Abstract
The American Cancer Society estimates that over 200,000 new breast cancer cases are diagnosed annually in the USA alone. Of these cases, the majority are invasive breast cancers and almost 70% are estrogen receptor-α positive. Therapies targeting the estrogen receptor-α are widely applied and include selective estrogen receptor modulators such as tamoxifen, a selective estrogen receptor downregulator such as Fulvestrant (Faslodex; FAS, ICI 182,780), or one of the third-generation aromatase inhibitors including letrozole or anastrozole. While these treatments reduce breast cancer mortality, many estrogen receptor-α-positive tumors eventually recur, highlighting the clinical significance of endocrine therapy resistance. The signaling leading to endocrine therapy resistance is poorly understood; however, preclinical studies have established an important role for autophagy in the acquired resistance phenotype. Autophagy is a cellular degradation process initiated in response to stress or nutrient deprivation, which attempts to restore metabolic homeostasis through the catabolic lysis of aggregated proteins, unfolded/misfolded proteins or damaged subcellular organelles. The duality of autophagy, which can be either pro-survival or pro-death, is well known. However, in the context of endocrine therapy resistance in breast cancer, the inhibition of autophagy can potentiate resensitization of previously antiestrogen resistant breast cancer cells. In this article, we discuss the complex and occasionally contradictory roles of autophagy in cancer and in resistance to endocrine therapies in breast cancer.
Keywords: 3-methyladenine, antiestrogen resistance, aromatase inhibitor, autophagy, bafilomycin A1, breast cancer, endoplasmic reticulum stress, fulvestrant, hydroxychloroquine, tamoxifen, unfolded protein response
Endocrine therapy resistance in estrogen receptor-α-positive (ER+) breast cancer, whether acquired or de novo, remains an important clinical problem. While adjuvant endocrine therapy reduces breast cancer mortality, many ER+ tumors will eventually recur. Mechanisms of antiestrogen resistance are still poorly understood; however, preclinical studies suggest that several druggable targets offer the potential to restore endocrine therapy sensitivity, such as key components of prosurvival autophagy signaling. Autophagy, or ‘self-eating’, is a mechanism by which a cell digests its own subcellular organelles or unfolded/misfolded/aggregated proteins. Under normal conditions, this provides a quality-control mechanism, removing damaged organelles and proteins. In response to a stressor, this autophagic digestion recovers energy in an attempt to maintain/restore metabolic homeostasis. Targeting autophagy through chemical inhibitors, such as hydroxychloroquine (HCQ) or 3-methyladenine (3-MA), or by RNAi targeting of beclin-1 can restore antiestrogen sensitivity in some resistant breast cancer cells. Several clinical trials have been initiated to investigate the role of autophagy in different cancer types, including metastatic breast cancer. One clinical trial of particular interest in this regard is the Preventing Invasive Breast Neoplasia with Chloroquine (PINC) study, which involves the inhibition of autophagy while concurrently treating with tamoxifen in patients with ductal carcinoma in situ of the breast.
The American Cancer Society (ACS) estimates that over 200,000 new cases of breast cancer are diagnosed in the USA each year [1]. Breast cancer remains the second highest killer of all cancers in women, second only to lung and bronchial cancer, with more than 40,000 reported deaths in women in the USA last year [1,2]. Over 1.15 million new cases of breast cancer are estimated to have been diagnosed worldwide last year, resulting in over 411,000 deaths in women. Breast cancer is the leading cause of cancer mortality in women worldwide [3]. Therapies targeting the ER are widely applied and include selective estrogen receptor modulators such as tamoxifen (TAM), a selective estrogen receptor downregulator such as Faslodex (FAS; fulvestrant [FAS], ICI 182,780), or one of the third-generation aromatase inhibitors (AIs) including letrozole or anastrozole. Antiestrogens are less toxic than cytotoxic chemotherapy and TAM has represented the ‘gold standard’ in first-line endocrine therapy for over 30 years [4]. More recently, AIs have begun to replace TAM as the first-line endocrine therapy of choice for ER+ postmenopausal breast cancer [5]. FAS, an antiestrogen drug lacking the agonist estrogenic affects of TAM, downregulates the ER through enhanced ubiquitin-mediated degradation of the receptor and has a different modality of action when compared with TAM [6,7]. In some patients, FAS is as effective as an AI [8]. While clinical studies demonstrate that adjuvant endocrine therapy reduces mortality, many ER+ breast tumors that initially respond to therapy develop acquired resistance [9–11]. For the most part, advanced ER+ breast cancer remains an incurable disease, highlighting the importance of understanding endocrine therapy resistance.
Two different types of antiestrogen resistance are generally described, de novo or intrinsic resistance and acquired resistance. A primary mechanism of de novo resistance to antiestrogen therapy is the lack of detectable ER expression [12,13]. Acquired resistance appears to occur through many different mechanisms, several of which involve changes in the ER including mutations, altered patterns of phosphorylation by growth factors and their downstream kinases, and altered expression of ER coregulators [13]. Much of our current understanding of antiestrogen resistance is based on studies focused on TAM resistance in experimental models of breast cancer. While these endocrine resistance studies have implicated many causative genes (reviewed in [13,14]), more recent studies associate autophagy and cell stress responses with endocrine resistance and thus open up a new area of research in this field (see recent reviews [15,16]).
Autophagy
Autophagy (macroautophagy) is a conserved evolutionary process that can enable cells to maintain homeostasis in unfavorable environmental conditions. An autophagic ‘self-eating’ allows the cell to recover energy from damaged or unnecessary subcellular components. However, if the insult is too severe and autophagy persists at a high level, it becomes pro-death; an autophagic cell death is often referred to as programmed cell death-2 (apoptosis is programmed cell death-1). Basal levels of autophagy help clear injured organelles or long-lived proteins; hypoxia, nutrient or growth factor deprivation, accumulation of misfolded or unfolded proteins in the endoplasmic reticulum or infection can each increase the extent of autophagy (reviewed in [17]).
The process of autophagy involves the segregation of cytoplasm and intracellular organelles in double membrane-bound structures called autophagosomes. Autophagosomes then fuse with lysosomes to form autolysosomes, facilitating the degradation of the sequestered cellular material by lysosomal hydrolases. Under starvation conditions, degraded organelles or proteins are recycled and converted into metabolic intermediates that can be used to fuel the cell. Under hypoxia, autophagy removes reactive oxygen species-generating mitochondria, thereby protecting the cell [18].
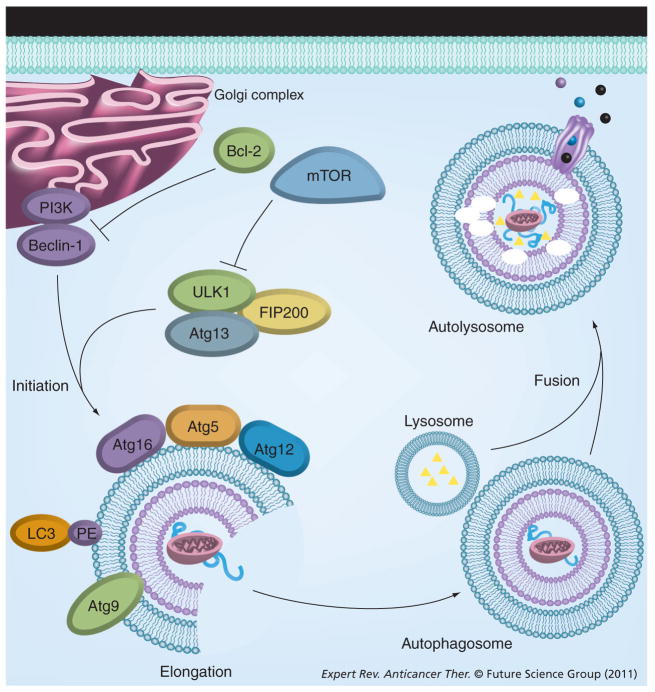
The autophagy-related family of proteins (Atg) comprise the distinct molecular machinery necessary for the induction and formation of autophagosomes, autophagosome-vesicle fusion, lysis and release of degraded molecules back into the cytosol (reviewed in [19]). Table 1 summarizes the primary autophagy-related genes and their effects on the autophagy pathway. The process of autophagy is best defined in yeast. Critical to the initiation of autophagy is the activation of Atg1 (mammalian homolog: Unc-51-like kinase [ULK]-1 and -2), which is negatively regulated by the serine/threonine protein kinase target of rapamycin (TOR) [20,21]. Under low-nutrient conditions where TOR is repressed, the kinase activity of Atg1 enables binding of Atg1 to Atg13 and Atg17 (mammalian homolog: focal adhesion kinase family-interacting protein of 200 kD; FIP200 and RB1CC1), thereby creating a scaffold for recruiting other Atg proteins [22]. Unlike yeast, mammalian cells can form stable ULK-Atg13-FIP200 complexes regardless of nutrient conditions.
Table 1.
Selected autophagy-related genes.
| Autophagy gene | HUGO gene symbol | Effect on autophagy |
|---|---|---|
| Atg3 | ATG3 | E2-like enzyme facilitates lipidation of LC3 |
| Atg4A, -B, -C, -D | ATG4A, ATG4B, ATG4C and ATG4D | Cleaves pro-LC3 to form LC3 |
| Atg5 | ATG5 | Forms a complex with Atg12-Atg16, resulting in lipidation of LC3 |
| Atg7 | ATG7 | E1-like enzyme activates Atg12 |
| Atg9A, -B | ATG9A and -B | Phagophore membrane expansion |
| Atg10 | ATG10 | E2-like enzyme facilitates the formation of Atg5-Atg12-Atg16 complex |
| Atg12 | ATG12 | Forms complex with Atg5-Atg16, resulting in lipidation of LC3 |
| Atg13 | ATG13 | Part of the initiation complex with ULK1, Atg101 and FIP200 |
| Atg16L1, -L2 | ATG16L1 and -2 | Forms a complex with Atg5-Atg12, resulting in lipidation of LC3 |
| Beclin-1 | BECN1 | Part of the initiation complex with Vps34 |
| Atg101 | C12orf44 | Part of the initiation complex with ULK1, FIP200 and Atg13 |
| Cathepsin B | CTSB | Lysosome enzyme |
| LAMP1, -2, -3 | LAMP1, -2 and -3 | Lysosome autophagosome fusion |
| LC3 (A, B or C) | MAP1LC3A, -B and -C | Phagophore membrane curvature and expansion |
| MTOR | MTOR | Inhibits ULK1 |
| PIK3C3 (Vps34) | PIK3C3 | Part of the initiation complex with beclin-1 |
| FIP200 | RB1CC1 | Part of the initiation complex with ULK1, Atg101 and Atg13 |
| p62 | SQSTM1 | Cargo recognition |
| Rab7 | RAB7A | Lysosome autophagosome fusion |
| ULK1 | ULK1 | Part of the initiation complex with Atg101, Atg13 and FIP200 |
HUGO: Human Genome Organisation; LAMP: Lysosomal membrane protein; LC: Light chain; ULK: Unc-51-like kinase.
In mammalian cells, autophagosome degradation is driven by p62/sequestosome-1 (SQSTM1), which binds directly to ubiquitinated proteins and microtubule-associated protein-1 (MAP1) light chain-3 (LC3), linking the ubiquitinated proteins to the autophagic machinery [23–25]. Formation of the autophagosome double membrane occurs de novo and not from either pre-existing organelles or by the annealing of single membranes, and requires the actions of vacuolar protein sorting 34 (Vps34), p150, Atg4 and beclin-1 (BECN1) [26–28]. Beclin-1 activity is regulated by B-cell lymphomia/leukemia-2 (Bcl-2) and is discussed more thoroughly later. LC3 determines autophagosome size and membrane curvature [19]; the Atg12/Atg5/Atg16 complex and the LC3-phosphatidylethanolamine (LC3-PE or LC3-II) complex participate in elongation of the autophagosome membrane. Atg9 is the only integral membrane protein identified in autophagosome formation, where it may function as a carrier of membrane materials. Atg9 is dependent on ULK1 and Atg13 for transportation from the trans-Golgi network to late endosomes [29–33]. The cysteine protease Atg4 cleaves pro-LC3 to expose a C-terminal glycine residue, enabling Atg12-Atg5 to conjugate LC3 to PE, via an amide bond; LC3-PE levels are often used as a measure of autophagy induction (reviewed in [34]). The early autophagosome fuses with a lysosome to form the late autolysosome. This fusion process is dependent upon the lysosomal membrane protein-2 (LAMP-2) and the small GTPase Rab7 [35,36]. After fusion of the lysosome, the resulting autolysosome degrades its protein/organelle load and inner membrane. In mammalian autophagy, degradation occurs through the actions of cathepsins B, D and L [17,37]. The resulting products of the catalytic degradation process are then transported to the cytosol and recycled. The process of autophagy described above is shown in Figure 1.
Figure 1. Cellular pathway of autophagy.
The PI3K complex mediates the initiation of the phagophore membrane, enveloping labeled cytosolic proteins organelles and fat. mTOR and Bcl-2 can inhibit the initiation of autophagy. The Atg12-Atg5-Atg16 complex, LC3 and the transmembrane Atg9 are recruited to the phagophore and are necessary for elongation of the double membrane. Lysosomes fuse with the autophagosome, creating the autolysosome. The resulting products of the catalytic degradation process are transported to the cytosol and recycled.
LC: Light chain-3.
Bcl-2 & the regulation of autophagy in breast cancer
The Bcl-2 family contains two distinct functional groups, the anti-apoptotic group that includes Bcl-2 and Bcl-XL, and the proapoptotic group including the Bax and Bak proteins. Bcl-2 is an antiapoptotic protein that exhibits oncogenic potential through its ability to regulate the intrinsic apoptotic pathway. The molecular activity of Bcl-2 involves binding to mitochondrial Bax, thereby preventing Bax activation, mitochondrial outer-membrane permeabilization and apoptosis. Bcl-2 is overexpressed in over 60% of breast tumors; overexpression of Bcl-2 correlates with chemotherapeutic and radiation resistance [38,39]. Moreover, a recent clinicopathological investigation that measured both beclin-1 and Bcl-2 in breast cancer tissue indicated that beclin-1 is inversely correlated with Bcl-2 expression. Increased Bcl-2 expression is associated with the estrogen receptor, increased histological grade and distant metastases [40]. These data highlight the role of Bcl-2 in breast cancer and resistance.
Beclin-1 was originally identified as a Bcl-2-interacting protein [41], the Bcl-2 family being a group of proteins containing Bcl-2 homology domains. Beclin-1 binds to Bcl-2, Bcl-W, Bcl-XL and Mcl-1, which results in the inhibition of autophagy [41–43]. Table 2 summarizes the Bcl-2 family members’ effects on apoptosis and autophagy. In several cell types, binding of Bcl-2 to beclin-1 inhibits the binding and activation of Vps34, decreasing Vps34-mediated PI3K activation and subsequently inhibiting autophagy. In cases of nutrient starvation, or when cells are treated with Bcl-2 inhibitors that reduce Bcl-2 protein levels, Bcl-2 and beclin-1 dissociate and autophagy is stimulated [41–43]. Conversely, proapoptotic Bcl-2 family members, such as Bad, Bik, BNIP3L, Noxa, Puma and BimEL, may induce autophagy by competitively binding to Bcl-2 family members and disrupting the interaction between beclin-1 and Bcl-2; thereby freeing beclin-1 [44–46]. A recent study into the role of Mcl-1 in autophagy reported that, in response to glucose deprivation and hypoxia, Mcl-1 is rapidly degraded and autophagy becomes activated. Furthermore, Mcl-1 overexpression prevents LC3-positive punctate formation, indicating a key regulatory role of Mcl-1 in autophagy [47].
Table 2.
Role of Bcl-2 family members on apoptosis and autophagy.
| Bcl-2 family member | HUGO gene symbol | Effect on apoptosis | Effect on autophagy |
|---|---|---|---|
| Bcl-2 | BCL2 | Antiapoptotic | Inhibits autophagy by binding to beclin-1 |
| Bcl-w | BCL2L2 | Antiapoptotic | Inhibits autophagy by binding to beclin-1 |
| Bcl-XL | BCL2L1 | Antiapoptotic | Inhibits autophagy by binding to beclin-1 |
| Mcl-1 | MCL1 | Antiapoptotic | Inhibits autophagy (to a lesser extent than Bcl-2, Bcl-w and Bcl-XL) by binding to beclin-1 |
| Bad | BAD | Proapoptotic | Promotes autophagy by competitively binding to Bcl-2, Bcl-w and Bcl-XL |
| t-Bid | BID | Proapoptotic | Promotes autophagy by competitively binding to Bcl-2, Bcl-w and Bcl-XL |
| BimEL | BCL2L11 | Proapoptotic | Promotes autophagy by competitively binding to Bcl-2, Bcl-w and Bcl-XL |
| Noxa | PMAIP1 | Proapoptotic | Promotes autophagy by competitively binding to Bcl-2, Bcl-w and Bcl-XL |
| Puma | BBC3 | Proapoptotic | Promotes autophagy by competitively binding to Bcl-2, Bcl-w and Bcl-XL |
| BNIP3L | BNIP3L | Proapoptotic | Promotes autophagy by competitively binding to Bcl-2, Bcl-w and Bcl-XL |
| Bik | BIK | Proapoptotic | Promotes autophagy by competitively binding to Bcl-2, Bcl-w and Bcl-XL |
| Bax | BAX | Proapoptotic | No effect |
| Bak | BAK1 | Proapoptotic | No effect |
HUGO: Human Genome Organisation.
Several Bcl-2 inhibitors are currently undergoing clinical trials. While the use of Bcl-2 inhibitors are predominately focused on leukemias and lymphomas, a potential role for these inhibitors in breast cancer is now evident [48]. For example, preclinical studies investigating the role of Bcl-2 in MCF-7 breast cancer cells show that silencing Bcl-2 by siRNA increases autophagy and cell death, highlighting the possible use of Bcl-2 inhibitors as a therapeutic strategy in breast cancer [49]. Gossypol, a BH3 mimetic isolated from cotton seeds, induces beclin-1-dependent and -independent autophagy, resulting in cytoprotection and survival of MCF-7 breast cancer cells [50]. These studies likely reflect an important role for Bcl-2 family members in the regulation of autophagy in breast cancer.
Unfolded protein response & the regulation of autophagy in breast cancer
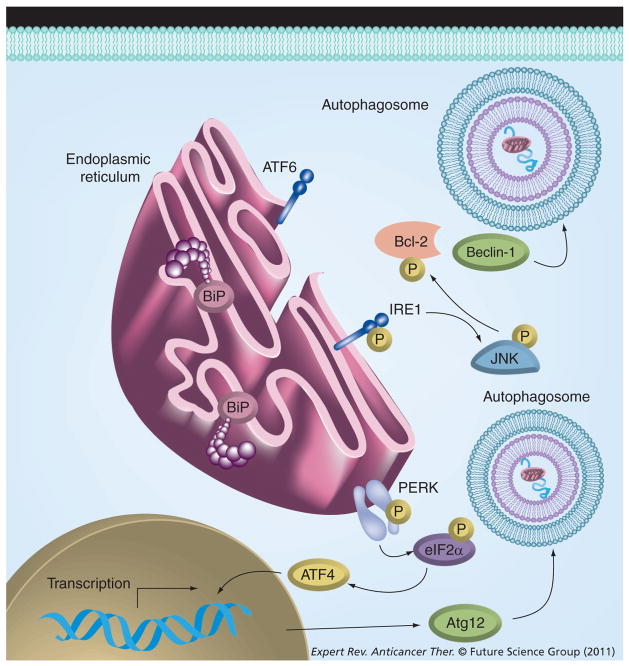
The unfolded protein response (UPR) pathway is activated in response to the accumulation of aggregated unfolded/mis-folded proteins within the endoplasmic reticulum (EnR) lumen. In response to this accumulation, the EnR protein chaperone glucose-regulated protein 78 (GRP78; BiP; HSPA5) is released from each of inositol requiring enzyme-1 (IRE1), activating transcription factor 6 (ATF6) and PKR-like endoplasmic reticulum kinase, enabling their respective activation. IRE1 dimerizes and becomes autophosphorylated, resulting in its activation and ability to perform the unconventional (cytosolic) splicing of the X-box binding protein-1 (XBP1) mRNA. XBP1 splicing creates the transcriptionally-active XBP1-S form [16,51], which can confer estrogen independence and antiestrogen resistance upon estrogen-dependent breast cancer cells [16,52] and is now known to be associated with a poor response to TAM [53].
Unfolded protein response stimulation promotes the activation of autophagy through different mechanisms. EnR stress results in phosphorylation of eIF2α by PKR-like endoplasmic reticulum kinase. Activated eIF2α increases ATF4 expression, which then increases the transcription of Atg12 and can thereby promote autophagy [54]. IRE1 activation also leads to the phosphorylation of c-Jun-terminal kinase, resulting in the phosphorylation of Bcl-2 at the T69, S70 and S87 residues in the unstructured loop of Bcl-2 [55]. Phosphorylation of Bcl-2 can cause dissociation of the Bcl-2/beclin-1 complex and thus may activate autophagy. Figure 2 illustrates the interaction between UPR signaling and autophagy. Calcium released from the EnR following stress also promotes autophagy by activating beclin-1 [17]. Therefore, the stimulation of UPR by mechanisms such as nutrient starvation, hypoxia or therapeutic drugs may result in the subsequent activation of a prosurvival autophagy.
Figure 2. Effect of the unfolded protein response on autophagy.
The unfolded protein response can activate autophagy through two distinct mechanisms. PERK activation leads to phosphorylation of eIF2α, resulting in increased ATF4 transcription. ATF4 promotes the transcription of Atg12, resulting in increased autophagy. Another mechanism of unfolded protein response-modulating autophagy is through activation of IRE1. IRE1 activates JNK, leading to the subsequent phosphorylation of Bcl-2. Bcl-2 phosphorylation prevents its binding to beclin-1, thereby promoting autophagy.
ATF: Activating transcription factor; IRE1: Inositol requiring enzyme-1; PERK: PKR-like endoplasmic reticulum kinase.
Dichotomy of autophagy in cancers
In cancer, autophagy can serve as either a ‘tumor suppressor’ or as a ‘tumor promoter’. Allelic loss of vital autophagy components, such as beclin-1, is often found in breast, ovarian and prostate cancers [56–59]. Moreover, inhibition of beclin-1 or deletion of Atg5 in immortalized epithelial kidney cells or breast cancer cell lines is associated with increased proliferation and tumorigenicity [60,61]. Genetically altered heterozygous beclin-1 knockout mice exhibit an increased incidence of hepatocellular carcinoma, lung adenocarcinoma and mammary hyperplasia [62]. Increased susceptibility to hepatitis B-induced hepatocellular carcinoma, when compared with their wild-type counterparts [63,64], is also reported. Brain tumors often have reduced beclin-1 compared with the normal surrounding tissue and reduced beclin-1 inversely correlates with malignancy [65]. Vps34 expression is also dysregulated in cancers. Vps34 overexpression in colon cancer cell lines reduced tumorigenicity, while heterozygous deletion of the Vps34 gene uvrag is often observed in colon tumors [66]. Knockdown of Atg4, the protease that cleaves LC3, increases the severity of chemically-induced fibrosarcomas in mice [67]. These data suggest that disruption of the autophagic process is a key event in tumorgenesis.
While direct modulation of the components of autophagy is observed in different cancers, mutations indirectly affecting autophagy are also reported. PI3K mutations are found in over 20% of breast cancers and 30% of colorectal cancers. These mutations may indirectly influence autophagy through the stimulation of mTOR, which would prevent ULK activity and inhibit autophagy [62]. Another possible autophagy-regulating event, p53 mutational inactivation, is observed in over 50% of all tumors [68]. Inactivating p53 mutations, mutation in the p53 activating kinases, overexpression of MDM2 that degrades p53 and loss of function of p14ARF, are each documented in various cancers and these result in a loss/reduction in p53 activity (reviewed in [45]). Nuclear p53 can affect autophagy through transactivation of death-associated protein kinase-1 (DAPK-1) and the lysosomal protein damage-regulated autophagy modulator [69]. DAPK-1 is commonly dysregulated in human tumors and has both proapoptotic and proautophagy activities. For example, DAPK-1 phosphorylates myosin light chain, promoting membrane blebbing and apoptosis [70]. DAPK-1 is also implicated in autophagy through its ability to bind MAPB1, which interacts with LC3 to inhibit autophagy [70,71]. Transactivation of DRAM by p53 can activate autophagy and is necessary for the execution of a DNA damage-induced p53-mediated cell death. Furthermore, p53 inhibits mTOR and can thus activate autophagy.
While the transcriptional activation of p53 promotes autophagy, cytosolic p53 can inhibit autophagy [72]. Preclinical studies show an increase in autophagy with an increased formation of LC3-containing autophagic vacuoles when p53 is knocked down, knocked out or otherwise inhibited [34,73]. Moreover, inhibition of nuclear transporters, resulting in the accumulation of p53 in the nucleus, prevents the inhibitory actions of p53 on autophagy. While the influence of p53 on autophagy is evident, the overall effect of p53 on the regulation of autophagy remains controversial. Function of the upstream activator of p53, p14ARF, is also lost in many cancers and the mitochondrial form of p14ARF is a potent stimulator of autophagy [74].
Defects in autophagy may also promote tumorgenesis. Impaired autophagy hinders the ability of a cell to survive stressful environmental conditions and can result in increased cell death [59,60]. While this may seem anti-tumorigenic at first, chronic cell death leads to a prolonged inflammatory response that can be oncogenic. Cancer-related inflammation is often considered the seventh hallmark of cancer [75]. For example, chronic cell death in the liver stimulates inflammation, increases organ damage and raises the risk of developing hepatocellular carcinoma. Necrotic cell death leads to the release of cellular debris, activating various cell-surface receptors on neighboring cells that can stimulate survival pathways and enhance cell growth [75,76]. Moreover, autophagy also limits genotoxic damage by reducing the formation of reactive oxygen species (ROS) and clearing damaged mitochondria [75,77]. When autophagy is impaired, damaged mitochondria remain in the cell, increasing ROS production and the associated protein, organelle and DNA damage. Oxidative damage from the accumulation of ROS may promote tumorgenesis, thereby supporting inadequate autophagy as a contributor to oncogenesis (reviewed in [76]).
While autophagy is implicated as a tumor supressor in early tumorgenesis, a growing body of evidence implicates autophagy as a tumor promoter in late stage cancers. The ability of the autophagic process to provide cellular resilience to stressors such as glucose deprivation and hypoxia, two common stresses experienced by tumors, enables long-term cell survival [45,57,60,78]. Autophagy allows cancer cells to ‘eat themselves’, progressively reducing in size, to conserve and provide nutrients for survival functions. Since some of these cancer cells retain the ability of self-renewal, they can return to their original size and proliferate given proper conditions. Thus, some cancer cells with intact autophagy may exhibit cellular dormancy (reviewed in [59]).
In summary, the oncogenic activity of autophagy stimulation may be stage dependent. Autophagy appears to a tumor suppressor in early tumorigenesis, perhaps because the cells cannot easily adapt to the increased elimination of subcellular organelles as an energy source. During progression, those cells that survive will likely have been able to adapt their physiology such that they can survive with the increased basal level of autophagy, which may now be providing sufficient energy for survival without exceeding a threshold where cell death becomes inevitable.
Autophagy in breast cancer
The role of autophagy in breast cancer, like other types of cancer, is an area of active investigation. Clinicopathological investigation of breast cancer tissue indicated three discernable LC3-positive patterns; diffuse cytoplasmic, cytoplasmic/juxtanuclear and dense round 5 μm ‘stone-like’ structures [79]. Diffuse cytoplasmic or cytoplasmic/juxtanuclear LC3 staining correlates directly to with estrogen and progesterone receptor expression in breast tissue. Moreover, the ‘stone-like’ LC3 stained phenotype is associated with high-grade tumors and a less favorable outcome, suggesting the more autophagic a tumor, the more aggressive the tumor [79].
Evidence also supporting the oncogenic activity of autophagy was obtained through 3D morphogenic assays investigating the role of beclin-1 on mammary acini formation. Immortalized mouse mammary epithelial cells with homozygous beclin-1 produce an atypical solid acini structure, owing to the autophagy-competent central acini epithelial cells having an increased capacity to survive anoikis and the hypoxic environment of the central lumen [61]. Conversely, heterozygous deletion of beclin-1 in Immortalized mouse mammary epithelial cells results in normal mammary lumen formation. The defect in autophagy resulting from beclin-1 deletion leads to necrosis of the central epithelial cells, allowing normal mammary lumen formation [78]. Morphogenesis assays capture the physiological context found in normal tissues to model adhesion signaling in acini formation. The ability of cells to form atypical solid acini parallels breast cancer’s invasion and metastatic potential in vivo; therefore, these data indicate a role of beclin-1 and autophagy in mammary acini development and in cancer progression. A role for beclin-1 in breast tumorigenesis is apparent in the significant variability of beclin-1 expression across different molecular subtypes; higher expression levels of beclin-1 are seen in the HER2-negative luminal-A or luminal-B breast cancers [80,81]. In combination with the mammary acini study, these data indicate that the role of beclin-1 in breast cancer may be more complex than that of just a tumor suppressor, consistent with data from other cancers.
In triple-negative breast cancer (ER negative, PR negative, HER2 negative), the effect of autophagy is just beginning to be elucidated. Recent studies report increased autophagic properties in the mitochondria of the metastatic triple-negative MDA-MB-231 when compared with the less metastatic MDA-MB-468 and noncancerous MCF7–10A cells [82]. However, investigation into the effect of phytochemical therapy and PPARγ ligands in triple-negative breast cancers shows an increase in cell death caused by autophagic activation and necrosis, suggesting there may be a threshold limitation between prosurvival and prodeath autophagy [83,84]. Various studies of the therapeutic response of experimental chemotherapies in breast cancer have implicated a prodeath role for autophagy. A lipid-modified estrogen derivative, developed to treat breast cancer independent of ER status, was shown to induce apoptosis and autophagy in the triple-negative MDA-MB-231 breast cancer cells [85]. Furthermore, this estrogenic compound interferes with mTOR activity, thereby inducing autophagy and promoting cell death. These observations suggest a possible therapeutic strategy for inhibiting triple-negative or ER-negative breast cancer growth through the stimulation of pro-death autophagy [85].
Autophagy in drug resistance
Preclinical studies using chemical inhibitors of autophagy (described in Table 3) or siRNA to knockdown vital autophagy genes demonstrate the role of autophagy in stress and chemotherapeutic sensitization of cancer cells. Most chemical inhibitors of autophagy lack specificity and often have off-target effects. Preclinical studies using these chemicals may benefit from siRNA knockdown of autophagy genes before concluding that the observed effects are due solely to autophagy inhibition. Inhibition of autophagy in glioblastoma, lung cancer, cervical cancer, prostate cancer, leukemia and breast cancer cells resensitized the cells to various therapeutic agents [86]. For example, upregulation of autophagy can protect cancer cells against various therapies including temozolomide, resveratrol, vitamin D3, anthocyanins, radiotherapy and TAM [16,76,86–89]. Treatment with temozolomide in malignant glioma cells stimulates autophagy without activating apoptosis and is associated with resistance to DNA-alkylating agents in some brain cancers [90]. These data suggest an important role of autophagy in promoting cancer therapeutic drug resistance.
Table 3.
Commonly used inhibitors of autophagy.
| Compound | Target and effect |
|---|---|
| Hydroxychloroquine or chloroquine | Lysosomal pH, prevents autophagosome–lysosome fusion |
| 3-methyladedine | Class III PI3K inhibition, prevents autophagosom formation |
| Wormatin | Class III PI3K inhibition, prevents autophagosome formation |
| LY294002 | Class III PI3K inhibition, prevents autophagosome formation |
| Bafilomycin A1 | Vacuolar ATPase inhibition, prevents autophagosome–lysosome fusion |
Breast cancer studies have also revealed a role of autophagy in resistance. Autophagy protects MCF-7 breast cancer cells against epirubicin-mediated cell death, and inhibition of autophagy through beclin-1 siRNA restored epirubicin effectiveness [91]. In addition, when treated with camptothecin or etoposide (DNA-damaging therapeutic drugs) autophagy can delay the onset of apoptotic cell death in breast cancer cells, an effect reversed by knockdown of the autophagy-dependent genes Atg7 and beclin-1 [44]. Furthermore, treatment of MCF-7 breast cancer cells with bortezomib, a proteasome inhibitor, results in a potent stimulation of autophagy and UPR. The authors speculate that the observed activation of UPR and autophagy is prosurvival, and therefore may explain the poor response to bortezomib in breast cancer patients [92]. HER2-targeted therapies, such as the monoclonal antibody herceptin and EGF receptor tyrosine kinase inhibitors, such as lapatinib, are sensitive to autophagy-mediated resistance [93,94]. Trastuzumab (herceptin) induces LC3-positive punctate formation in SKBr3 cells (HER2-amplified breast cancer cell line). Inhibition of autophagy by 3-MA and LY294002 increases cell death in response to trastuzumab, suggesting autophagy as a cytoprotective response [94]. Moreover, inhibition of autophagy restores EGF receptor-mediated cell death in lapatinib-resistant BT-474 cells (HER2 amplified breast cancer cell line) [93]. These data suggest that targeting autophagy can be sufficient to restore chemotherapeutic drug sensitivity and promote breast cancer cell death.
Studies have also investigated the outcome of autophagy stimulation in antiestrogen therapy. Bursch et al. treated MCF-7 estrogen-dependent ER+ breast cancer cells with the antiestrogens TAM and ICI, and found that dying cells showed increased cytosolic autophagosome formation [95]. These authors concluded that autophagy, stimulated by antiestrogens in MCF-7 cells, resulted in active cell death. However, more recently Samaddar et al. suggest that this conclusion more likely reflects cells’ failed attempts at survival. Samaddar et al. demonstrated that in the surviving MCF-7 cellular population (~70%) after antiestrogen treatment, there was an increase in autophagosome formation. This group also hypothesized that whether autophagy promotes survival or cell death may be dependent on the number of autophagosomes in each cell, resulting in a threshold limit. Inhibiting autophagosome formation via 3-methyladenine (3-MA) or beclin-1 siRNA significantly enhanced antiestrogen-induced cell death in MCF-7 cells, further suggesting a prosurvival role of autophagy in anti-estrogen therapy [96]. Qadir et al. used siRNA with Atg5, beclin-1 and Atg7 to inhibit autophagy in MCF-7, T47D and TAM-resistant MCF7-HER2 cell lines, and reported that concurrent knockdown of autophagy and treatment with TAM resulted in increased mitochondrial-mediated apoptotic cell death and overall reduced cell viability [97]. Moreover, we have shown that inhibition of autophagy through beclin-1 shRNA or 3-MA treatment in the ICI resistant, TAM cross-resistant MCF7/LCC9 breast cancer cells partially restored antiestrogen therapy effectiveness [89].
Increased responsiveness of resistant breast cancer cells to anti-estrogen therapy requires concurrent inhibition of both Bcl-2 and beclin-1. Dual inhibition of Bcl-2 by the chemical inhibitor, YC137 and beclin-1 knockdown increases apoptosis and decreases cell survival in response to antiestrogen therapy [89]. These data illustrate that inhibition of autophagy pathway, coupled with Bcl-2 inactivation, is more detrimental to antiestrogen resistant breast cancer cell survival than the individual inhibition of either pathway alone. Thus, dual targeting of synergistic molecular pathways may be beneficial to resensitizing antiestrogen-resistant breast cancers. Investigating the role of estrogen signaling in breast cancer cells by beclin-1 has shown that overexpression of beclin-1 results in decreased growth in response in estrogen, with a decrease in estrogen-regulated genes including c-myc, c-fos and egr 1. These decreases in estrogenic growth by beclin-1 overexpression may appear anti-tumorigenic but beclin-1 overexpression in breast cancer cells also leads to a loss of sensitivity to the antiestrogens raloxifene and TAM, further implying a role for autophagy in promoting antiestrogen resistance [98]. Recent preclinical studies into possible drug combinations to overcome autophagy-mediated TAM resistance suggest a possible therapeutic benefit of combining histone deacetylase inhibitors or proteasomal inhibitors with antiestrogens [99,100]. Treatment of antiestrogen-resistant T47D and MCF7 ER+ breast cancer cells with bortezomib produces a potent induction of cell death and an inhibition of autophagy as measured by p62 and LC3 accumulation, suggesting the benefit of targeting autophagy in antiestrogen-resistant breast cancer [99]. Taken together, these data clearly support a role for autophagy in the promotion of antiestrogen resistance in breast cancer.
Since the weight of preclinical data indicates that inhibiting autophagy resensitizes some resistant cancer to specific therapies, it is not surprising that clinical trials targeting autophagy have recently been initiated (Table 4). Since it has been used extensively for the treatment of malaria, safety data for the use of HCQ in humans is already accessible. Clinical trials have been initiated using HCQ in combination with gefitinib in lung cancer, with docetaxel in prostate cancer, with temozolomide in glioblastoma multiforme, with ixabepilone in metastatic breast cancer, and with bortezomib in multiple myeloma. Of particular interest in ER+ breast cancer is a study in ductal carcinoma in situ, in which patients will receive TAM, chloroquine or a combination of both for 3 months before surgical removal of the tumor. It will be of great interest to see whether inhibition of autophagy in combination with TAM treatment reduces the growth and invasiveness of these breast tumors. The results of the clinical trials listed in Table 4 should hold promising answers to some of the questions pertaining to the role of autophagy in cancer. Moreover, several groups have recently investigated possible small molecular regulators of autophagy through LC3-GFP imaged-based high-throughput screening [101,102]. It is interesting to note that several of these autophagy modulators, either autophagy inducers or autophagy inhibitors, are already US FDA approved for the treatment of various diseases including cardiovascular disorders, schizophrenia and irritable bowel syndrome.
Table 4.
Selected ongoing clinical trials inhibiting autophagy in cancer.
| Cancer type | Treatment | Study phase | ClinicalTrials.gov identifier [201] |
|---|---|---|---|
| Glioblastoma multiforme | Hydroxychloroquine, radiation, temozolomide | I/II | NCT00486603 |
| Multiple myeloma | Hydroxychloroquine, bortezomib | I/II | NCT00568880 |
| Advanced non-small-cell lung cancer | Hydroxychloroquine, carboplatin, paclitaxel, bevacizumab | I/II | NCT00728845 |
| Breast (metastatic) | Hydroxychloroquine, ixabepilone | I/II | NCT00765765 |
| Colorectal (metastatic) | Hydroxychloroquine, capecitabine, oxaliplatin, bevacizumab | II | NCT01006369 |
| Prostate | Hydroxychloroquine | II | NCT00726596 |
| Renal cell carcinoma | Hydroxychloroquine, surgery | I | NCT01144169 |
| Breast (DCIS) | Chloroquine, tamoxifen | I/II | NCT01023477 |
| Prostate (metastatic) | Hydroxychloroquine, docetaxel | I/II | NCT00786682 |
| Lung | Hydroxychloroquine, gefitinib | I/II | NCT00809237 |
DCIS: Ductal carcinoma in situ.
Expert commentary
Targeting autophagy, particularly when it is acting in a survival mode, has significant potential to lead to the development of novel agents and therapeutic regimens. Existing data already suggest that this could be beneficial in combination with both cytotoxic chemotherapy and with endocrine therapy in some cancers. While it is difficult to predict the outcomes of early trials using ‘first-generation’ inhibitors such as HCQ, the field is ripe for the development of more specific inhibitors or combinations of new inhibitors. Outcomes from the early trials indicated in Table 4 should begin to offer powerful new insights into these exciting opportunities.
Longer term success in targeting autophagy may require the development of a greater understanding of the signaling that both regulates and executes autophagy. While the basic machinery for its execution is defined in normal systems such as yeast, whether this provides an adequate definition of how autophagy signaling is present or altered in different human cancers remains unclear. Perhaps the greatest opportunity will lie in the identification of cancer-specific modifications in the regulatory signaling, rather than in the execution machinery. Such knowledge may best be obtained by the development of useful computational and/or mathematical models of the signaling-control mechanisms [15].
Five-year view
Greater detail on the control signaling of autophagy will likely emerge and provide new insights into how the extent and duration of prosurvival autophagy are regulated to allow cancer cells to survive for prolonged periods in the presence of natural (nutrient deprivation or immunologic suppression of growth leading to dormancy) or imposed (therapeutic intervention leading to resistance) stress. Data from clinical trials will show some evidence for the activity of autophagy inhibitors, although the full value of this will await a better understanding of the redundancy in the signals controlling autophagy and the development of combination regimens that address this redundancy. Considering the complexity of this mechanism, systems biology-based approaches will generate the most useful insights, and initial computational and/or mathematical models of autophagy regulation and execution will emerge. While the true clinical potential will likely take longer than 5 years to realize, in part owing to the time needed for clinical follow-up and adequate outcome measures, interest and excitement in this field seems certain to rise substantially within the next 5 years.
Key issues.
With over 70% of all breast cancer cases being estrogen receptor-α-positive, endocrine therapy remains the primary treatment for these breast cancer patients.
Many breast tumors that initially respond to antiestrogen treatments eventually develop acquired resistance; preventing and overcoming antiestrogen resistance remain important clinical goals.
Autophagy, the processes of ‘self-eating’, can enable cell survival in adverse environmental conditions, including nutrient deprivation and hypoxia.
Several cancer therapies induce autophagy, such as radiation, temozolomide, cytotoxic drugs, antiestrogens and aromatase inhibitors.
Inhibitors of autophagy restore antiestrogen sensitivity in endocrine-resistant breast cancer cells growing in vitro.
Clinical trials involving autophagy inhibitors in combination with endocrine or cytotoxic therapies are now being initiated to study the role of autophagy in the survival and progression of cancers.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
Katherine Clark is the recipient of an NIH training grant (grant no. 5-T32-CA009686). This research was supported in part by awards from the Department of Defense Breast Cancer Research Program (BC073977) and from the US Department of Health and Human Services (R01-CA131465; U54-CA149147 and 9XS194 In Silico Research Centers of Excellence) to Robert Clarke. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Center MM, Ward E, et al. Cancer occurrence. Methods Mol Biol. 2009;471:3–29. doi: 10.1007/978-1-59745-416-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Fernandez LM. Use of statistics to assess the global burden of breast cancer. Breast J. 2006;12(Suppl 1):S70–S80. doi: 10.1111/j.1075-122X.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 4.Clarke M, Collins R, Davies C, et al. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351(9114):1451–1467. [PubMed] [Google Scholar]
- 5.Doggrell SA. Is long-term adjuvant treatment of breast cancer with anastrozole indicated? Expert Opin Pharmacother. 2008;9(9):1619–1622. doi: 10.1517/14656566.9.9.1619. [DOI] [PubMed] [Google Scholar]
- 6.Howell A. Pure oestrogen antagonists for the treatment of advanced breast cancer. Endocr Relat Cancer. 2006;13(3):689–706. doi: 10.1677/erc.1.00846. [DOI] [PubMed] [Google Scholar]
- 7.Howell A. Fulvestrant (‘Faslodex’): current and future role in breast cancer management. Crit Rev Oncol Hematol. 2006;57(3):265–273. doi: 10.1016/j.critrevonc.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Robertson JF. Fulvestrant (Faslodex) – how to make a good drug better. Oncologist. 2007;12(7):774–784. doi: 10.1634/theoncologist.12-7-774. [DOI] [PubMed] [Google Scholar]
- 9.Highlights from the Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) 2005–2006 worldwide overview. Breast Cancer Res Treat. 2006;100:S19. No authors listed. [Google Scholar]
- 10.Abe O, Abe R, Enomoto K, et al. Polychemotherapy for early breast cancer: an overview of the randomised trials. Lancet. 1998;352(9132):930–942. [PubMed] [Google Scholar]
- 11.Clarke R, Skaar TC, Bouker KB, et al. Molecular and pharmacological aspects of antiestrogen resistance. J Steroid Biochem Mol Biol. 2001;76(1–5):71–84. doi: 10.1016/s0960-0760(00)00193-x. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez MC, Detre S, Johnston S, et al. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor HER-2, and p38 mitogen-activated protein kinase. J Clin Oncol. 2005;23(11):2469–2476. doi: 10.1200/JCO.2005.01.172. [DOI] [PubMed] [Google Scholar]
- 13.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 14.Clarke R, Leonessa F, Welch JN, et al. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharm Rev. 2001;53(1):25–71. [PubMed] [Google Scholar]
- 15•.Clarke R, Shajahan AN, Wang Y, et al. Endoplasmic reticulum stress, the unfolded protein response, and gene network modeling in antiestrogen resistant breast cancer. Hormone Mol Biol Clin Invest. 2011;5(1):35–44. doi: 10.1515/hmbci.2010.073. Gives a broad overview on the molecular mechanisms of endocrine resistance in breast cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Clarke R, Shajahan AN, Riggins RB, et al. Gene network signaling in hormone responsiveness modifies apoptosis and autophagy in breast cancer cells. J Steroid Biochem Mol Biol. 2009;114(1–2):8–20. doi: 10.1016/j.jsbmb.2008.12.023. Provides a current perspective on endocrine-resistance mechanisms, such as the unfolded protein response in the endoplasmic reticulum, and the use of gene network modeling to target resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.He CC, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Ann Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. Recent review on gene network modeling of autophagy and apoptosis in antiestrogen resistant breast cancers. This article provides an excellent background on autophagy and gene network models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pursiheimo JP, Rantanen K, Heikkinen PT, et al. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene. 2009;28(3):334–344. doi: 10.1038/onc.2008.392. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 20.Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung CH, Jun CB, Ro SH, et al. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjorkoy G, Lamark T, Pankiv S, et al. Monitoring autophagic degradation of P62/Sqstm1. Methods Enzymol Autophagy Mammalian Syst. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 25.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 26.Criollo A, Maiuri MC, Tasdemir E, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differentiation. 2007;14(5):1029–1039. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 27.Kihara A, Noda T, Ishihara N, et al. Two distinct Vps34 phosphatidylinositol 3–kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Peng ZL. Autophagy, beclin 1, and their relation to oncogenesis. Labmedicine. 2008;39(5):287–290. [Google Scholar]
- 29.Chang CY, Huang WP. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol Biol Cell. 2007;18(3):919–929. doi: 10.1091/mbc.E06-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He CC, Baba M, Klionsky DJ. Double duty of Atg9 self-association in autophagosome biogenesis. Autophagy. 2009;5(3):385–387. doi: 10.4161/auto.5.3.7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legakis JE, Yen WL, He CC, et al. Autophagosome formation involves cycling of ATG9. Autophagy. 2006;2(4):334. [Google Scholar]
- 32.Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3(5):422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- 33.Yen WL, Legakis JE, Nair U, et al. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007;18(2):581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. Extensive review on autophagy and proper assays for monitoring autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jager S, Bucci C, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117(20):4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406(6798):902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 37.Uchiyama Y. Autophagic cell death and its execution by lysosomal cathepsins. Arch Histol Cytol. 2001;64(3):233–246. doi: 10.1679/aohc.64.233. [DOI] [PubMed] [Google Scholar]
- 38.Kitada S, Takayama S, De Riel K, et al. Reversal of chemoresistance of lymphoma cells by antisense-mediated reduction of bcl-2 gene expression. Antisense Res Dev. 1994;4(2):71–79. doi: 10.1089/ard.1994.4.71. [DOI] [PubMed] [Google Scholar]
- 39.Reed JC, Kitada S, Takayama S, et al. Regulation of chemoresistance by the bcl-2 oncoprotein in non-Hodgkin’s lymphoma and lymphocytic leukemia cell lines. Ann Oncol. 1994;5(Suppl 1):61–65. doi: 10.1093/annonc/5.suppl_1.s61. [DOI] [PubMed] [Google Scholar]
- 40.Won KY, Kim GY, Kim YW, et al. Clinicopathologic correlation of beclin-1 and bcl-2 expression in human breast cancer. Hum Pathol. 2010;41(1):107–112. doi: 10.1016/j.humpath.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Erlich S, Mizrachy L, Segev O, et al. Differential interactions between Beclin 1 and bcl-2 family members. Autophagy. 2007;3(6):561–568. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 42.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X-L and a BH3-like domain in Beclin-1. Embo J. 2007;26(10):2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pattingre S, Tassa A, Qu XP, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Abedin MJ, Wang D, McDonnell MA, et al. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differentiation. 2007;14(3):500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 45.Morselli E, Galluzzi L, Kepp O, et al. Anti- and pro-tumor functions of autophagy. Biochim Biophys Acta Mol Cell Res. 2009;1793(9):1524–1532. doi: 10.1016/j.bbamcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Elgendy M, Sheridan C, Brumatti G, et al. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42(1):23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 47.Germain M, Nguyen AP, Le Grand JN, et al. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. Embo J. 2011;30(2):395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Brien SM, Claxton DF, Crump M, et al. Phase I study of obatoclax mesylate (GX15-070), a small molecule pan-Bcl-2 family antagonist, in patients with advanced chronic lymphocytic leukemia. Blood. 2009;113(2):299–305. doi: 10.1182/blood-2008-02-137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akar U, Chaves-Reyez A, Barria M, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4(5):669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- 50.Gao P, Bauvy C, Souquere S, et al. The Bcl-2 homology domain 3 mimetic gossypol induces both Beclin 1-dependent and Beclin 1-independent cytoprotective autophagy in cancer cells. J Biol Chem. 2010;285(33):25570–25581. doi: 10.1074/jbc.M110.118125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shajahan AN, Riggins RB, Clarke R. The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 2009;22(5):241–246. doi: 10.1358/dnp.2009.22.5.1378631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez BP, Riggins RB, Shajahan AN, et al. Human X-box binding protein-1 confers both estrogen independence and antiestrogen resistance in breast cancer cell lines. FASEB J. 2007;21(14):4013–4027. doi: 10.1096/fj.06-7990com. [DOI] [PubMed] [Google Scholar]
- 53.Davies MP, Barraclough DL, Stewart C, et al. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer. 2008;123(1):85–88. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]
- 54.Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26(24):9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei YJ, Pattingre S, Sinha S, et al. JNK1-mediated phosphorylation of BcI-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brech A, Ahlquist T, Lothe RA, et al. Autophagy in tumour suppression and promotion. Mol Oncol. 2009;3(4):366–375. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta Mol Cell Res. 2009;1793(9):1516–1523. doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 59•.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15(17):5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. Comprehensive review of the role of autophagy in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy. 2007;3(6):610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenberg-Lerner A, Bialik S, Simon HU, et al. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differentiation. 2009;16(7):966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 63.Qu XP, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yue ZY, Jin SK, Yang CW, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miracco C, Cosci E, Oliveri G, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30(2):429–436. [PubMed] [Google Scholar]
- 66.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8(7):688–694. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 67.Marino G, Salvador-Montoliu N, Fueyo A, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/Autophagin-3. J Biol Chem. 2007;282(25):18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 68.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 69.Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy. 2007;3(1):72–74. doi: 10.4161/auto.3438. [DOI] [PubMed] [Google Scholar]
- 70.Harrison B, Kraus M, Burch L, et al. DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J Biol Chem. 2008;283(15):9999–10014. doi: 10.1074/jbc.M706040200. [DOI] [PubMed] [Google Scholar]
- 71.Wang QJ, Ding Y, Kohtz DS, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26(31):8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lemasters JJ, Nieminen AL, Qian T, et al. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366(1–2):177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 73.Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10(6):676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reef S, Zalckvar E, Shifman O, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22(4):463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 75.Colotta F, Allavena P, Sica A, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 76.White E. Role of autophagy in cancer and therapy. EJC Suppl. 2008;6(12):16. [Google Scholar]
- 77.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21(13):1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sivridis E, Koukourakis MI, Zois CE, et al. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am J Pathol. 2010;176(5):2477–2489. doi: 10.2353/ajpath.2010.090049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Negri T, Tarantino E, Orsenigo M, et al. Chromosome band 17q21 in breast cancer: significant association between beclin 1 loss and HER2/NEU amplification. Genes Chromosomes Cancer. 2010;49(10):901–909. doi: 10.1002/gcc.20798. [DOI] [PubMed] [Google Scholar]
- 81.Vazquez-Martin A, Cufi S, Oliveras-Ferraros C, et al. Expression status of the autophagy-regulatory gene ATG6/BECN1 in ERBB2-positive breast carcinomas: bypassing ERBB2-induced oncogenic senescence to regulate the efficacy of ERBB2-targeted therapies. Genes Chromosomes Cancer. 2011;50(4):284–290. doi: 10.1002/gcc.20846. [DOI] [PubMed] [Google Scholar]
- 82.Tu YF, Kaipparettu BA, Ma Y, et al. Mitochondria of highly metastatic breast cancer cell line MDA-MB-231 exhibits increased autophagic properties. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbabio.2011.04.015. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 83.Vanderlaag K, Su Y, Frankel AE, et al. 1, 1-Bis(3′-indolyl)-1-(p-substituted phenyl) methanes induce autophagic cell death in estrogen receptor negative breast cancer. BMC Cancer. 2010;10:669. doi: 10.1186/1471-2407-10-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou J, Zhang W, Liang B, et al. PPARgamma activation induces autophagy in breast cancer cells. Int J Biochem Cell Biol. 2009;41(11):2334–2342. doi: 10.1016/j.biocel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sinha S, Roy S, Reddy BS, et al. A lipid-modified estrogen derivative that treats breast cancer independent of estrogen receptor expression through simultaneous induction of autophagy and apoptosis. Mol Cancer Res. 2011;9(3):364–374. doi: 10.1158/1541-7786.MCR-10-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen S, Rehman S, Zhang W, et al. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010;1806:220–229. doi: 10.1016/j.bbcan.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 87.Kondo Y, Kanzawa T, Sawaya R, et al. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5(9):726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 88.Turcotte S, Giaccia AJ. Targeting cancer cells through autophagy for anticancer therapy. Current Opin Cell Biol. 2010;22(2):246–251. doi: 10.1016/j.ceb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crawford AC, Riggins RB, Shajahan AN, et al. Co-inhibition of BCL-W and BCL2 restores antiestrogen sensitivity through BECN1 and promotes an autophagy-associated necrosis. Plos One. 2010;5(1):e8604. doi: 10.1371/journal.pone.0008604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ito H, Daido S, Kanzawa T, et al. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26(5):1401–1410. [PubMed] [Google Scholar]
- 91.Sun WL, Chen J, Wang YP, et al. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7(10):1033–1042. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 92.Milani M, Rzymski T, Mellor HR, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with bortezomib. Cancer Res. 2009;69(10):4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 93.Chen S, Li X, Feng J, et al. Autophagy facilitates the lapatinib resistance of HER2 positive breast cancer cells. Med Hypotheses. 2011;77(2):206–208. doi: 10.1016/j.mehy.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 94.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. Plos One. 2009;4(7):e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95•.Bursch W, Ellinger A, Kienzl H, et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17(8):1595–1607. doi: 10.1093/carcin/17.8.1595. First study to show a role of macroautophagy in antiestrogen therapy resistance in breast cancer. [DOI] [PubMed] [Google Scholar]
- 96••.Samaddar JS, Gaddy VT, Duplantier J, et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7(9):2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. First study to show that macroautophagy stimulation protected breast cancer cells against tamoxifen-mediated cell death and promoted antiestrogen resistance. [DOI] [PubMed] [Google Scholar]
- 97••.Qadir MA, Kwok B, Dragowska WH, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112(3):389–403. doi: 10.1007/s10549-007-9873-4. Showed that macroautophagy inhibition promoted tamoxifen resistance breast cancer cells to antiestrogen therapy and enhanced cell death through mitochondria depolarization. [DOI] [PubMed] [Google Scholar]
- 98.John S, Nayvelt I, Hsu HC, et al. Regulation of estrogenic effects by Beclin 1 in breast cancer cells. Cancer Res. 2008;68(19):7855–7863. doi: 10.1158/0008-5472.CAN-07-5875. [DOI] [PubMed] [Google Scholar]
- 99.Periyasamy-Thandavan S, Jackson WH, Samaddar JS, et al. Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress and induces caspase-dependent cell death in antiestrogen-sensitive and resistant ER+ breast cancer cells. Autophagy. 2010;6(1):19–35. doi: 10.4161/auto.6.1.10323. [DOI] [PubMed] [Google Scholar]
- 100.Thomas S, Thurn KT, Bicaku E, et al. Addition of a histone deacetylase inhibitor redirects tamoxifen-treated breast cancer cells into apoptosis, which is opposed by the induction of autophagy. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1364-y. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shu CW, Madiraju C, Zhai D, et al. High-throughput fluorescence assay for small-molecule inhibitors of autophagins/Atg4. J Biomol Screen. 2011;16(2):174–182. doi: 10.1177/1087057110392996. [DOI] [PubMed] [Google Scholar]
- 102.Zhang L, Yu J, Pan H, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA. 2007;104(48):19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 201.ClinicalTrials.gov. http://clinicaltrials.gov.