Abstract
The re-colonization of aquatic habitats by angiosperms has presented a difficult challenge to plants whose long evolutionary history primarily reflects adaptations to terrestrial conditions. Many aquatics must complete vital stages of their life cycle on the water surface by means of floating or emergent leaves and flowers. Only a few species, mainly within the order Alismatales, are able to complete all aspects of their life cycle including pollination, entirely underwater. Water-pollinated Alismatales include seagrasses and water nymphs (Najas), the latter being the only freshwater genus in the family Hydrocharitaceae with subsurface water-pollination. We have determined the complete nucleotide sequence of the plastid genome of Najas flexilis. The plastid genome of N. flexilis is a circular AT-rich DNA molecule of 156 kb, which displays a quadripartite structure with two inverted repeats (IR) separating the large single copy (LSC) from the small single copy (SSC) regions. In N. flexilis, as in other Alismatales, the rps19 and trnH genes are localized in the LSC region instead of within the IR regions as in other monocots. However, the N. flexilis plastid genome presents some anomalous modifications. The size of the SSC region is only one third of that reported for closely related species. The number of genes in the plastid is considerably less. Both features are due to loss of the eleven ndh genes in the Najas flexilis plastid. In angiosperms, the absence of ndh genes has been related mainly to the loss of photosynthetic function in parasitic plants. The ndh genes encode the NAD(P)H dehydrogenase complex, believed essential in terrestrial environments, where it increases photosynthetic efficiency in variable light intensities. The modified structure of the N. flexilis plastid genome suggests that adaptation to submersed environments, where light is scarce, has involved the loss of the NDH complex in at least some photosynthetic angiosperms.
Introduction
Chloroplasts evolved from prokaryotic photosynthetic endosymbionts [1] as cell organelles that maintain their own genetic material in a double stranded DNA molecule ranging in size from 35 to 217 kb [2]. Compared to their cyanobacterium-like ancestors, plastid genomes have experienced a dramatic reduction in gene number from the +3 000 once present in free-living Cyanobacteria to only 120–250 genes in photosynthetic eukaryotes. The plastid genes that have been retained encode products necessary for photosynthetic and housekeeping functions. During photosynthetic eukaryote evolution, cyanobacterial genes were transferred from the endosymbiont to the host nucleus or were lost entirely, in instances where the function of those genes was no longer essential [3]. The process of gene transfer has not stopped [4] but continues as a constant flood of plastid and mitochondrial genome fragments to the nucleus, where organelle DNA can be integrated as functional genes. However, over time, such genes usually are pseudogenized and lost, with only a small proportion of the transferred DNA integrated into functional areas and being conserved [4]. Red algal plastids retain the highest number of genes of any other group of photosynthetic eukaryotes (232–252) [3]. In contrast, the chloroplast of land plants (Embryophyta), and of their ancestral green algae (Chlorophyta), retains only 120 genes. It usually consists of two copies of an inverted repeat (IRa, IRb) that separate a large single copy region (LSC) from a small single copy region (SSC). While missing in some algae (Glaucophyta, Rhodophyta), green plant plastids are rich in repeated regions and possess editing mechanisms [3].
Key photosynthetic elements are encoded in plastid genomes, such as photosystem I and II genes, RuBisCO and thylakoid NAD(P)H dehydrogenase. Independent of any former function of ndh genes in Cyanobacteria, ndh genes are essential for photosynthesis in land plants [5]. Lost in other algal divisions, the ndh genes probably were essential in the adaptation of green algae to the fluctuating conditions of shoreline environments [5] The eleven plastid ndh genes together with four nuclear genes (nhdL, ndhM, ndhN, and ndhO) encode the thylakoid NAD(P)H dehydrogenase complex which functions mainly in the electron transfer from NADH to plastoquinone, which protects the cell against photooxidative-related stress and maintains optimal rates of cyclic photophosphorylation [5]. In land plants, small changes in any of the ndh genes significantly decrease net photosynthesis [6]. As a consequence of such strong selective pressure, the ndh genes are highly conserved across all vascular plant divisions [7].
In angiosperms, ndh loss in plastomes is associated primarily with heterotrophic (i.e., parasitic) plants [8]. The plastid genome of non-photosynthetic organisms undergoes severe rearrangements and deletions that lead to losses of both photosynthetic and chlororespiratory genes, which no longer are needed to maintain metabolic functions. Convergent (homoplasious) losses of ndh genes are evident among unrelated parasitic plants; such as Epifagus (Lamiales) [9], Cuscuta (Solanales) [10], [11] or the mycotrophic orchid Neottia [12]. Pseudogenization and loss of ndh genes in the parasitic bryophyte Aneura mirabilis [13] further substantiates the relationship between parasitism and ndh loss. Due to the mutual interaction between symbiotic fungi and orchids [14], it is understandable that the lack of functional ndh genes is widespread within the Orchidaceae, even green-leaved orchids [15]–[17]. Recent data suggest pseudogenization of ndhB and even complete loss of ndhF for some taxa in the order Alismatales (seagrasses and water nymphs) [18]. Homoplasic loss of ndh genes in aquatic angioperms might be related to potential adaptations to the constraints of the underwater environment [19]. The ndh genes are also absent in some photosynthetic gymnosperms [20]. The shared loss of ndh genes in Gnetales and Pinaceae is regarded as a rare synapomorphic event, which provides support to the gnepine hypothesis [8]. While the loss of ndh genes in Gnetales and Pines occurred early in the evolution of land plants, ndh loss in some Erodium (Geraniaceae) species provides an example of particularly recent loss [21]. The possible explanation for ndh genes loss in Gnetales, pines and some Erodium species is difficult to elucidate.
Recolonization of aquatic environments by land plants has occurred up to 100 independent times, comprising up to 2% of the approximate 350,000 angiosperm species [22]. These recolonization events presumably would have required many physiological, metabolic and reproductive adaptations essential to accommodate the broad and novel conditions of aquatic habitats, which range from warm and sunny shallow waters to cool and dark deep waters. The monocot order Alismatales includes ∼4500 extant species in 13 families, which includes several that are predominantly aquatic (see [18], [22] for phylogeny). The order encompass various wetland and aquatic life-forms ranging from emergent, floating-leaved, free-floating and submersed, only the latter life-form fully adapted to the aquatic environment. Additionally, a diverse array of reproductive strategies representing different degrees of adaptation to the aquatic environment is evident and includes anemophily, entomophily, self-pollination, unusual water-facilitated types involving detached, floating flowers, and both surface and subsurface water-pollination [23]. Complete adaptation to submersed life, including water-pollination (hydrophily), is quite rare, being present in only 130 angiosperm species [22]. The greatest concentration of hydrophilous species occurs in the Alismatales, most notably in seagrasses and water nymphs. The genus Najas L. (water nymph) includes up to 40 species, all of them completely submersed and hydrophilous. It is the only freshwater genus in the family Hydrocharitaceae with subsurface water-pollination. Najas exhibits a number of morphological adaptations associated with the successful colonization of deep water habitats, such as a lack of stomata and reduced leaves formed by only two layers of epidermal cells [24]. However physiological adaptations to maximize underwater photosynthesis have remained obscure.
Arguably, the low light levels found in submersed aquatic environments should increase selective pressures on some features, while reducing the adaptive potential of others. Recent work has indicated that such selection has induced changes even in genes extremely conserved across plant evolution. For example, the 25-nucleotides of the intercistronic spacer region in the photosynthesis-related genes psaA-psaB is highly conserved through plant evolution, from green algae to land plants. Yet, parasitic plants, where photosynthetic genes are lost or pseudogenized, and aquatics, which are under different selective pressures than terrestrial plants, accumulate most of the variants detected in that plastid region [19]. In particular, the genus Najas possesses a unique spacer configuration that likely regulates transcription of these genes in response to temperature [19].
To provide additional information on the plastid genome structure of fully submersed water-pollinated aquatic angiosperms, we have sequenced the complete plastid genome of Najas flexilis (Willd.) Rostk. & Schmidt (nodding waternymph). We have analyzed the structure and gene content of the Najas flexilis plastid genome and have compared the Najas flexilis plastome to those available for other monocots and angiosperm species. We have designed specific primers to evaluate problematic areas such as the reduced SSC region, and investigated their cross-transferability in other Najas species. Modifications in the chloroplast of Najas flexilis reported here may provide insight on the importance of the plastid NAD(P)H dehydrogenase during land colonization by photosynthetic eukaryotes.
Materials and Methods
1. DNA Extraction and Sequencing
Plant material was collected from two sites that potentially reflect genetic diversity in Najas flexilis. Najas flexilis ‘canadensis’ was collected in Scotland, Lower Glenastle Loch (Islay, Scotland; 18 August 2010) permit issued by the Scottish Natural Heritage (License number 11012) and Najas flexilis ‘flexilis’ was collected in the USA (Connecticut), from the public access site at West Side Pond with permission from the CT DEEP (20 September 2010). Fresh material of each sample was stored in saturated NaCl-CTAB solution [25]. Prior to DNA extraction, leaves of a single plant were washed in double-distilled water and carefully cleaned under a dissecting microscope to remove epiphytes from the plant tissue surface.
Total DNA was extracted using standard procedures [26], re-suspended in 30 µl of 1X TE buffer and checked in a 2% agarose gel. DNA concentration was determined by measuring the optical density at 260 nm and 280 nm in a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Asheville, NC, USA). Molecular confirmation of the identity of each accession was performed by sequencing the ITS, trnK- matK, and rbcL regions as described previously [23].
Genomic DNA (500 ng) was sheared by nebulization, subjected to 454 library preparation and shotgun sequencing using the Genome Sequencer (GS) FLX Titanium pyrosequencing platform (454 Life Science Corporation, Branford, CT, USA) at the in-house facility (Center for Applied Genetics and Technology) at the University of Connecticut. Two independent runs (1/16 picotiter plate each) were performed with each library to a total of 1/4 picotiter plate for the species. The sequencing runs provided a total of 198 152 raw reads of which 80 858 were assigned successfully to the Scottish material and 116 118 to the American material. Reads were trimmed in CLC Genomics Workbench 5.1 (CLC bio, Aarhus, Denmark) under the following criteria: quality score 0.05, mismatches 2. Reads under 70 nt or over 800 nt were discarded. Average length for each of the runs was 388.5 and 301.9. As the same libraries were used in the second run, this length decay was expected. Over 95% of the reads presented a PHRED score over 25.
2. Genome Assembly and Annotation
Out of 191 597 raw reads that were above the quality threshold, 84 739 reads were assembled in 10 979 contigs (aprox 5 mill nt) using CLC Genomic Workbench. Contigs were filtered against the Lemna minor complete plastid sequence (NC_010109) [27]. Additional identification of chloroplast contigs was performed using the blastn algorithm in Blast2Go [28]. A total of 51 contigs with an average length of 2 306 nt corresponded to the Najas flexilis plastid genome. The longest contig extended to 24.5 kb and included most of the inverted repeat region. Average sequencing depth was 12-fold, reaching maximum numbers in the IR region (44-fold). Total coverage of the combined contigs was 117 603 nucleotides leaving 6% of the plastid genome unresolved when only one IR region was included. Where possible, gaps were closed between de novo contigs using consensus sequences of newly assembled reads in Geneious version 5.6 (Biomatters; available from http://www.geneious.com/). To maximize the number of reads assembled the concatenated Najas flexilis contigs were used as new references for filtering reads prior to de novo re-assemble of contigs.
Sixteen areas of the scaffold were checked by PCR and ABI sequencing (Table S1). These areas included gaps or low coverage areas, and structurally conflicting features, such as junctions and areas in the SSC. Primers were designed using Primer 3 [29] as implemented in Geneious. Duplicate PCR mixes were prepared for each primer set and included 1 U Titanium Taq (Clontech), 1x reaction buffer, 0.2 um of each primer, and 5–15 ng of DNA. One of the reaction mixes included the PCR additive betaine (Affymetrix) to a final concentration of 1 uM. Optimal annealing temperatures for each primer set were tested by gradient PCR (annealing temperature ranging from 52 C to 58 C) for each primer set and PCR mix. At least two PCR reactions were sequenced for each primer combination.
The fully sequenced Najas flexilis genome was annotated in Geneious by using the ORF finder plug-in along with comparison to other annotated plastid genomes. Annotations were checked using DOGMA (Dual Organellar GenoMe Annotator) [30] and additional ORF searches were performed with Glimmer 3 [31] and ORF Finder (T. & R. Tatusov) both available through the NCBI website.
3. Examination of Genome Structure
Mauve [32] was used to perform multiple genome alignments and explore the large-scale structure of the plastid genome of Najas flexilis. Available plastid genomes of species belonging to the Alismatales were aligned with Progressive Mauve algorithm using Acorus as an outgroup. Similar alignments were performed using broader phylogenetic groups (i.e., available monocots or angiosperms).
An additional whole-genome analysis was conducted using the CGView Comparison Tool [33]. The plastome of Najas flexilis was compared to those of other available angiosperms, using representatives of the major groups spanning the entire angiosperm phylogeny as outlined in [34]. Two different sets of blast maps were generated based on DNA-DNA and CDS analysis. In these maps, the sequence identity was represented by a different color scheme related to the BLAST hits (blastn and blastp). Additional features such as identification of GC rich areas in the reference genome or GC Skew index were also included. As a means of visual comparison, an analysis using the same parameters was also run for the closely related species Elodea canadensis (JQ310743) [35].
Results and Discussion
1. Najas flexilis Plastid Genome
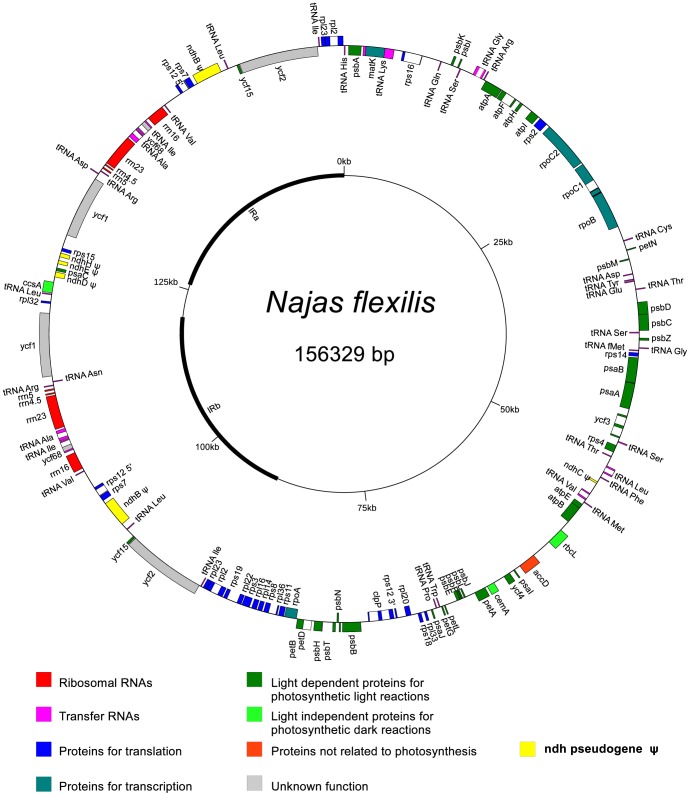
The determined sequence for the Najas flexilis plastid genome is 156 329 bp (Fig. 1, GenBank accession JX978472). It displays a circular quadripartite structure, with the large single copy region (LSC; 88 668 bp, 56.72% of the total genome) separated from the small single copy region (SSC; 5 266 bp, 3.36%) by two inverted repeat regions (IRb and IRa; 31 198 bp, 19.95% each). Average GC content is 38.2%, similar to those reported for such closely related monocots, as Smilax china (37.25%) [36], Elodea canadensis (37%) [35], and members of the Araceae family (35.2% to 35.9%) [27], [37]. The IR regions encode, among other genes, ribosomal gene cluster and therefore they display a higher GC content than either the small or large single copy areas (42.9% vs. 30.4% and 35.4%).
Figure 1. The Najas flexilis chloroplast genome.
Inner circle, schematic representation of the quadripartite structure of the genome, IR regions outlined in black. Outer circle, gene organization in the genome. Genes shown outside the circle are transcribed counterclockwise; genes in the inside are transcribed clockwise. Genes are color-coded by function.
The plastome of Najas flexilis codes for 103 unique genes, sixteen of which are duplicated in the inverted repeat regions (Table 1 and Fig. 1). Seventeen genes present discontinuous reading frames; 14 have a single intron and two genes have two introns. The rps12 gene is transpliced with exon 1 coded in the LSC region and exons 2 and 3 in the IRs. In the closely related species Elodea canadensis, the rpl16 is interrupted by a 1 kb intron [35] and the two exons expand for 10 and 400 nt respectively. In Najas flexilis, the rpl16 intron is not present. Maximum likelihood trees of the coding region of rpl16 (nt/aa) successfully placed Najas flexilis in the same clade as Elodea and separate from the Spirodela-Lemna-Wolffia-Wolffiella clade (both 99%–100% bootstrap support, data not shown), showing that neither the sequence nor protein encoded by the rps16 gene had been altered excessively by the intron loss.
Table 1. Genes encoded in the Najas flexilis chloroplast.
| Function | Gene Class | Genes | Number |
| Proteins not related to photosynthesis | Acetyl-CoA carboxylasecarboxyltransferase beta subunit | accD | 1 |
| Light independent proteins | Membrane protein | cemA | 1 |
| Cytocrome c biogenesis | ccsA | 1 | |
| Rubisco | rbcL | 1 | |
| Light dependent proteins | ATP synthase | atpA, atpF *, atpH, atpI | 4 |
| ndh Genes | |||
| ndhBΨx2, ndhCΨ, ndhDΨ, ndhEΨ, ndhHΨ | |||
| Photosystem I assembly factor | yfc3 **, ycf4 | 2 | |
| Cytocrome | petA, petB *, petD *, petG, petN, petL | 6 | |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 | |
| Photosystem II | psbA, psbB, psbC, psbD, psaE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 | |
| Genetic apparatus | Maturase | matK | 1 |
| Proteins for transcription | RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | 4 |
| Proteins for translation | ATP-dependant protease | clpP ** | 1 |
| Translation initiation factor A | infA | 1 | |
| Ribosomal protein (large) | rpl2 * x2, rpl14, rpl16, rpl20, rpl22, rpl23 * x2, rpl32,rpl33, rpl36 | 10 | |
| Ribosomal protein (small) | rps2, rps3, rps4, rps7x2, rps8, rps11, rps12 **x2, rps14, rps15, rps16 *, rps18, rps19 | 12 | |
| Structural RNAs | Ribosomal RNAs | rRNA 4.5Sx2, rRNA 5Sx2, rRNA 16Sx2, rRNA 23Sx2 | 4 |
| Transfer RNAs | tRNA-His(GUG), tRNA-Lys(UUU)*, tRNA-Gln(UUG), tRNA-Ser(GCU), tRNA-Gly(UCC)*, tRNA-Arg(UCU), tRNA-Cys(GCA), tRNA-Asp(GUC), tRNA-Tyr(GUA), tRNA-Glu(UUC), tRNA-Thr(GGU), tRNA-Ser(UGA), tRNA-Gly(UCC), tRNA-fMet(CAU), tRNA-Ser(GGA), tRNA-Thr(UGU), tRNA-Leu(UAA)*, tRNA-Phe(GAA), tRNA-Val(UAC)*, tRNA-Met(CAU), tRNA-Trp(CCA), tRNA-Pro(UGG), tRNA-Ile(CAU)x2, tRNA-Leu(CAA)x2, tRNA-Val(GAC)*x2, tRNA-Ile(GAU)*x2, tRNA-Ala(UGC)*x2, tRNA-Arg(ACG)x2, tRNA-Asn(GUU)x2, tRNA-Leu(AUG) | 30 | |
| Conserved reading frames | ycf1x2, ycf2x2, yfc15x2, ycf68x2 | 4 | |
| 103 |
gen with one intron;
gene with two introns; crossed missing;
pseudogene.
The 103 genes are grouped by function.
Out of the 103 genes encoded in the Najas flexilis plastid, four correspond to ribosomal RNAs, 30 to transfer RNA, and 69 are protein-coding genes. Usually, nearly 80 protein-coding genes are present in the angiosperm plastid genome [38]. The lower number of functional protein-coding genes in Najas flexilis is exclusively caused by the lack of functionality of the ndh genes (Fig. 1). Small fragments of truncated ndh genes were detected in the LSC region (ndhC) and in the SSC (ndhD, E and H) while only ndhB, in the repeat regions, maintained the expected gene size. However, the non-functional role of this gene was clearly indicated by the presence of stop codons and indels that cause frame shifts.
2. Najas flexilis Plastid Genome Maintains a Quadripartite Structure
Comparison of overall genomic structure of the Najas flexilis plastid using Mauve alignment in a broad phylogenetic comparison (from Amborella to Asterales) showed that it is collinear with the plastid genome reported for the angiosperm Amborella, sister to all other extant angiosperms [39] or other monocots, such as Acorus [40], Lemna [27] or Elodea [35] (data not shown). The plastome of N. flexilis has not suffered structural rearrangements affecting gene order and overall homology aside from loss of the ndh genes. Additional analyses using different phylogenetic groups (i.e., Alismatales and other monocots) produced the same result (Fig. S1).
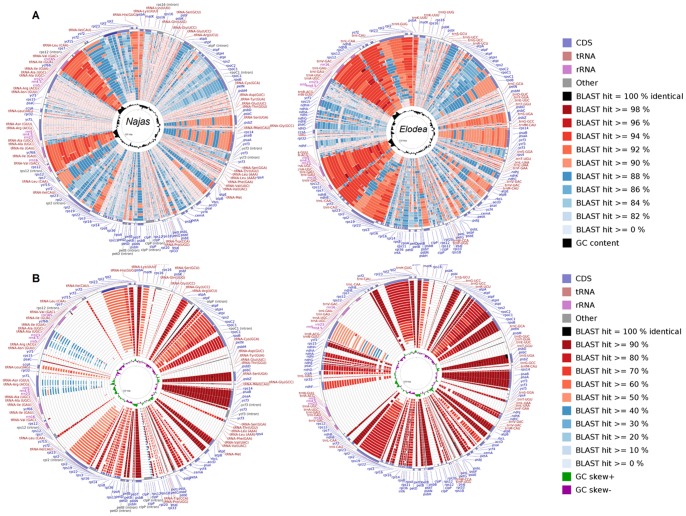
Comparison of the plastid genomes of Najas flexilis and Elodea canadensis reflected other shared structural features aside from gene collinearity. Both species shared equivalent distributional patterns of GC islands or areas where G and C are distributed unevenly between the DNA strains (Fig. 2). However, in a broader phylogenetic context, Najas flexilis is highly divergent. Blast analysis showed lower overall sequence identity to other angiosperms than did the closely related Elodea canandensis, when both were compared within the same broad phylogenetic representation (Fig. 2). Although Elodea canadensis presents 90% sequence similarity across the entire region, in N. flexilis this level of high similarity is restricted to the IR regions, ribosomal gene clusters, tRNAs and the junction area, where rpl2 is encoded. In the Najas plastid, protein-coding sequences that show high similarity to other angiosperms are related to photosynthesis. Genes encoding Photosystem I and II, ATP synthase or RuBisCO are highly conserved both at the nucleotide and protein level. Some other genes are highly dissimilar, such as clpP, ycf1 and ycf2.
Figure 2. BLAST comparison of the Najas flexilis and Elodea canadensis chloroplast genomes.
Najas flexilis (left) and Elodea canadensis (right) are compared to other monocots and selected species representing overall angiosperm diversity. The analyses were generated using blastn (A, DNA comparison) and blastp (B, CDS comparison) in CGViewer. In the upper part of the figure (A), schematic representation of the GC content is provided for each species. In the lower one (B), the values of GC Skew index are represented in green and purple. In each genome comparison, genes are color-coded by function. Blast hit values are color-coded by percentage of similarity. In the CDS analysis the bar height is proportional to the similarity value. For each figure, the specific order of the genome rings is determined by the similarity to the reference genome. This sorting emphasizes sequence divergence trends for CDS or sequences in the reference genome. Similarity was determined as defined in CGViewer, by a heuristic considering the total number of genome bases contributing to the hits and their scores [29]. Note: data for some included taxa were downloaded from Genbank: (in alphabetical order) Acorus, Amborella, Calycanthus, Ceratophyllum, Chloranthus, Colocasia, Dioscorea, Drimys, Elaeis, Elodea, Fragaria, Illicium, Jacobea, Lemna, Magnolia, Najas, Nymphaea, Oryza, Phalaenopsis, Piper, Ranunculus, Smilax, Spirodela, Triticum, Wolffia, Wolfiella, and Zea.
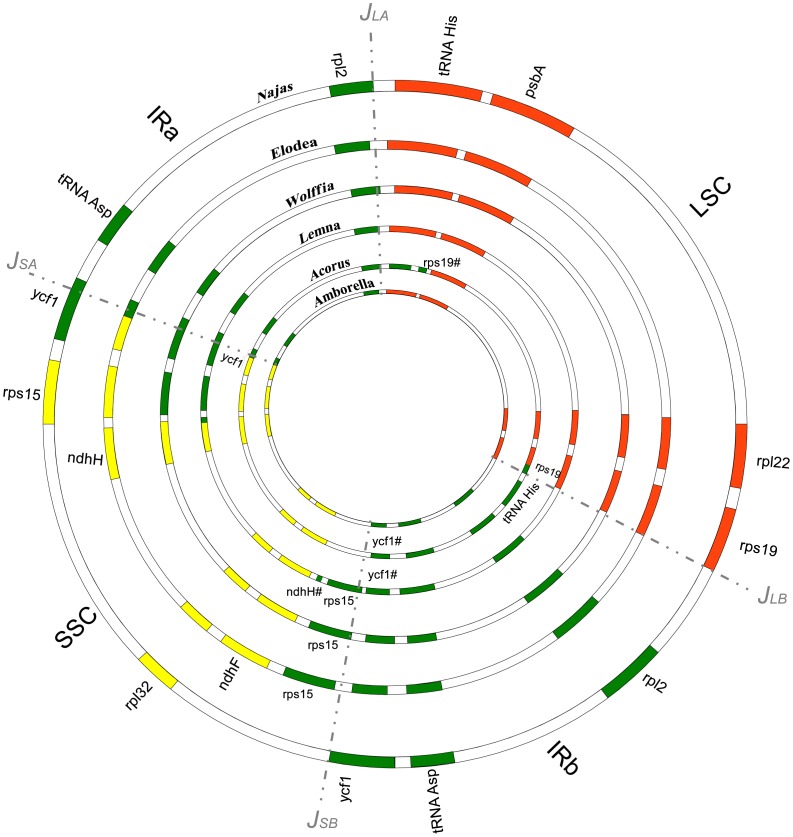
The quadripartite structure of the plastid genome is characterized by the existence of highly dynamic junctions between the IR and single copy regions (JLA, JSA, JSB, JLB). Events that cause the expansion and contraction of the IR regions account for most of the size variation among genomes of different taxa [41]. Size changes are caused by translation of the junctions and inclusion or exclusion of genes in the repeat regions. Plastid junctions have been suggested as evolutionary markers for elucidating relationships among taxa [41]. The plastid genome of Najas flexilis shares the general organization described for the Alismatales [27], [35], [37]. Most monocots, including Acorus, are characterized by the presence of a trnH gene in each repeat region [40]. The JLA junction is situated downstream of the start codon of rps19, producing either a truncated or complete rps19 gene in the IRs. Analysis of the interspacer region of the trnH-rps19 cluster has suggested that this feature evolved from a single event in monocots [41]. The order Alismatales differs from other monocots at the JLA and JLB areas, as the genes rps19 and trnH are localized in the LSC region. This configuration is consistent with that reported for Amborella [39] or Nymphea [42], well supported sister taxa to all other angiosperms [34]. However, analysis of the intergenic spacers confirmed that these sequences in Alismatales are more similar to other monocots than to any other group of angiosperms and might have resulted from a separate contraction [40]. The JLA and JLB junctions in the Najas flexilis plastid present no differences to those previously reported for Alismatales (Fig. 3) [27], [35], [37].
Figure 3. Comparison of expansion and contraction patterns that lead to junction changes between the IR and single copy regions (JLA, JSA, JSB, JLB).
The chloroplast genome of Najas flexilis is compared to those available for Alismatales, Acorus, sister to all other monocots, and Amborella, sister to all other angiosperms. Genes involved in IR expansions are color-coded. Green, genes in the IR regions; orange, LSC; and yellow, SSC. Dashed lines indicate the expected junction area. #, truncated gene.
On the other hand, the position of the SSC and the IR junctions (JSA and JSB) differs within different families in the Alismatales. The family Araceae, including Lemna or Wolffia, is characterized by the inclusion of rps15 in the IRs [27], [37] whereas Najas and Elodea (Hydrocharitaceae) maintain rps15 in the SSC region, a shared trait with Acorus and Amborella, which are respectively sister to all other monocots, and all other angiosperms. The JSA in Elodea canadensis is within ycf1, producing a truncated gene in the IR. In Najas flexilis, JSA is sited downstream of rps15, resulting in the presence of two full copies of ycf1 in its plastid genome (Fig. 3).
3. ndh Genes are Lost in the Najas flexilis Plastid
Plastid genes can be ascribed to light-dependent or light-independent photosynthetic pathways, to non photosynthetic related functions and to the genetic apparatus [38]. The number of genes encoded in plastid genomes of photosynthetic plants typically is 100–120, nearly 80 of these genes are translated into proteins. Monocots present little over 110 genes (Acorus 112 [40]; Lemna 112 [27]; Phoenix 112 [43]; Elodea 113 [35]; Hordeum, Sorghum and Agrostis 113 [44]; Smilax 114 [36]). Usually, differences in the number of genes reported are related to the presence of the ORFs ycf15 and ycf68. The function of these genes, both present in Najas flexilis (Table 1), still remains unclear, although they have been suggested to act as transcriptional regulators [38].
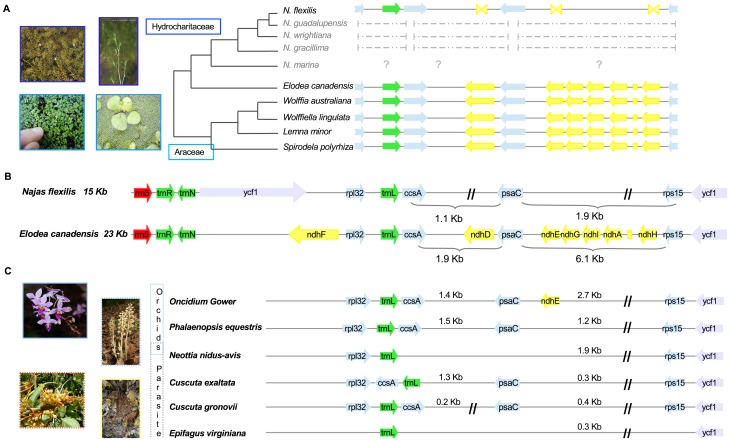
The Najas flexilis plastome encodes the complete set of structural ribosomal RNA genes and 30 transfer RNAs. All protein-coding genes related to transcription, translation and intron maturation are intact. While the groups of genes involved in synthesis of Photosystem I and II, ATP synthase, and the cytochrome b6/f complex are intact, none of the genes encoding the NAD(P)H dehydrogenase complex is functional (Table 1). Out of the 11 ndh genes in the plastome, six are missing (ndhJ, ndhK, ndhF, ndhG, ndhI, ndhA) and only pseudogenized or truncated sequences of the other five can be found (ndhCΨ, ndhBΨx2, ndhDΨ, ndhEΨ, ndhHΨ) in Najas flexilis. The loss or pseudogenization of the ndh genes is responsible for the small size of the SSC (Fig. 4B) region (5 266), which is less than one third of that reported for Elodea canadensis (17 810 bp) [35]. In this study, the fate of the nuclear-encoded ndh genes ndhL, ndhM, ndhN, and ndhO is not explored.
Figure 4. Loss of ndh genes in the small single copy region (SSC).
The ndh genes are represented by yellow arrows or truncated yellow arrows; blue, protein-coding genes; green, tRNA; red, rRNA; grey, genes of unknown function. Parallel lines indicate areas reduced in size. A. Comparison of genes detected in the SSC regions in different Alismatales. The tree is a consensus of those published elsewhere and is included only to illustrate phylogenetic relationships among the species. All members of the family Araceae (light blue) possess a complete set of ndh genes. In the family Hydrocharitaceae (dark blue), the ndh genes are retained in Elodea canadensis but missing in Najas. The dashed lines represent sequences retrieved by targeted-PCR in other Najas species. B. Schema of the IR/SSC region in Najas flexilis and Elodea canadensis. C. SSC regions in orchids (blue) and parasitic plants (dashed blue, Orchidaceae; dashed orange, Solanales; dashed yellow, Lamiales). The ‘loss as a suite’ pattern in these groups is equivalent to the example reported here for Najas flexilis.
To confirm the lack of ndh genes in the plastid genome of Najas flexilis, specific primers were designed surrounding predicted areas for gene encoding (Table 2). PCR and sequencing of the amplified fragments in Najas flexilis confirmed in each case partial or total loss of ndh genes. Amplified fragment size corresponding to the LSC region, where ndhJ-C genes are encoded, was one third shorter than expected by comparison with the same region in the Elodea plastid genome. Only 159 nts were identified as part of a truncated ndhC (Elodea, Lemna 363 nt), while total loss of the ndhJ and ndhI genes was confirmed. Similar results were confirmed for the SSC region, most notable being a psaC-rps15 reduction (Fig. 4A). General gene organization in this area is highly conserved across the green plant phylogeny. It usually expands over 6 kb encoding the ndhH-D operon, including six ndh genes and psaC. In Najas flexilis, psaC is just 1.9 kb away from rps15, while in Elodea this distance is 6.1 kb (Fig. 4B). This size reduction is caused by alteration of the ndh genes in the ndhH-D operon where only truncated ndhH, ndhE and ndhD genes were detected. Not even small fragments of the three other ndh genes (ndhA, ndhI, and ndhG) were detected by DOGMA.
Table 2. Primer sets to amplify the regions containing ndh genes.
| Chloroplast region | Primer name | Gene | Sequence | TM | Position | Product size(Elodea) | Product size (Najas) | Missing genes (Najas) | |
| LSC-Leu | tRNA-Leu-1F | tRNA-Leu(UAA) | Forward | TACACGGCCAAGGAATCTCG | 59.8 | 51740 | >2400 | 831 | ndhJ, ndhK, ndhCΨ |
| tRNA-Leu-1C-F | tRNA-Phe(GAA) | Forward | AGGGTCGAGTCAGGATAGCT | 59.4 | 51960 | >2100 | 611 | ||
| tRNA-Leu-1R | ndhC | Reverse | GGGTATTTGTCACCGGGGTT | 60 | 52570 | ||||
| IR b_SSC | NF_IRb_SSC_3F | past ycf1 | Forward | CTTCGGAAGAAATCCATTGGGCAAAAA | 57.02 | 119590 | ? | 324 | ndhF |
| NF_IRb_SSC_3R | rpl32 | Reverse | TGGAACTGCCATTTCAAAGCGACT | 57.58 | 119914 | ||||
| SSC_rpl32 | NF_SSC_rpl32_1F | rpl32 | Forward | TTGGCTGCGGTTAAAGCTTTTTCTTT | 57.02 | 119977 | >1350 | 1269 | none |
| NF_SSC_rpl32_1R | ccsA | Reverse | CCACTGCGACTGTAGAGCAGGC | 59.55 | 121246 | ||||
| SSC_ccsA | NF_SSC_ccsA_1F | ccsA | Forward | GCCTGCTCTACAGTCGCAGTGG | 59.55 | 121225 | >2400 | 1688 | ndhDΨ |
| NF_SSC_ccsA_1R | psaC | Reverse | TGCTTCCGCGCCAAGAACAGA | 59.32 | 122913 | ||||
| SSC_psaC | NF_SSC_psaC_1F | psaC | Forward | GTCCTCTGTTCTTGGCGCGGA | 59.13 | 122889 | >6400 | 1763 | ndhEΨ, ndhG, ndhI, ndhA, ndhHΨ |
| NF_SSC_psaC_1R | rps15 | Reverse | CGAAGGATTTTAGGAAAACGCCAACG | 57.58 | 124652 |
pseudogene.
Primers are transferable among different Najas species.
Loss of the ndh genes is the main cause of the extremely small size of the SSC region across the Najas genus (Fig. 4A). Successful amplification was achieved in other Najas species using the same primer sets described for N. flexilis (Table 2). Unfortunately, due to high divergence, amplification of the SSC region was not possible in N. marina, the only species in the subgenus Najas (see [23] for a revised phylogeny of the genus Najas). PCR amplification and sequencing of the SSC region in other Najas species, such as N. guadalupensis, N. wrightiana (section Americanae) and N. gracillima (section Euvaginatae), indicate that loss of ndh genes and a reduced SSC are traits shared across the genus Najas. The size of amplified fragments in other Najas species matches that reported above for the ndh-defective Najas flexilis. Alignment of the sequences and comparison with the Najas flexilis plastid genome allowed the unequivocal identification of the fragments as intergenic regions in the SSC (Fig. 4A). In each case, the reduced size of the amplified fragments and posterior sequence analysis strongly suggest loss of the plastome ndh genes across the Najas genus and the maintenance of non-ndh genes in the region. Loss of ndh genes in all Najas species probably follows the ‘loss as a suite’ pattern reported for Pinaceae, Gnetales, orchids and parasitic species [8]–[17], [20] (Fig. 4C).
Loss of ndh genes in the genus Erodium (Gerananiaceae) has been defined as the most recent (5–15.45 MYA) and phylogenetically restricted amongst photosynthetic seed plants [21]. This loss was reported only in species belonging to the ‘long-branch clade’ (LBC) while complete plastid genome sequences of other Erodium species confirmed the presence of functional ndh genes in other clades. In the Alismatales, loss of the ndh genes is a homoplastic feature. PCR-targeted analysis detected alterations in ndhF and ndhB in seagrasses of the tepaloid clade (Posidonia and Amphibiolis) and in members of the petaloid clade (Najas flexilis and Thalassia) suggesting that loss of ndh genes has occurred several times in the Alismatales [18]. Sequencing of the Najas flexilis plastome confirms that the lack of amplification of ndhF in Najas was caused by the complete loss of the gene. Maximum Likelihood tree (data not shown) of the ndhB gene successfully resolve the Hydrocharitaceae family with an equivalent topology to that obtained with 17 plastid genes [18]. Najas and Thalassia pseudogenized ndhB genes share a common origin with those functional ndhB genes in the Elodea canadensis (and possibly Hydrocharis and Stradiotes) plastome. The pseudogenization of ndhB in the petaloid clade is independent of the event in the Posidonia-Amphibiolis clade (tepaloid).
4. Implications of the Loss of the NDH Complex in Aquatic Angiosperms
The plastome ndh genes, equivalent to those present in the mitochondria, encode the NAD(P)H dehydrogenase complex located in the stromal thylakoid. The main function of this complex is electron transfer from NADH to plastoquinone, and plays an essential role in land plant photosynthesis by protecting against photooxidative-related stress and maintaining optimal rates of cyclic photophosphorylation [5]. Strong selection acting on the ndh genes would explain the high conservation evident across large phylogenetic distances, even including the angiosperm-gymnosperm divergence [7], [8]. Previously, ndh genes have been reported as rich in editing sites [45], [46]. Although post-trancriptional editing in chloroplasts is not fully understood, it is evident that it is a highly specific process [47]. Under controlled conditions, editing of most of the C targets might not be essential for plant survival [reviewed in [48]. It can be argued however that, in less optimal conditions, individuals capable of altering their transcripts to conserved amino acids might present a selective advantage [47]. Functional characterization of the NDH complex has shown that even a single amino acid change in one of the genes can decrease net photosynthesis by up to 20% under field conditions but has no effect under milder conditions (e.g. fluorescent light) [6]. This lack of effect under certain environmental conditions might provide an explanation for the accumulation of editing sites in the ndh genes of flowering plants. RNA editing mechanisms would have allowed the rescue of ndh genes after episodes of dispensability during plant evolution [5].
In Cyanobacteria, ndh genes may encode several complexes formed by different NDH subunits. It has been suggested that these complexes could have different genetic regulation and different functions such as cyclic electron transport in Photosystem I, respiration and CO2 uptake [5]. As an endosymbiont derived from a free-living cyanobacterium, the plastid genome has suffered gene reduction and has readjusted its functions during the evolution of photosynthetic organisms [38]. Some of the forces driving those changes have been related to land colonization with photosynthetic organisms needing to adapt to new environments characterized by high light intensities. These new conditions forced the photosynthetic-related genes to acquire new roles as a means of coping with fluctuating conditions of the terrestrial environment. The ndh genes may have acquired new functions to improve photosynthetic performance under rapidly fluctuating terrestrial conditions [5]. Most of the algae (red, golden, brown) have lost the ndh genes. Excluding the exceptional case of the genus Nephroselmis, only the Charophyte algae, which include the presumed ancestor of land plants (Embryophytes) and usually inhabit waters of fluctuating shoreline environments, maintain the set of ndh genes [5].
During the evolution of land plants, aquatic environments have been re-colonized repeatedly. Aquatic plants are presently represented in roughly 17% of angiosperm families as the result of 100 independent evolutionary origins [22]. Adaptation to the aquatic environment is a difficult task and it involves major changes in morphology, physiology, and even reproductive strategies. Many aquatic species deal with water-associated problems of low light intensity or pollination by performing some of their functions on the surface, using floating or emergent leaves and flowers. Few species of angiosperms, however, have been able to solve all associated problems to enable a return to the completely submersed state [22]. For angiosperms, colonization of the aquatic environment requires the acquisition of traits equivalent to those present in algae, such as underwater reproduction. At the same time, selective forces to retain features specifically tied to terrestrial habitation disappear. While in land plants a completely functional ndh complex might provide adaptive advantages to deal with fluctuating light-intensities [6], submersed aquatics have to re-adapt their entire photosynthetic apparatus to low light conditions [18], [19]. The Najas flexilis plastid genome indicates that the relaxed pressure on some photosynthetic genes, as a consequence of the return to the aquatic environment, can lead to extreme genetic modifications or losses. As in deep water algae, the NDH complex, once essential for land-colonization, appears to be dispensable in at least some submersed angiosperms like Najas. Further studies of aquatic plant chloroplast genomes should determine whether the loss of these genes is characteristic of hydrophytes in general.
Supporting Information
Mauve alignment of the chloroplast genome structures among the order Alismatales using Acorus americanus as the reference. The color-coded boxes represent genome segments. In the diagram the lower boxes represent the genes transcribed in reverse direction. Genes are color-coded: white, protein coding genes; green, tRNA; red rRNA; pink boxes, IR. Numbers above the boxes indicate nucleotide positions from the origin.
(TIF)
Primer sets used in the chloroplast genome of Najas flexilis for amplification and Sanger sequencing of assembly gaps, low coverage areas and junctions.
(XLS)
Acknowledgments
We thank the Center for Applied Genetics and Technology (University of Connecticut) for the use of their facilities and to C. Obergfell, R. O’Neill, and L. Strausbaugh for their technical assistance and support. We thank our reviewers, Sean Graham and anonymous reviewer, for improving this manuscript. Photos in Figure 4; Authors: Ursula King, UCONN Live Collections. Authors under under CCAL Share-Alike 3.0: Christian Fischer, Eric Guinther, Atomx, BerndH, Silvae, Cody Hough.
Funding Statement
Portions of this research were funded by grants from the National Science Foundation (NSF DEB–0841658), Fulbright Foundation and Spanish MEC, Irish Research Council for Science, Engineering and Technology (IRCSET) and University of Connecticut CLAS. Publication costs were provided by the University of Connecticut Open Access Author Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bonen L, Doolittle W (1975) On the prokaryotic nature of red algal chloroplasts. Proc Natl Acad Sci U S A 72: 2310–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ravi V, Khurana JP, Tyagia K, Khurana P (2008) An update on chloroplast genomes. Plant Syst Evol 271: 101–122 doi:–––10.1007/s00606–007–0608–0 [Google Scholar]
- 3. Green BR (2011) Chloroplast genomes of photosynthetic eukaryotes. Plant J 66: 34–44 doi:–10.1111/j.1365–313X.2011.04541.x [DOI] [PubMed] [Google Scholar]
- 4. Kleine T, Maier UG, Leister D (2009) DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60: 115–138 doi:10.1146/annurev.arplant.043008.092119 [DOI] [PubMed] [Google Scholar]
- 5. Martín M, Sabater B (2010) Plastid ndh genes in plant evolution. Plant Physiol Biochem 48: 636–645 doi:10.1016/j.plaphy.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 6. Martín M, Funk HT, Serrot PH, Poltnigg P, Sabater B (2009) Functional characterization of the thylakoid Ndh complex phosphorylation by site-directed mutations in the ndhF gene. Biochim et Biophys Acta 1787: 920–928 doi:10.1016/j.bbabio.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 7. Neyland R, Urbatsch L (1996) The ndhF chloroplast gene detected in all vascular plant divisions. Planta 200: 273–277. [DOI] [PubMed] [Google Scholar]
- 8. Werner T, Braukmann A, Kuzmina M, Stefanović S, Braukmann T, et al. (2009) Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr Genet 55: 323–337 doi:–––10.1007/s00294–009–0249–7 [DOI] [PubMed] [Google Scholar]
- 9. Wolfe KH, Morden CW, Palmer JD (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A 89: 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haberhausen G, Zetsche K (1994) Functional loss of all ndh genes in an otherwise relatively unaltered plastid genome of the holoparasitic flowering plant Cuscuta reflexa . Plant Mol Biol 24: 217–222. [DOI] [PubMed] [Google Scholar]
- 11. Funk HT, Berg S, Krupinska K, Maier UG, Krause K (2007) Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii . BMC Plant Biol 7: 45 doi:10.1186/1471-2229-7-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Logacheva MD, Schelkunov MI, Penin AA (2011) Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis . Genome Biol and Evol 3: 1296–1303 doi:10.1093/gbe/evr102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wickett NJ, Zhang Y, Hansen SK, Roper JM, Kuehl JV, et al. (2008) Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis . Mol Biol Evol 25: 393–401 doi:10.1093/molbev/msm267 [DOI] [PubMed] [Google Scholar]
- 14. Cameron DD, Leake JR, Read DJ (2006) Mutualistic mycorrhiza in orchids: evidence from plant-fungus carbon and nitrogen transfers in the green-leaved terrestrial orchid Goodyera repens . New Phytol 171: 405–416 doi:–10.1111/j.1469–8137.2006.01767.x [DOI] [PubMed] [Google Scholar]
- 15. Chang CC, Lin H-C, Lin I-P, Chow T-Y, Chen H-H, et al. (2006) The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol 23: 279–291 doi:10.1093/molbev/msj029 [DOI] [PubMed] [Google Scholar]
- 16. Wu F-H, Chan M-T, Liao D-C, Hsu C-T, Lee Y-W, et al. (2010) Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol 10: 68 doi:–––10.1186/1471–2229–10–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pan I-C, Liao D-C, Wu F-H, Daniell H, Singh ND, et al. (2012) Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PloS one 7: e34738 doi:10.1371/journal.pone.0034738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iles WJD, Smith SY, Graham SW (2013) A well-supported phylogenetic framework for the monocot order Alismatales reveals multiple losses of the plastid NADH dehydrogenase complex and a strong long-branch effect. In: Mayo P, Wilkin JS, editors. Early Events in Monocot Evolution. Cambridge University Press. p.1–28.
- 19. Peredo EL, Les DH, King UM, Benoit LK (2012) Extreme conservation of the psaA/psaB intercistronic spacer reveals a translational motif coincident with the evolution of land plants. J Mol Evol 75: 184–197 doi:10.1007/s00239-012-9526-z [DOI] [PubMed] [Google Scholar]
- 20. Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, et al. (1994) Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii . Proc Natl Acad Sci U S A 91: 9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blazier CJ, Guisinger MM, Jansen RK (2011) Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol Biol 76: 263–272 doi:–––10.1007/s11103–011–9753–5 [DOI] [PubMed] [Google Scholar]
- 22. Les D, Cleland M, Waycott M (1997) Phylogenetic studies in Alismatidae, II: evolution of marine angiosperms (seagrasses) and hydrophily. Syst Bot 22: 443–463. [Google Scholar]
- 23. Les DH, Sheldon SP, Tippery NP (2010) Hybridization in hydrophiles: natural interspecific hybrids in Najas (Hydrocharitaceae). Syst Bot 35: 736–744 doi:10.1600/036364410X539826 [Google Scholar]
- 24.Sculthorpe C (1967) The biology of aquatic vascular plants. London: Edward Arnold.
- 25. Rogstad S (1992) Saturated NaCl-CTAB solution as a means of field preservation of leaves for DNA analyses. Taxon 41: 701–708. [Google Scholar]
- 26. Doyle J (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 13–15. [Google Scholar]
- 27. Mardanov A V, Ravin N V, Kuznetsov BB, Samigullin TH, Antonov AS, et al. (2008) Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol 66: 555–564 doi:–––10.1007/s00239–008–9091–7 [DOI] [PubMed] [Google Scholar]
- 28. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 doi:10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 29.Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ. 365–386. [DOI] [PubMed]
- 30. Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255 doi:10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 31. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27: 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one 5: e11147 doi:10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grant JR, Arantes AS, Stothard P (2012) Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics 13: 202 doi:–––10.1186/1471–2164–13–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, et al. (2011) Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot 98: 704–730 doi:10.3732/ajb.1000404 [DOI] [PubMed] [Google Scholar]
- 35. Huotari T, Korpelainen H (2012) Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene 508: 96–105 doi:10.1016/j.gene.2012.07.020 [DOI] [PubMed] [Google Scholar]
- 36. Liu J, Qi Z-C, Zhao Y-P, Fu C-X, Jenny Xiang Q-Y (2012) Complete cpDNA genome sequence of Smilax china and phylogenetic placement of Liliales - influences of gene partitions and taxon sampling. Mol Phyl Evol 64: 545–562 doi:10.1016/j.ympev.2012.05.010 [DOI] [PubMed] [Google Scholar]
- 37. Wang W, Messing J (2011) High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PloS one 6: e24670 doi:10.1371/journal.pone.0024670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76: 273–297 doi:–––10.1007/s11103–011–9762–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH (2003) Analysis of the Amborella trichopoda chloroplast genome sequence suggests that amborella is not a basal angiosperm. Mol Biol Evol 20: 1499–1505 doi:10.1093/molbev/msg159 [DOI] [PubMed] [Google Scholar]
- 40. Goremykin V V, Holland B, Hirsch-Ernst KI, Hellwig FH (2005) Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol 22: 1813–1822 doi:10.1093/molbev/msi173 [DOI] [PubMed] [Google Scholar]
- 41. Wang R-J, Cheng C-L, Chang C-C, Wu C-L, Su T-M, et al. (2008) Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol 8: 36 doi:10.1186/1471-2148-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goremykin V V, Hirsch-Ernst KI, Wölfl S, Hellwig FH (2004) The chloroplast genome of Nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm. Mol Biol Evol 21: 1445–1454 doi:10.1093/molbev/msh147 [DOI] [PubMed] [Google Scholar]
- 43. Yang M, Zhang X, Liu G, Yin Y, Chen K, et al. (2010) The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PloS one 5: e12762 doi:10.1371/journal.pone.0012762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saski C, Lee S-B, Fjellheim S, Guda C, Jansen RK, et al. (2007) Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet 115: 571–590 doi:–––10.1007/s00122–007–0567–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hammani K, Okuda K, Tanz SK, Chateigner-Boutin A-L, Shikanai T, et al. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 doi:10.1105/tpc.109.071472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grosche C, Funk HT, Maier UG, Zauner S (2012) The chloroplast genome of Pellia endiviifolia: gene content, RNA editing pattern and the origin of chloroplast editing. Genome Biol Evol 4: 1349–1357 doi:10.1093/gbe/evs114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boussardon C, Salone V, Avon A, Berthomé R, Hammani K, et al. (2012) Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24: 3684–3694 doi:10.1105/tpc.112.099507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Ann Rev Plant Biol 61: 125–155 doi:10.1146/annurev-arplant-042809-112242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mauve alignment of the chloroplast genome structures among the order Alismatales using Acorus americanus as the reference. The color-coded boxes represent genome segments. In the diagram the lower boxes represent the genes transcribed in reverse direction. Genes are color-coded: white, protein coding genes; green, tRNA; red rRNA; pink boxes, IR. Numbers above the boxes indicate nucleotide positions from the origin.
(TIF)
Primer sets used in the chloroplast genome of Najas flexilis for amplification and Sanger sequencing of assembly gaps, low coverage areas and junctions.
(XLS)