their overexpression has been associated with genomic instability (5, 6). Stress-induced transcription of satellites in cultured cells has also been linked to the activation of retroelements encoding RNA polymerase activity such as long interspersed nuclear element 1 (LINE-1) (L1TD1) (7, 8). The global expression of repetitive ncRNAs in primary tumors has not been analyzed owing to the bias of microarray platforms toward annotated coding sequences and the specific exclusion of repeat sequences from standard analytic programs.
We used a next-generation digital gene expression (DGE) method (9) to obtain a comprehensive view of the transcriptome of primary tumors. We first evaluated mouse pancreatic ductal adenocarcinomas (PDACs) generated through pancreas-targeted expression of activated Kras and loss of Tp53 (10). These tumors are histopathological and genetic mimics of human PDAC, which almost universally display mutations in the KRAS oncogene and show frequent loss of the TP53 tumor suppressor gene. Notably, 47% of transcripts sequenced in the first PDAC (468,359 transcripts per million; tpm) were not annotated and mapped to the major mouse satellite, which contributes to only 0.02 to 0.4% of transcripts in normal pancreas or liver. In the tumor, satellite reads were found in both sense and antisense directions and were absent from purified polyadenylated RNA. The number of transcripts was >100 times that of normal tissue and 3600 times as abundant in the tumor as mRNA transcripts of the Gapdh (glyceraldehyde-3-phosphate dehydrogenase) housekeeping gene. We extended DGE analysis to additional mouse tumors with diverse genotypes: Increased satellite expression was noted in 7 of 9 PDACs, 2 of 3 colon cancers, and 2 of 2 lung cancers (range 12,236 to 160,186 tpm) (Fig. 1A and table S1). In primary tumors over-expressing satellites, the composite distribution of all RNA reads among coding, ribosomal, and other nc transcripts differed significantly from that of normal tissues (Fig. 1B), suggesting that the cellular transcriptional machinery is affected by the massive expression of satellites. Genomic amplification of satellites did not account for the exceptional abundance of these transcripts, as determined by next-generation DNA digital copy number variation analysis, implicating transcriptional derepression of heterochromatin as a possible driving mechanism (table S2).
Fig. 1.
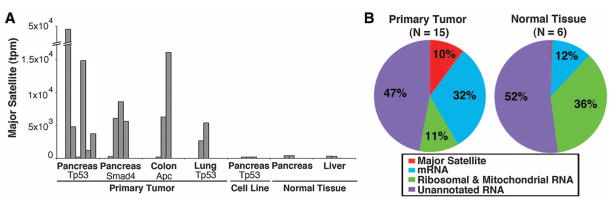
Massive expression of major satellites in mouse pancreatic tumors. (A) Expression of major satellite in primary tumors, cell lines, and normal tissues, presented as transcripts per million of aligned genomic reads. All tumors and cell lines have KrasG12D; deleted genes are listed individually (Tp53, Smad4, and Apc). (B) Graphical representation of sequence read contributions from major satellites, averaged among all primary tumors versus normal tissues (pancreas and liver). “Unannotated RNA” indicates reads that aligned to the mouse genome, but not to the mouse reference transcriptome.
Northern blots of mouse PDACs demonstrated that the major satellite-derived transcripts ranged from 100 base pairs (bp) to 5 kbp (Fig. 2A), consistent with the proposed cleavage of the primary transcript by Dicer1 (11), whose expression is 2.6 times higher in mouse pancreatic tumors with increased satellite expression (P = 0.0006, t test). Immortalized cell lines established from three satellite-overexpressing PDACs displayed minimal expression of satellites (range 173 to 433 tpm), suggesting either negative selection pressure or reestablishment of satellite silencing mechanisms under in vitro conditions. Treatment with 5-azacytidine (AZA) led to massive reexpression of satellites, supporting DNA methylation as a potential mechanism for satellite silencing in vitro (Fig. 2, A and B). Inoculation of an established PDAC cell line (CL3) into nude mice to generate subcutaneous tumors (n = 5) resulted in reexpression of satellites, suggesting that these loci become derepressed in vivo (Fig. 2C). Most normal adult mouse tissues, except lung, showed minimal expression of satellites, but the uncleaved 5-kbp satellite transcript was expressed in embryonic tissues (fig. S1). Thus, the aberrant expression of satellites in primary pancreatic tumors does not appear to simply recapitulate an embryonic cell fate, but possibly reflects altered processing of the primary 5-kbp satellite transcript.
Fig. 2.
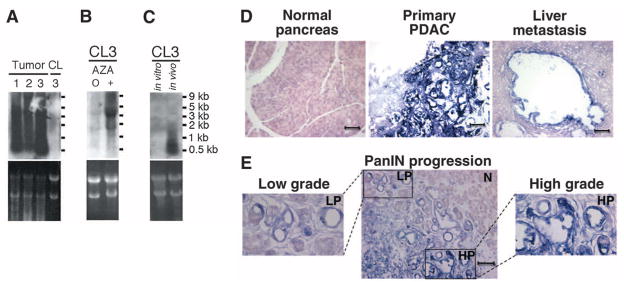
Expression patterns of major satellites in tumors and normal mouse tissues. Northern blot analyses: (A) Three KrasG12D, Tp53lox/+ pancreatic primary tumors (Tumors 1 to 3) and a stable cell line (CL3) derived from Tumor 3. (B) CL3 before (0) and after (+) treatment with AZA. (C) CL3 cells cultured in vitro and grown as subcutaneous tumors in vivo. (D and E) RNA-ISH with major satellite probe (purple stain): (D) Normal pancreas, primary PDAC, and liver metastasis. (E) Preneoplastic low-grade PanIN (LP) lesion adjacent to high-grade PanIN (HP) and normal pancreas (N). Higher magnification (400×) of inset low-grade (left) and high-grade (right) PanIN lesions. All images are at 200× magnification (scale bar, 100 μm).
RNA in situ hybridization (RNA-ISH) showed high levels of mouse major satellite expression in all cells within primary tumors and metastases (Fig. 2D). Notably, elevated satellite expression was evident in early preneoplastic low-grade pancreatic intraepithelial neoplasia (PanIN), and it increased further upon transition to high-grade PanIN (Fig. 2E). Clearly defined metastatic lesions to the liver were strongly positive by RNA-ISH, as were small clusters of PDAC cells within the liver parenchyma, that otherwise would not have been detected by histopathological analysis (fig. S2). Low-level diffuse expression was evident in mouse embryonic liver and lung (fig. S3), but no normal adult or embryonic tissues demonstrated satellite expression comparable to that in tumor cells.
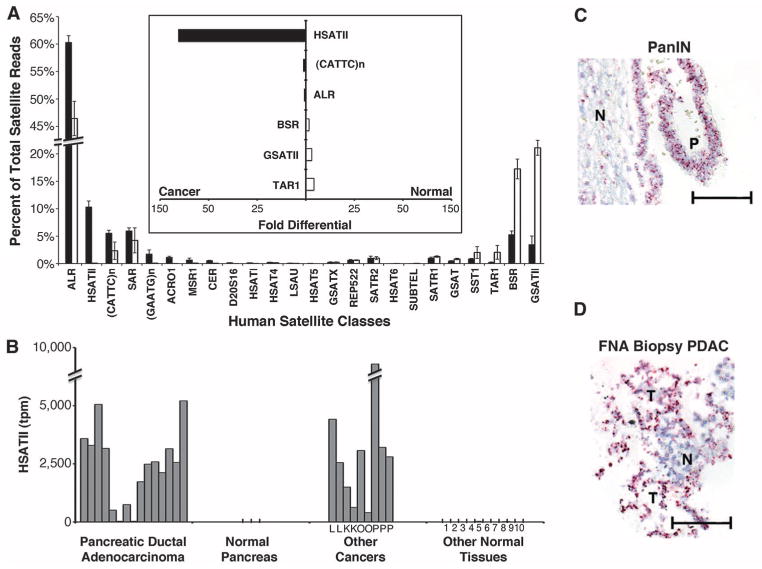
To investigate whether human tumors also overexpress satellite ncRNAs, we extended the DGE analysis to various human malignancies with a particular focus on PDAC. We first measured the total amount of all satellite transcripts: Analysis of 15 PDACs showed a median 21-fold increased expression compared with normal pancreas, but some other normal human tissues also had measurable levels of total satellite expression (fig. S4 and table S3). However, subdivision of human satellites among their multiple classes (4) revealed major differences between tumors and all normal tissues (Fig. 3A). The greatest differential expression in cancer was in the pericentromeric satellite HSATII (mean 2416 tpm; 10.3% of satellite reads), which was undetectable in normal human pancreas and had minimal expression in other normal tissues (131-fold differential expression; Fig. 3, A and B). In contrast, normal tissues had a high representation of GSATII, beta satellite (BSR), and TAR1, although these satellite classes constitute a small minority of satellite reads in pancreatic cancer. The most abundant class of normally expressed human satellites, alpha (ALR) (12), was expressed at 294 tpm in normal human pancreas, but constituted on average 12,535 tpm in PDACs (60.3% of satellite reads; 43-fold differential expression). Thus, whereas the over-expression of human ALR was comparable to that of mouse major satellites, the less abundant HSATII showed exceptional specificity for human PDAC. High levels of HSATII were also observed in other human cancers, including lung (2 of 2), kidney (2 of 2), ovarian (2 of 2), and prostate (3 of 3), indicating that this may be a shared feature of various carcinomas (mean 2820 tpm; Fig. 3B).
Fig. 3.
Overexpression of satellites in human cancers. (A) Breakdown of satellite classes as a percentage of total satellites in human PDAC (black, n = 15) and normal human tissues (white, n = 12). Satellites are ordered from highest in tumors to highest in normal tissue (left to right). Error bars represent SEM. Inset (bar graph, center) shows the differential expression of selected satellite classes enriched in cancer (left, black bars) or normal tissue (right, white bars). (B) HSATII expression in human PDAC, normal pancreas, other cancers (L, lung; K, kidney; O, ovarian; P, prostate), and normal human tissues (1, fetal brain; 2, adult brain; 3, colon; 4, fetal liver; 5, adult liver; 6, lung; 7, kidney; 8, placenta; 9, prostate; and 10, uterus) quantitated by DGE. Satellite expression is shown as transcripts per million (tpm) aligned to human genome. (C and D) RNA-ISH with HSATII probe (red stain): (C) human PanIN (P) and normal adjacent tissue (N). (D) EUS-FNA biopsy of confirmed tumor (T) and normal adjacent tissue (N). All images are at 200× magnification (scale bar, 100 μm).
RNA-ISH analysis of human tissues showed differential expression of HSATII in PDAC and PanIN (n = 4) compared to normal adjacent tissue as well as in chronic pancreatitis (n = 8) (Fig. 3C and fig. S5). When we applied this assay to clinical samples [endoscopic ultrasound-guided fine-needle aspirates (EUS-FNA) of pancreatic masses], HSATII-positive cells were identified in 10 of 10 cases confirmed to have pancreatic cancer at the time of surgical resection, including two cases in which the FNA histopathology was non-diagnostic (Fig. 3D). These initial results suggest that HSATII merits further study as a potential cancer biomarker.
To identify other transcripts co-regulated with satellites in tumors, we performed linear regression analysis in both mouse (major satellite) and human (ALR satellite) (fig. S6). Using a linear correlation cutoff of R > 0.85, we created two sets of highly correlative genes (mouse: 297 genes, table S4; human: 539 genes, table S5), which we refer to as satellite correlated genes (SCGs). Mouse and human SCGs were enriched for transposable elements, with the autonomous retrotransposon LINE-1 having the highest expression level in tumors (Fig. 4A). In addition to transposons, a subset of cellular mRNAs showed high correlation with the expression of satellites across diverse tissues. Absence of a shared transcriptional silencing mechanism may contribute to derepression of both LINE-1 and satellites, but the increased expression of diverse mRNAs is less readily explained. LINE-1 insertion upstream of transcriptional start sites of cellular transcripts has recently been implicated in gene regulation (13–15), leading usto testthe proximity ofgenomic LINE-1insertions to the SCGs. In mouse, there was a marked correlation between SCGs and their distance to LINE-1 genomic insertions (Fig. 4B). A similar measurable effect was evident with human SCGs, albeit dampened most likely by the heterogeneity of LINE-1 insertions in the human genome (16–18) (fig. S7). Together, these observations suggest that tumor-associated derepression of satellites is highly correlated with increased expression of LINE-1, along with a subset of cellular genes in close proximity to this retrotransposon.
Fig. 4.
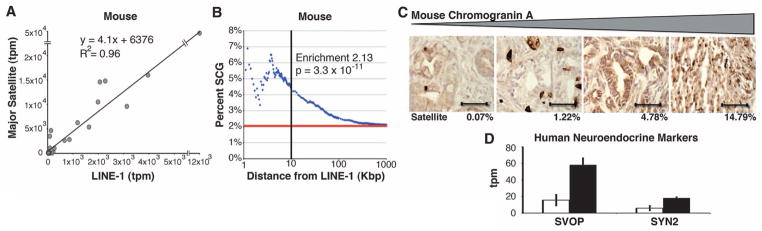
Correlation of satellite expression with LINE-1 and cellular transcriptional profile. (A) Linear correlation of mouse major satellite to LINE-1 expression. (B) Fraction of mouse SCGs (blue) versus predicted (red) transcriptional start sites within a given distance of a LINE-1. Enrichment calculations were done at a distance of 10 kbp (black line). (C) Immunohistochemistry of mouse PDAC for the neuroendocrine marker chromogranin A. Tumors are depicted as a function of increasing chromogranin A staining (brown), with the relative level of major satellite expression noted for each tumor (bottom; percentage of all transcripts). Images are at 200× magnification (scale bar, 100 μm). (D) Differentially expressed (Q < 0.05) neuroendocrine genes in human PDACs with high (black) versus low (white) ALR satellite levels. Error bars represent SEM.
Of cellular transcripts that constitute SCGs, 190 of 297 mouse SCGs and 206 of 539 human SCGs are recognized by the DAVID gene ontology program (19, 20); in both species, the transcripts are highly enriched for genes implicated in neural cell fates and germ or stem cell pathways (table S6). Neuroendocrine differentiation has been described in a variety of epithelial malignancies, including pancreatic cancer (21), and it is correlated with increased aggressiveness in prostate cancer (22). In mouse PDACs, we observed a marked correlation between the level of satellite expression and the number of carcinoma cells staining for the neuroendocrine marker chromogranin A (Fig. 4C), whereas in human PDACs, the neuroendocrine markers synaptic vesicle 2–related protein (SVOP) and synapsin 2 (SYN) were associated with high ALR satellite expression (Fig. 4D and table S7). Together these data suggest that a global alteration in expression of heterochromatic ncRNAs may affect a known cellular differentiation program implicated in cancer.
In summary, we have identified the massive generation of bidirectional ncRNAs from the major satellite in mouse tumor models and from ALR and HSATII satellites in human pancreatic and other epithelial cancers. The discovery of satellite repeat overexpression was made possible by the development of next-generation DGE approaches, which provide a quantitative and sequence-specific measure of highly repetitive sequences that are excluded from traditional analytic programs. Indeed, BLAST sequence matching of satellite sequences in both mouse and human tumors first identified sequences in the recently completed parasite genomes (see supporting online text). Although further analyses are required to explore the mechanism and consequences of aberrant expression of satellites in cancer tissues, we hypothesize that it likely results from a general derepression of chromosomal marks affecting both satellites and LINE-1 retrotransposons, with proximity to LINE-1 activation affecting the expression of cellular genes enriched for neuroendocrine specification. Current evidence indicates that both DNA methylation and histone H3 lysine 9 (H3K9) trimethylation are critical for the maintenance of satellite repression (14) and that dysregulation of these epigenetic marks is linked to carcinogenesis. Targeted DGE analysis of all known epigenetic regulators (23) in mouse and human tumors showed distinct expression patterns, but no single consistent abnormality (tables S8 to S10). Finally, the potential importance of satellite and LINE-1 deregulation as a consistent biomarker in diverse epithelial cancers merits further clinical testing.
Supplementary Material
Acknowledgments
This work was supported by a Pancreatic Cancer Action Network–American Association for Cancer Research Fellowship and the Warshaw Institute for Pancreatic Cancer Research (D.T.T.); Fond.Veronesi (G.C.); Howard Hughes Medical Institute (D.A.H. and M.N.R.); and National Cancer Institute CA129933 (D.A.H). We thank T. Raz, P. Kapranov, E. Giladi, and J. Whetstine for helpful discussions and K. Haigis and K. Wong for providing mouse colon and lung tumors, respectively. Massachusetts General Hospital and the authors (D.A.H., D.L., S.M., D.T.T.) have filed a patent application relating to detection of satellite and LINE sequences in human cancers.
Footnotes
References and Notes
- 1.Berretta J, Morillon A. EMBO Rep. 2009;10:973. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacquier A. Nat Rev Genet. 2009;10:833. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 3.Guenatri M, Bailly D, Maison C, Almouzni G. J Cell Biol. 2004;166:493. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurka J, et al. Cytogenet Genome Res. 2005;110:462. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 5.Bouzinba-Segard H, Guais A, Francastel C. Proc Natl Acad Sci USA. 2006;103:8709. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valgardsdottir R, et al. Nucleic Acids Res. 2008;36:423. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ugarkovic D. EMBO Rep. 2005;6:1035. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carone DM, et al. Chromosoma. 2009;118:113. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 9.Lipson D, et al. Nat Biotechnol. 2009;27:652. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 10.Bardeesy N, et al. Proc Natl Acad Sci USA. 2006;103:5947. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanellopoulou C, et al. Genes Dev. 2005;19:489. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada T, et al. Cell. 2007;131:1287. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 13.Kuwabara T, et al. Nat Neurosci. 2009;12:1097. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey JA, Carrel L, Chakravarti A, Eichler EE. Proc Natl Acad Sci USA. 2000;97:6634. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montoya-Durango DE, et al. Mutat Res. 2009;665:20. doi: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck CR, et al. Cell. 2010;141:1159. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CR, et al. Cell. 2010;141:1171. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iskow RC, et al. Cell. 2010;141:1253. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennis G, Jr, et al. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Nat Protoc. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Tezel E, Nagasaka T, Nomoto S, Sugimoto H, Nakao A. Cancer. 2000;89:2230. doi: 10.1002/1097-0142(20001201)89:11<2230::aid-cncr11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 22.Cindolo L, Cantile M, Vacherot F, Terry S, de la Taille A. Urol Int. 2007;79:287. doi: 10.1159/000109711. [DOI] [PubMed] [Google Scholar]
- 23.Cloos PA, Christensen J, Agger K, Helin K. Genes Dev. 2008;22:1115. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.