Abstract
Introduction
Wnt signalling has been implicated in stem cell regulation however its role in breast cancer stem cell regulation remains unclear.
Methods
We used a panel of normal and breast cancer cell lines to assess Wnt pathway gene and protein expression, and for the investigation of Wnt signalling within stem cell-enriched populations, mRNA and protein expression was analysed after the selection of anoikis-resistant cells. Finally, cell lines and patient-derived samples were used to investigate Wnt pathway effects on stem cell activity in vitro.
Results
Wnt pathway signalling increased in cancer compared to normal breast and in both cell lines and patient samples, expression of Wnt pathway genes correlated with estrogen receptor (ER) expression. Furthermore, specific Wnt pathway genes were predictive for recurrence within subtypes of breast cancer. Canonical Wnt pathway genes were increased in breast cancer stem cell-enriched populations in comparison to normal breast stem cell-enriched populations. Furthermore in cell lines, the ligand Wnt3a increased whilst the inhibitor DKK1 reduced mammosphere formation with the greatest inhibitory effects observed in ER+ve breast cancer cell lines. In patient-derived metastatic breast cancer samples, only ER-ve mammospheres were responsive to the ligand Wnt3a. However, the inhibitor DKK1 efficiently inhibited both ER+ve and ER-ve breast cancer but not normal mammosphere formation, suggesting that the Wnt pathway is aberrantly activated in breast cancer mammospheres.
Conclusions
Collectively, these data highlight differential Wnt signalling in breast cancer subtypes and activity in patient-derived metastatic cancer stem-like cells indicating a potential for Wnt-targeted treatment in breast cancers.
Introduction
WNT proteins are a family of secreted, glycosylated, and palmitoylated peptides which function in diverse processes such as embryonic induction, generation of cell polarity, and cell fate specification [1]. Aberrant activation of Wnt signaling has been described in a number of human cancers including colorectal cancer, ovarian cancer and breast cancer [2], [3], [4], [5], [6], [7]. β-catenin expression has been associated with poor prognosis in breast cancer patients in a number of studies [8], [9] and is enriched in basal-like breast cancer [6]. Furthermore loss of negative pathway regulators such as the extracellular inhibitor of WNT signaling, secreted Frizzled-related protein 1 (sFRP1), is found in many breast tumors and is associated with poor prognosis [3], [10]. Down regulation of the inhibitor Dickkopf 1 (DKK1) in a lung metastases derived MCF7-LM cell line demonstrates the importance of Wnt regulation in the metastatic process in breast cancer [11]. Collectively these data suggest that WNT pathway de-regulation within the breast contributes to cancer formation and metastasis.
Recent studies suggest breast cancer initiation and recurrence may be regulated by a small population of cells within the tumor, either stem cells or cells that exhibit stem-like properties [12]. Transplantation experiments using immunocomprimised mice, showed that as few as 100 human breast cancer cells with the cell surface markers CD44+CD24−/low were tumorigenic and could be serially passaged to generate new tumours [13].
Cells isolated from human breast cancers marked by CD44+CD24−/low lineage are anoikis-resistant and capable of self-renewal as mammosphere (MS) colonies providing a link between MS and cell surface markers that enrich for tumorgenic cells [14], [15].
Expression of Wnt1 in human mammary epithelial cells increases stem cell self renewal, resistance to apoptosis and failure to senesce [16]. More recent work using the MMTV-WNT-1 mouse model has identified an expanded mammary stem cell (SC) pool from a population of committed luminal progenitors indicating that Wnt-1 activation induces the appearance of aberrant progenitor cells, and suggest that both mammary stem and progenitor cells can serve as the cellular targets of WNT-1-induced tumorigenesis [17].
WNT pathway activation increases radio resistance of progenitor cells in the mouse mammary gland and human breast cancer cell lines [18], [19], which implicates the WNT pathway in resistance to current anti-cancer therapies, potentially through the regulation of stem and progenitor cell populations.
In this study we investigated the WNT pathway both in whole cell populations and stem-like cells of breast cell lines and patient-derived metastatic breast cancer samples. WNT pathway gene expression correlates with estrogen receptor (ER) expression and molecular sub-type, and some genes predict prognosis. WNT signalling was found to be activated in breast cancer stem-like cells compared to normal stem-like cells. Finally, we show that WNT pathway inhibition preferentially reduces stem-like cell activity in patient-derived metastatic breast cancer compared to normal cells. Collectively, these data suggest potential of the WNT-targeted therapeutics in breast cancer.
Methods
Cell Lines
Six normal breast (HB4A, MRSV4.4, MRSV1.7, MCF10A, MCF10F, 226L-U19), six ER+ve breast cancer (BT474, MCF7, MCF7-HER2-18, MDA-MB361, T47D and ZR75-1) and five ER-ve cancer (BT20, Hs578T, MDA-MB231, MDA-MB468 and SKBR3) cell lines were used in this study.
Primary Normal Human Breast Cells
Histologically confirmed normal breast tissue was obtained from 3 premenopausal patients undergoing fibroadenoma excision or breast reduction surgery (n = 3; mean age 38). Normal breast tissue was prepared and purified as previously described [20]. Briefly, tissue samples were dissected into 3- to 5-mm cubes and digested for 16–18 hours at 37°C in serum-free Dulbecco’s modified Eagle Medium (DMEM; Gibco) containing 200 U/mL type 3 collagenase (Worthington Biochemical Corporation), 5 mg/ml pronase (Roche) and 10x antibiotic/antimycotic (Invitrogen). The cells were then washed, collected by centrifugation at 1000 g and filtered to obtain a single-cell suspension, which was verified microscopically. CD45 positive cells were removed using anti-CD45 magnetic beads (Miltenyi). CD45 negative cells were then collected and resuspended in mammosphere culture medium.
Patient-derived Metastatic Breast Cancer Cells
Samples were collected from patients (ER+ve n = 3 and ER-ve n = 3) with metastatic breast cancer (Table 1). Tumour samples were prepared and purified as previously described [15]. Briefly, metastatic fluid was collected and centrifuged at 2000 g from 5 minutes and responded in PBS. Blood cells were removed by centrifugation through Lymphoprep solution (Axis Shield), followed by removal of CD45 positive cells using anti-CD45 magnetic beads (Miltenyi). CD45 negative cells were then collected and resuspended in mammosphere culture medium.
Table 1. Primary metastatic breast cancer samples.
| Sample ID | Histology | Source | Grade | ER | PgR | Her2 |
| BB3RC30 | IDC | PE | 2 | Pos | Pos | UN |
| BB3RC33 | ILC | AS | 2 | Pos | Pos | Pos |
| BB3RC36 | IDC | PE | 2 | Pos | Pos | UN |
| BB3RC37 | IDC | PE | UN | Neg | Neg | Neg |
| BB3RC38 | IDC | AS | 3 | Neg | Neg | Neg |
| BB3RC39 | IDC | PE | 3 | Neg | Neg | Neg |
Details of primary breast cancer samples used in this study. UN unknown, PE pleural effusion, AS Ascites sample IDC invasive ductal carcinoma, ILC invasive lobular carcinoma, ER oestrogen receptor alpha, PgR progesterone receptor, Neg negative, Pos positive.
Ethics Statement
Any experimental research reported in the manuscript has been performed with the approval of an appropriate ethics committee and in compliance with the Helsinki Declaration. All samples were collected with informed written consent. Normal breast tissue was collected with approval from South Manchester Research Ethics Committee (COREC#05/1403/159). Patient derived metastatic breast cancer cells were collected with approval from Tameside and Glossop Local Research Ethics Committee (COREC # 05/Q1402/25).
Mammosphere Culture
A single cell suspension was created from cell lines grown in monolayer culture using enzymatic digestion (Trypsin EDTA) followed by manual disaggregation (25 Gauge needle). Patient-derived breast cells did not require the addition of trypsin, a single cell suspension was created using manual disaggregation alone. Single cells from cell lines and patient derived cells were plated at a density of 500 cells/cm2 in non-adherent conditions, in culture flasks coated with (2-hydroxyethylmethacrylate) (poly-HEMA [Sigma]). Culture of cells in these conditions allows the survival of stem-like cells and subsequent spheroid growth from a single cell using both cell lines and primary tissue samples. Mammospheres grow at a similar rate when plated as single cells or at higher densities indicating that mammospheres are truly clonal structures and not formed through aggregation [21], [22].
Anoikis resistant cells
were collected after 12 hrs (RNA) or 24 hrs (protein) in non-adherent, mammosphere culture. As described above, single cell suspensions were obtained and cultured in non-adherent conditions at a density of 500 cells/cm2. Anoikis resistant cells were then collected by centrifugation at 1800 rpm for 2 minutes. Prior to RNA/protein extraction cells were incubated with 100 ul of Dead cell Removal micro-beads and dead cells removed using an MS column and MACS Seperator (Miltenyi Biotech). Mammospheres (MS) were counted after culture for 7 days in MS medium (DMEM: F12 medium supplemented with B27 without vitamin A [diluted 1∶ 50; Gibco]) and mammary epithelial growth medium aliquot of gentamicin/amphotericin-B and recombinant human epidermal growth factor (EGF), (SingleQuot) (Lorne Laboratories). Spheres over 50 µM were counted and the percentage of cells plated which formed spheres was calculated and is referred to as the percentage mammosphere formation units (%MFU). All cells were maintained in a humidified incubator at 37°C at an atmospheric pressure in 5% (v/v) carbon dioxide/air.
Wnt pathway inhibition
MCF10a, MCF7, MDA-MB-231, primary human normal breast cells and primary human invasive breast cancer cells were plated into MS culture and treated with a single dose of human recombinant DKK1 (R and D systems) at increasing concentrations (0–100ng/ml).
Wnt pathway activation
MCF10a, MCF7, MDA-MB-231, Patient-derived normal and metastatic breast cancer cells were plated into MS culture and treated with a single dose of mouse recombinant WNT3A (R and D systems) at increasing concentrations (0–50 ng/ml).
RNA Extraction and Real-time Quantitative PCR Assays of Breast Cell Lines
Total RNA was extracted from the sixteen cell lines harvested at log phase, using the RNeasy® Plus (Qiagen) kit following the manufacturer’s protocol. For real-time quantitative PCR (qPCR), mRNA was reverse-transcribed into cDNA using an ABI RT kit (Applied Biosystems). qPCR primers and probes were designed using Roche Probe Finder Design Software (Roche Applied Sciences) and reactions performed using the ABI PRISM 7900 Sequence Detection System instrument and software (Applied Biosystems). The reactions were incubated in a 384-well optical plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 10 min. Experiments were performed in triplicate for each sample. Gene expression was normalized to internal control SDHA (succinate dehydrogenase), YWHAZ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide).
RNA Extraction of Monolayer and AR Breast Cells
Cells were collected from 226L-U19, HB4a, MCF10a, MCF7, T47D and SKBR3 cell lines cultured in monolayer (pre-incubated with MS media for 12 hrs) and from viable AR cells. RNA was extracted using the RNeasy® Plus Mini (Qiagen) following the manufacturers protocol. In addition to the gDNA eliminator spin column provided in the RNeasy kit, an on-column DNase digestion was performed (RNase-Free DNase Set; Qiagen) to ensure maximum removal of DNA. RNA was eluted and the concentration measured using a GeneQuant machine (Amersham Biosciences). RNA integrity was assessed by microanalysis (Agilent Bioanalyser) and amplified using the WT-Ovation™ Pico RNA Amplification System (NuGEN) following the manufacturers protocol which employs SPI™ amplification (linear isothermal DNA amplification process) to amplify the RNA about 15,000-fold.
Custom Gene Expression Microarray Analysis of Monolayer and AR Breast Cells
Custom microarray chips were designed using Agilent technology. cDNA was fluorescently labelled using the FL-Ovation™ cDNA Fluorescent Module kit (NuGEN) with a single tag (Cy™3) which incorporates into the cDNA. The fluorescently tagged cDNA was loaded onto a microarray slides and the Agilent microarray chips attached. Slides were incubated at 65°C for 40 hours in a hybridisation oven to allow hybridisation to occur. The chips were then washed and scanned using an Agilent scanner.
Primary Breast Cancer Microarray
Affymetrix gene expression data representing a total of 1107 primary breast tumors from six previously published microarray studies [23], [24], [25], [26], [27], [28] were integrated as described previously using ComBat [29] to remove batch effects [30]. Centroid prediction [31] was used to assign the tumors from each dataset to the five Norway/Stanford subtypes (Basal, Luminal A, Luminal B, ERBB2 and Normal-like [32].
Gene Expression Analysis
Centred average linkage clustering of cell lines, monolayer/anoikis resistant cells and integrated tumour datasets was performed using Cluster [33] and heatmaps generated using TreeView programs as described previously [34].
Accession Numbers
Genbank accession numbers from NCBI, of genes used in microarray are provided below. [Genbank: NM_004655, NM_001904, NM_012242, NM_000125, NM_012193, NM_003507, NM_003508, NM_002093, NM_030915, NM_001130713, NM_002335, NM_002336, NM_003012, NM_003013, NM_003014, NM_005985, NM_001083962, NM_001204869, NM_003881, NM_005430, NM_025216, NM_003394, NM_030761, NM_001256105, NM_030775].
Protein Analysis
Western blotting was carried out as previously described [35]. Antibodies used in this study were anti unphoshorylated B-catenin (Upstate, Millipore) AXIN2 (Cell signalling) LEF1 (Cell signalling) and DKK1 (abcam) and B-ACTIN (abcam).
Statistical Analyses
Normally distributed data was analysed using analysis of variance (ANOVA) to determine significant differences of DKK1 treatment followed by individual comparisons to control (0 ng/ml) using the independent T-test. Data which deviated from the normal distribution was analysed using Kruskall-wallis followed by individual comparisons to control (0 ng/ml) using the Mann Whitney U test. Gene expression: data was analysed using two tailed students’t-tests.
Results
Activation of Wnt Signalling in Breast Cancer Cell Lines
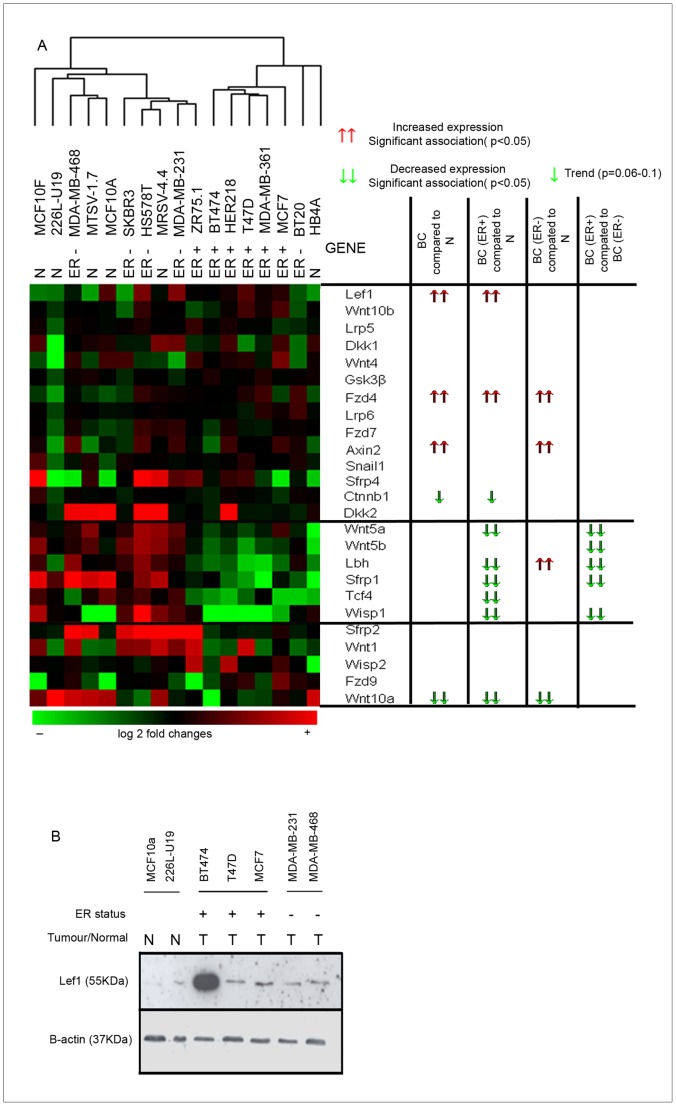
Comprehensive analysis of WNT pathway mRNA expression across a large panel of breast cell lines was performed, which revealed cell type specific gene expression within the cell lines tested (Figure 1A). Well established downstream targets, AXIN2 and LEF1 (Figure 1A) showed higher expression in breast cancer cell lines suggesting activation of canonical WNT signalling. Consistent with its gene expression level, LEF1 protein was higher in five breast cancer cell lines compared to two normal cell lines (MCF10A and 226LU19), providing further evidence that WNT signalling is active in breast cancer cell lines (Figure 1B). Interestingly, we observed high levels of LEF1 in the HER2 expressing cell line BT474. HER2 is known to activate β-catenin activity which may account for this increase in the downstream target LEF1 [36]. A WNT receptor and a WNT ligand were also dysregulated in breast cancer cell lines, FZD4 showed higher expression in breast cancer cell lines whilst WNT10A expression was decreased. The expression pattern of WNT pathway genes largely clustered the cell lines by estrogen receptor-α (ER) status; ER+ve cell lines predominately expressed the downstream target LEF1 whilst ER-ve cell lines express another downstream gene AXIN2 (Figure 1A). As reported by others, WNT5A, LBH, WISP1 and TCF4 were expressed at lower levels in ER+ cell lines compared to ER-ve cancer but also compared to immortalised normal breast cell lines (Figure 1A), perhaps reflecting their ER-ve status. Independent of ER status, overall WNT pathway signalling assessed by the canonical downstream target genes AXIN2 and LEF1 is higher in breast cancer cell lines compared to normal breast cell lines.
Figure 1. Wnt signalling in normal breast cell lines (N), ER-ve (T−) and ER+ve (T+) breast cancer cell lines.
A) mRNA expression was normalised to house keeper genes. Cluster analysis was performed and data displayed in a heatmap: decreased (green) or increased (red) expression compared to the mean mRNA expression. (Red) ↑↑ Indicates significant increased expression (<0.05) (red) ↑ indicates a trend towards increased expression. (Green) Indicates significant increased expression (<0.05) (green) indicated a trend towards increased expression. B) Protein expression of Lef 1 and B-actin (housekeeper) in normal breast cell lines (N), ER-ve (T−) and ER+ve (T+) breast cancer cell lines.
Prognostic Value of WNT Expression in Breast Cancer
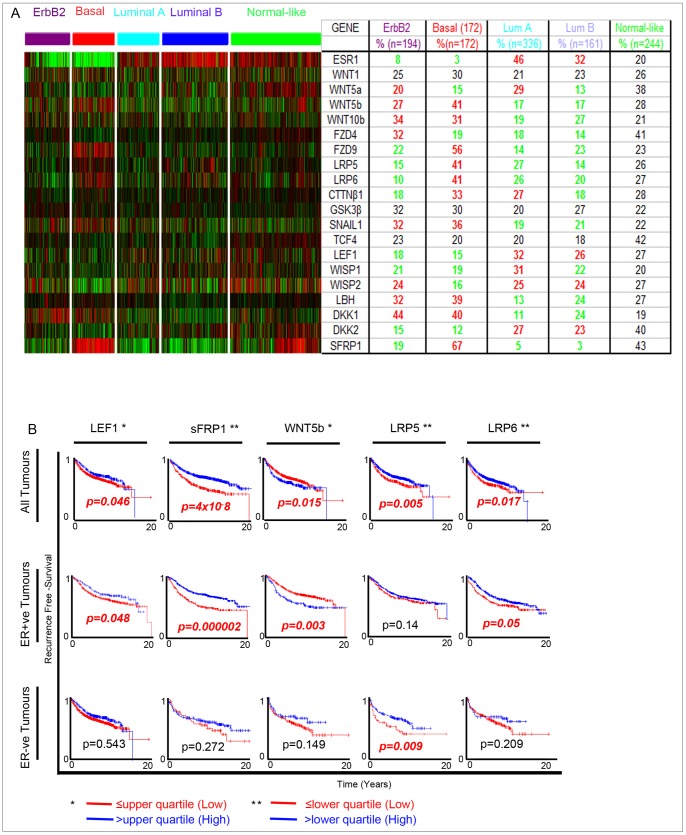
To assess the clinical importance of WNT pathway genes, we performed analysis of a large cohort of breast cancer tumours comprising six publically available gene expression data set [30]. Analysis of WNT signalling gene expression within defined subtypes of breast cancer showed variation in gene expression across the subgroups. Confirming the results observed using cell lines (Figure 1) we observed higher expression of LEF1, suggesting WNT activation in Luminal, ER+ve breast cancer patients (Figure 2A). A number of genes were differentially expressed between Luminal and Basal/ErbB2 breast cancer, such as WNT5B, WNT10B and DKK1. A number of genes were highly expressed specifically within the basal subtype of breast cancer, LRP5, LRP6, and sFRP1 (Figure 2A). Analysis of these genes showed significant associations to recurrence free survival within breast cancer patients. The most significant association was with sFRP1, where decreased expression predicts early recurrence in ER+ve breast cancer, most likely due to lack of Wnt inhibition. Lower levels of Lef1 were also predictive of recurrence in breast cancer within the ER+ve subgroup. It might be expected that higher levels of Lef1 would correlate with recurrence, however lower Lef1 expression may identify a subgroup of ER+ve tumours that have a basal-type phenotype, based upon their Wnt gene expression. This subgroup within ER+ve tumours can also be observed with analysis of Wnt5b, where high expression predicts early recurrence, and where high expression is a marker of the basal-type phenotype. LRP5 and LRP6 although demonstrating a very similar expression profile within breast tumours have very different effects on recurrence. Low expression of LRP6 in ER+ve tumours was predictive of early recurrence, whilst low levels of LRP5 was predictive of early recurrence in ER-ve breast cancer (Figure 2B). LRP5/6 are co-receptors for WNT ligands, previously thought to act similarly to activate downstream signalling. Recent research suggests that they may actually function differently, dependant upon physiological conditions and the type and availability of WNT ligand [37], [38]. Further investigations will be needed to determine their precise roles in breast cancer.
Figure 2. Analysis of mRNA expression in primary human breast cancer samples.
Expression data was combined from multiple studies into a combined cohort of ERBB2 (n = 194) Basal (172) Luminal A (n = 336) and Luminal B (161) and normal-like (n = 244) subtypes of breast cancer. A) mRNA expression data was displayed in a heatmap: decreased (green) or increased (red) expression compared to the mean mRNA expression. % of samples with high gene expression are detailed and denoted as green when expression is decreased and red when expression is increased. B) Kaplan Meier plots Wnt gene expression show the years of recurrence free survival, where red indicates lower gene expression and blue indicates higher gene expression. * Red ≤upper quartile: Blue >upper quartile ** Red ≤lower quartile: Blue >lower quartile. P values were generated and significant results were highlighted in red.
Wnt Pathway Activation in Breast Cancer Stem-like Cells
Numerous reports implicate Wnt signalling in breast stem cell activity [19], [39], [40]. To investigate this further, we employed the mammosphere culture technique to enrich for cells with stem-like activity, breast cancer stem-like cells (BCSCs). Anoikis resistant (AR) cells are enriched for stem-like activity and have increased tumour forming capacity in mice [15]. Comparison of MCF7 AR cells to adherent monolayer cells showed an increase in active WNT signalling in AR cells was confirmed by increased expression of activated beta-catenin protein, both downstream targets AXIN2, LEF1, and decreased expression of DKK1 protein (Figure 3A and Figure S2).
Figure 3. Gene expression analysis of Wnt signalling in monolayer (Mono) and anoikis resistant (AR) cells of normal breast cell lines (N), ER-ve and ER+ve breast cancer cell lines.
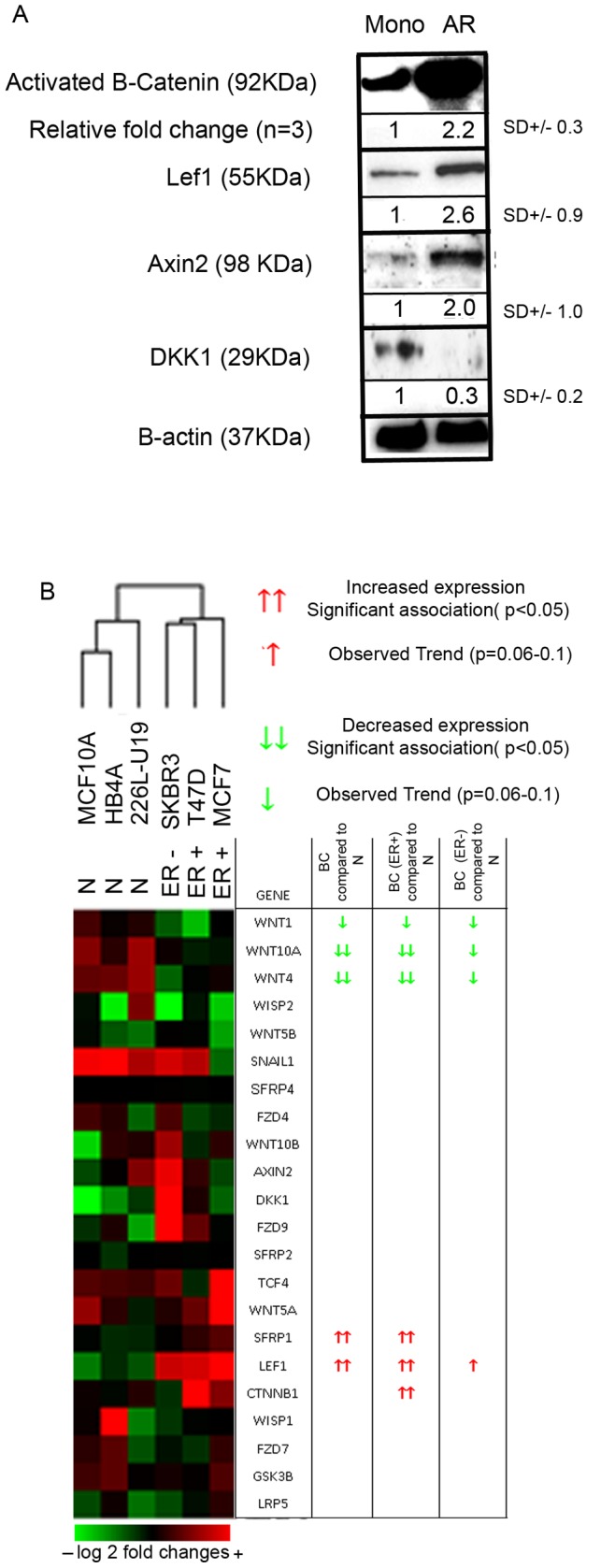
A) Protein expression of activated B-catenin (unphosphorylated), Lef1, Axin2, DKK1 and B-actin (housekeeper) in MCF7 monolayer and AR cells. B) Cluster analysis was performed using the fold change in expression between Mono and AR cells. Data is displayed in a heatmap represented by either decreased (green) or increased (red) expression compared to the mean mRNA expression. (Red) ↑↑ Indicates significant increased expression (<0.05) (red) ↑ indicates a trend towards increased expression. (Green) Indicates significant increased expression (<0.05) (green) indicated a trend towards increased expression.
Wnt Signalling is Suppressed in Normal Breast Stem-like Cells
We next investigated WNT signalling gene expression using a custom Agilent microarray using AR-derived and monolayer cultured populations from 3 immortalised normal and three breast cancer cell lines. AR-derived populations contain stem-like cells, which has been validated by in vivo tumour initiation [15;Ablett et al submitted]. Gene expression analysis confirmed results in Figure 1 that there are higher levels of WNT signalling in breast cancer cell lines compared to normal breast cell lines when grown in either monolayer or AR culture (Figure S1). Fold change differences between monolayer and AR cells (enriched for BCSCs) for each cell line were calculated and displayed as a heatmap (Figure 3B). Importantly, analysis of the change in gene expression between monolayer cultured and AR cells (enriched for BCSCs) showed differences between breast cancer and normal cell lines (Figure 3B). WNT ligands WNT1, WNT10a and WNT4 were expressed at higher levels in normal AR cells compared to monolayer cells, however in the breast cancer cell lines, their expression was lower in the AR cells compared to monolayer cells. sFRP1 and β-catenin were highly expressed in BCSCs from ER+ve breast cancer cell lines (MCF7 and T47D). No increase in expression was observed in the ER-ve breast cancer cell line tested (SK3RB) suggesting that canonical WNT signalling in BCSCs may correlate with ER (Figure 3B). Most importantly, we found higher levels of the WNT signalling gene LEF1 in BCSCs compared to monolayer cells, irrespective of ER expression, while normal breast stem cell populations showed lower levels compared to normal monolayer cells (Figure 3B). The data demonstrates specific upregulation of WNT pathway signalling in a population enriched for BCSCs.
Modulation of Wnt Pathway Signalling Affects Stem Cell-like Activity
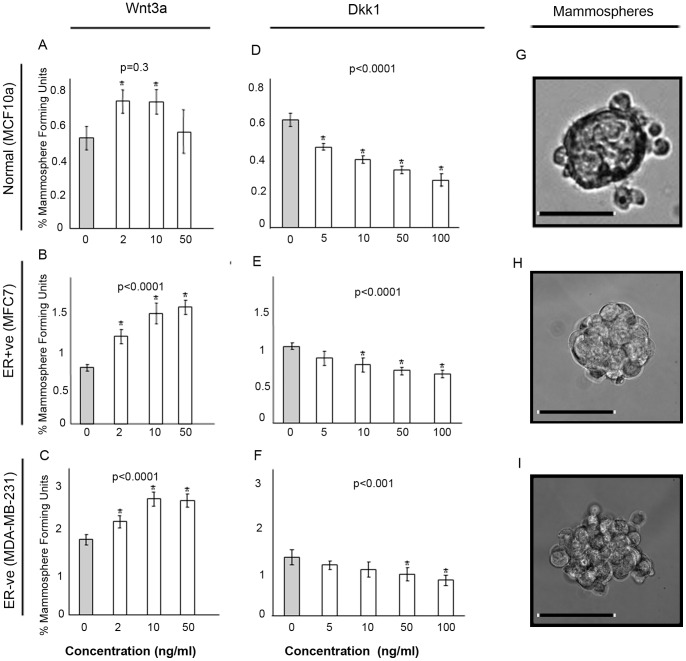
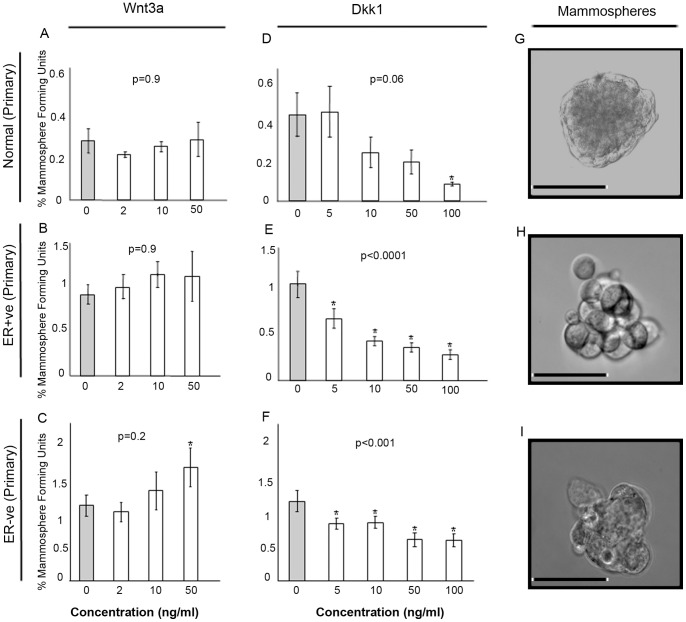
To test the potential of targeting the WNT pathway we investigated the effects of WNT pathway modulation in cell lines. Cells were cultured as mammospheres (MS) with increasing concentrations of either DKK1 to inhibit or WNT3A to activate the WNT pathway. Recombinant WNT3A treatment significantly increased the number of MS in both normal (MCF10A) and breast cancer (MCF7 and MDA-MB-231) cell lines (Figure 4A–C). DKK1 treatment significantly decreased the number of MS in all cell lines tested with the most significant decrease observed using the highest concentration (100 ng/ml) of DKK1 (Figure 4D–F). Using patient-derived cell samples from both ER+ve (n = 3) and ER-ve (n = 3) breast cancer and normal breast tissue (n = 3) we found that WNT3A caused a significant increase in MS only in the ER-ve sample (Figure 5C). Inhibition of WNT signalling using DDK1 showed that low dose (5 ng/ml) treatment was sufficient to decrease mammosphere formation in primary breast cancer cells whilst leaving the normal cells of the breast unaffected. Only high concentrations (100 ng/ml) of DKK1 were sufficient to inhibit MS formation in normal primary breast cells (Figure 5D–F).
Figure 4. Modulation of Wnt signalling in normal and breast cancer cell lines.
Single cells were plated in non-adherent conditions and treated with increasing concentrations of either Wnt3a (0–50 ng/ml) or DKK1 (0–100 ng/ml) and cultured for 7 days and number of mammospheres counted. Wnt3a treatments are displayed in the left panel and DKK1 treatments in the right panel. Light grey bars represent untreated control A) MCF10a cells (Wnt3a) B) MCF7 cells (Wnt3a) C) MDA-MB-231 cells (Wnt3a) D) MCF10a cells (DKK1) E) MCF7 cells (DKK1) F) MDA-MB-231 cells (DKK1). Data is expressed as % mammosphere formation units. P values were generated by ANOVA. Asterisks mark individual comparisons which reached statistical significance * <0.01 ** <0.001 generated by a T-test. G) Image of a MCF10a mammosphere H) Image of an MCF7 mammosphere I) Image of an MDA-MB-231 mammosphere. Scale bar represents 50 µM.
Figure 5. Modulation of Wnt signalling in normal and primary breast cancer samples (Normal n = 3; ER+ve n = 3; ER-ve n = 3).
Single cells were plated in non-adherent conditions and treated with increasing concentrations of either Wnt3a (0–50 ng/ml) or DKK1 (0–100 ng/ml) and cultured for 7 days and number of mammospheres counted. Wnt3a treatments are displayed in the left panel and DKK1 treatments in the right panel. Light grey bars represent untreated control A) primary normal breast cells (Wnt3a) B) ER+ve primary breast cancer cells (Wnt3a) C) ER-ve primary breast cancer cells (Wnt3a) D) primary normal breast cells (DKK1) E) ER+ve primary breast cancer cells (DKK1) F) ER−ve primary breast cancer cells (DKK1). Data is expressed as % mammosphere formation units. P values were generated by ANOVA. Asterisks mark individual comparisons which reached statistical significance * >0.01 ** >0.001 generated by a T-test. G) Image of a normal primary mammosphere H) Image of an ER positive primary tumour mammosphere I) Image of an ER negative primary tumour mammosphere. Scale bar represents 50 µM.
Discussion
Our results are consistent with existing data [16], [41] which suggest activation of WNT signalling in breast cancer through the increased expression of key signalling components and downstream target such as LEF1 at both the mRNA and protein level. We observed higher levels of WNT signalling in breast cancer cell lines correlating with ER expression. Although both classical downstream targets AXIN2 and LEF1 were on average expressed at higher levels in breast cancer cell lines, ER-ve cell lines expressed the highest levels of AXIN2 whereas the major downstream target activated in ER+ve cell lines was LEF1 demonstrating the subtle differences in WNT signalling that can occur.
We also observed a lack of expression of a subset of genes including WNT5A mRNA in ER+ve breast cancer cell lines. Recent studies suggest that WNT5A plays a critical role in malignant progression although loss of WNT5A protein has been linked with poor prognosis in breast cancer and correlated with the loss of ER expression [42]. This is contradictory to our observations of higher WNT5A mRNA expression in ER-ve cell lines but reported data suggests an inverse relationship between mRNA levels and expression of WNT5A protein [43], [44], [45].
Other genes that were inversely correlated with ER expression included LBH, WISP1, and TCF4. LBH is a newly identified target of the WNT/β-CATENIN signalling pathway expressed at abnormally high levels in poorly differentiated, basal-subtype, ER-ve [46]. WISP1 is overexpressed in breast cancer and was associated with advanced clinical features [47]. TCF4 has been reported to interact with ER signalling [48].
We next examined Wnt pathway gene expression in 1107 primary breast cancer tumours and found differences across subtypes of breast cancer. Many were associated with both basal and ERBB2 sub-types such as WNT5B and others specifically in basal cancers such as LRP5/6 and sFRP1. Lower levels of sFRP1 or LRP6 predict an early recurrence in ER+ve breast cancer whilst higher levels of WNT5B expression predict early recurrence. LRP5 was the only gene that was predictive in ER-ve breast cancer where low levels predict early recurrence. The downstream target LEF1, which was upregulated in ER+ve cell lines, was also increased in Luminal sub-type patient samples and lower levels predicted recurrence in ER+ve patients. These analyses demonstrate the importance of Wnt signalling in breast cancer, and highlight a subgroup of ER+ve patients with a basal-type phenotype based upon their Wnt expression and decreased recurrence free survival.
Our data has shown that the WNT pathway activation is significantly higher in populations enriched for BCSCs, while populations enriched for normal stem-like cells have lower levels of WNT signalling. Furthermore, our data suggests that normal breast cancer stem cells express different WNT ligands (WNT4 and WNT10) than BCSCs when compared to their monolayer populations. Little is known about the role of WNT10A in breast cancer but Wnt4 is essential for normal progesterone induced mammary gland development [49]. A subset of genes such as sFRP1 and β-CATENIN, despite being lower in ER+ compared to ER-ve cell lines, was higher in BCSCs of ER+ve cells. This suggests that BCSCs from ER+ve cell lines share a similar expression profile to cells from an ER−ve cell line supporting the growing hypothesis of a cell hierarchy in which stem cells in ER+ve tumours are ER-ve [50]. Finally we established that the WNT pathway regulates MS formation. WNT3A treatment caused a significant increase in MS in the MCF7 and MDA-MB-231 cancer cell lines while MCF10a immortalised normal breast cells were less sensitive.
To demonstrate biologically the clinical relevance of WNT modulation, we used patient-derived breast samples from both normal and cancerous tissue and compared them to cell lines. Patient-derived samples responded less sensitively to WNT modulation and only ER−ve patient-derived breast cancer cells responded to WNT activation. Treatment with the WNT inhibitor DKK1 successfully decreased MS in both ER+ve and ER-ve patient-derived samples with most significant reduction in ER+ve cells. The data suggest that moderate DKK1 concentrations were sufficient to inhibit MS and therefore stem cell-like activity in both ER+ve and ER−ve tumours whilst not affecting the activity within normal breast tissue. This agrees with the recent in vivo studies of Gurney et al (2012), who show that a therapeutic antibody to the FZD receptor can prevent cancer stem cell activity without toxicity in normal stem cells [51].
Conclusions
In conclusion, we indentified a number of novel WNT signalling components in BCSC signalling. LEF1 has shown the most consistent activation in breast cancer cell lines and increased expression in BCSCs in both ER−ve and ER+ve patient breast cancer samples. Finally, we show that WNT pathway inhibition preferentially reduces stem-like cell activity in patient-derived metastatic breast cancer compared to normal cells. Collectively, these data suggest potential of the WNT-targeted therapeutics in breast cancer.
Supporting Information
Gene expression analysis of Wnt signalling in monolayer (MO) and anoikis resistant (AR) cells of normal breast cell lines (N), ER positive (ER+) and ER negative (ER−) breast cancer cell lines. B) Cluster analysis was performed using gene expression data of cells in MO and AR. Data is displayed in a heatmap represented by either decreased (green) or increased (red) expression compared to the mean mRNA expression. (Red) ↑↑ Indicates significant increased expression (<0.05), (red) ↑ indicates a trend towards increased expression. (Green) Indicates significant increased expression (<0.05), (green) indicated a trend towards increased expression.
(TIF)
Full membranes used to detect Wnt signalling protein expression. Protein was quantified and gel loading volumes determined to achieve equal loading of 50 ug. Multiple gels were loaded concurrently with protein and probed for activated B-catenin (unphosphorylated), Lef1, Axin2, DKK1 and B-actin (housekeeper) in MCF7 monolayer (Day1 mono) and AR cells (Day 1 MS). B-actin was probed using Gel 1. * marks the band that represents the protein of interest.
(TIF)
Funding Statement
The study was supported by Breast Cancer Campaign (R.B. Clarke) (http://www.breastcancercampaign.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810. [DOI] [PubMed] [Google Scholar]
- 2.Kaler P, Godasi BN, Augenlicht L, Klampfer L (2009) The NF-kappaB/AKT-dependent Induction of Wnt Signaling in Colon Cancer Cells by Macrophages and IL-1beta. Cancer Microenviron. [DOI] [PMC free article] [PubMed]
- 3. Klopocki E, Kristiansen G, Wild PJ, Klaman I, Castanos-Velez E, et al. (2004) Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol 25: 641–649. [PubMed] [Google Scholar]
- 4. Rask K, Nilsson A, Brannstrom M, Carlsson P, Hellberg P, et al. (2003) Wnt-signalling pathway in ovarian epithelial tumours: increased expression of beta-catenin and GSK3beta. Br J Cancer 89: 1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Su HY, Lai HC, Lin YW, Liu CY, Chen CK, et al. (2010) Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int J Cancer 127: 555–567. [DOI] [PubMed] [Google Scholar]
- 6. Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, et al. (2010) Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol 176: 2911–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang J, Li Y, Liu Q, Lu W, Bu G (2010) Wnt signaling activation and mammary gland hyperplasia in MMTV-LRP6 transgenic mice: implication for breast cancer tumorigenesis. Oncogene 29: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lopez-Knowles E, Zardawi SJ, McNeil CM, Millar EK, Crea P, et al. (2010) Cytoplasmic localization of beta-catenin is a marker of poor outcome in breast cancer patients. Cancer Epidemiol Biomarkers Prev 19: 301–309. [DOI] [PubMed] [Google Scholar]
- 9. Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, et al. (2000) Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A 97: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, et al. (2008) Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis 29: 991–998. [DOI] [PubMed] [Google Scholar]
- 11. Zhou XL, Qin XR, Zhang XD, Ye LH (2010) Downregulation of Dickkopf-1 is responsible for high proliferation of breast cancer cells via losing control of Wnt/beta-catenin signaling. Acta Pharmacol Sin 31: 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kakarala M, Wicha MS (2008) Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol 26: 2813–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, et al. (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 65: 5506–5511. [DOI] [PubMed] [Google Scholar]
- 15. Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, et al. (2010) Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res 70: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, et al. (2006) Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci U S A 103: 3799–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, et al. (2008) The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res 68: 7711–7717. [DOI] [PubMed] [Google Scholar]
- 18. Chen MS, Woodward WA, Behbod F, Peddibhotla S, Alfaro MP, et al. (2007) Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J Cell Sci 120: 468–477. [DOI] [PubMed] [Google Scholar]
- 19. Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, et al. (2007) WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A 104: 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clarke RB, Spence K, Anderson E, Howell A, Okano H, et al. (2005) A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol 277: 443–456. [DOI] [PubMed] [Google Scholar]
- 21. Shaw FL, Harrison H, Spence K, Ablett MP, Simoes BM, et al. (2012) A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J Mammary Gland Biol Neoplasia 17: 111–117. [DOI] [PubMed] [Google Scholar]
- 22. Singh JK, Farnie G, Bundred NJ, Simoes BM, Shergill A, et al. (2013) Targeting CXCR1/2 significantly reduces breast cancer stem cell activity and increases the efficacy of inhibiting HER2 via HER2-dependent and -independent mechanisms. Clin Cancer Res 19: 643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, et al. (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541. [DOI] [PubMed] [Google Scholar]
- 24. Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, et al. (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 13: 3207–3214. [DOI] [PubMed] [Google Scholar]
- 25. Ivshina AV, George J, Senko O, Mow B, Putti TC, et al. (2006) Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res 66: 10292–10301. [DOI] [PubMed] [Google Scholar]
- 26. Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, et al. (2005) Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res 7: R953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, et al. (2006) Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98: 262–272. [DOI] [PubMed] [Google Scholar]
- 28. Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, et al. (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365: 671–679. [DOI] [PubMed] [Google Scholar]
- 29. Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127. [DOI] [PubMed] [Google Scholar]
- 30. Sims AH, Smethurst GJ, Hey Y, Okoniewski MJ, Pepper SD, et al. (2008) The removal of multiplicative, systematic bias allows integration of breast cancer gene expression datasets - improving meta-analysis and prediction of prognosis. BMC Med Genomics 1: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Calza S, Hall P, Auer G, Bjohle J, Klaar S, et al. (2006) Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res 8: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, et al. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100: 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98: 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, et al. (2007) Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst 99: 616–627. [DOI] [PubMed] [Google Scholar]
- 36. Khalil S, Tan GA, Giri DD, Zhou XK, Howe LR (2012) Activation status of Wnt/ss-catenin signaling in normal and neoplastic breast tissues: relationship to HER2/neu expression in human and mouse. PLoS One 7: e33421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goel S, Chin EN, Fakhraldeen SA, Berry SM, Beebe DJ, et al. (2009) Both LRP5 and LRP6 receptors are required to respond to physiological Wnt ligands in mammary epithelial cells and fibroblasts. J Biol Chem 287: 16454–16466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. MacDonald BT, Semenov MV, Huang H, He X (2011) Dissecting molecular differences between Wnt coreceptors LRP5 and LRP6. PLoS One 6: e23537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collu GM, Brennan K (2007) Cooperation between Wnt and Notch signalling in human breast cancer. Breast Cancer Res 9: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Amerongen R, Bowman AN, Nusse R (2012) Developmental Stage and Time Dictate the Fate of Wnt/beta-Catenin-Responsive Stem Cells in the Mammary Gland. Cell Stem Cell 11: 387–400. [DOI] [PubMed] [Google Scholar]
- 41. Howe LR, Brown AM (2004) Wnt signaling and breast cancer. Cancer Biol Ther 3: 36–41. [DOI] [PubMed] [Google Scholar]
- 42. Jonsson M, Dejmek J, Bendahl PO, Andersson T (2002) Loss of Wnt-5a protein is associated with early relapse in invasive ductal breast carcinomas. Cancer Res 62: 409–416. [PubMed] [Google Scholar]
- 43. Dejmek J, Dejmek A, Safholm A, Sjolander A, Andersson T (2005) Wnt-5a protein expression in primary dukes B colon cancers identifies a subgroup of patients with good prognosis. Cancer Res 65: 9142–9146. [DOI] [PubMed] [Google Scholar]
- 44. Dejmek J, Leandersson K, Manjer J, Bjartell A, Emdin SO, et al. (2005) Expression and signaling activity of Wnt-5a/discoidin domain receptor-1 and Syk plays distinct but decisive roles in breast cancer patient survival. Clin Cancer Res 11: 520–528. [PubMed] [Google Scholar]
- 45. Leandersson K, Riesbeck K, Andersson T (2006) Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res 34: 3988–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ (2010) The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol 30: 4267–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP (2001) Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res 61: 8917–8923. [PubMed] [Google Scholar]
- 48. El-Tanani M, Fernig DG, Barraclough R, Green C, Rudland P (2001) Differential modulation of transcriptional activity of estrogen receptors by direct protein-protein interactions with the T cell factor family of transcription factors. J Biol Chem 276: 41675–41682. [DOI] [PubMed] [Google Scholar]
- 49. Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, et al. (2000) Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev 14: 650–654. [PMC free article] [PubMed] [Google Scholar]
- 50. Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, et al. (2007) Molecular definition of breast tumor heterogeneity. Cancer Cell 11: 259–273. [DOI] [PubMed] [Google Scholar]
- 51. Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, et al. (2012) Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A 109: 11717–11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression analysis of Wnt signalling in monolayer (MO) and anoikis resistant (AR) cells of normal breast cell lines (N), ER positive (ER+) and ER negative (ER−) breast cancer cell lines. B) Cluster analysis was performed using gene expression data of cells in MO and AR. Data is displayed in a heatmap represented by either decreased (green) or increased (red) expression compared to the mean mRNA expression. (Red) ↑↑ Indicates significant increased expression (<0.05), (red) ↑ indicates a trend towards increased expression. (Green) Indicates significant increased expression (<0.05), (green) indicated a trend towards increased expression.
(TIF)
Full membranes used to detect Wnt signalling protein expression. Protein was quantified and gel loading volumes determined to achieve equal loading of 50 ug. Multiple gels were loaded concurrently with protein and probed for activated B-catenin (unphosphorylated), Lef1, Axin2, DKK1 and B-actin (housekeeper) in MCF7 monolayer (Day1 mono) and AR cells (Day 1 MS). B-actin was probed using Gel 1. * marks the band that represents the protein of interest.
(TIF)