Abstract
Methotrexate (MTX) has emerged as first-line therapy for early moderate to severe rheumatoid arthritis (RA), but individual variation in treatment response remains unexplained. We tested the associations between 863 known pharmacogenetic variants and MTX response in 471 TEAR Trial participants with early RA. Efficacy and toxicity were modeled using multiple regression, adjusted for demographic and clinical covariates. Penalized regression models were used to test joint associations of markers and/or covariates with the outcomes. The strongest genetic associations with efficacy were in CHST11 (five markers with P <0.003), encoding carbohydrate (chondroitin 4) sulfotransferase 11. Top markers associated with MTX toxicity were in the cytochrome p450 genes CYP20A1 and CYP39A1, solute carrier genes SLC22A2 and SLC7A7, and the mitochondrial aldehyde dehydrogenase gene ALDH2. The selected markers explained a consistently higher proportion of variation in toxicity than efficacy. These findings could inform future development of personalized therapeutic approaches.
Keywords: Methotrexate, rheumatoid arthritis, pharmacogenetics
Introduction
Methotrexate (MTX) is a disease-modifying anti-rheumatic drug (DMARD) commonly used as a first-line agent in the treatment of rheumatoid arthritis (RA).1,2 There is considerable inter-individual heterogeneity in response to MTX, both in treatment efficacy and associated toxicity. While smoking, female sex, younger age, longer disease duration, and the presence of serum rheumatoid factor (RF) are associated with poor response to MTX therapy, they explain a rather low proportion of response variability, suggesting a role for pharmacogenetic factors.3,4 Consistent with that hypothesis, several reports identified polymorphisms in the MTX metabolic pathway to be significantly associated with treatment outcomes.5,6 However, much of the currently available genetic evidence remains conflicting and limited to a small number of candidate genes involved in the mechanism of MTX action, i.e. the major histocompatibility locus including HLA–DRB1 and LTA–TNF as well as purine, folate, and adenosine pathways.7–10
Genetic determinants of MTX response can be evaluated early in the course of RA, when pharmacologic intervention confers maximum benefits. Thus, identification and validation of new markers has important clinical implications, including development of personalized therapeutic approaches that result in greater efficacy and lower toxicity. To that end, this study used the Affymetrix Drug Metabolism, Excretion, and Transport (DMET) Plus Platform to comprehensively examine the role of genetic variation in key pharmacokinetic pathways in MTX efficacy and toxicity among participants of the Treatment of Early Aggressive Rheumatoid Arthritis Trial (TEAR). The early RA phenotype of the TEAR cohort is uniquely suited for exploring pharmacogenetic associations as improvements associated with DMARD therapy are on average greater for participants with early rather than advanced disease, potentially increasing statistical power and informing future clinical interventions.11
Patients and Methods
Study population
The TEAR Trial (n=755), described in previous publications,12, 13, 14 is a 2-year, double-blind, active control, multicenter Phase IV clinical trial in patients with early (<3 years disease duration) RA characterized by an “aggressive” clinical phenotype, defined as autoantibody positivity or the presence of erosions on radiographs of hands and feet. Using a 2 × 2 factorial design, the TEAR Trial compared two treatment strategies (early intensive therapy versus step-up therapy) and two combinations of medications (etanercept (ETN) plus MTX versus hydroxychloroquine (HCQ) plus sulfasalazine (SSZ) plus MTX). After 24 weeks of treatment, participants randomized to MTX monotherapy were “stepped up” to either oral triple therapy or MTX+ETN if their DAS28 exceeded 3.2. All participants received a daily 1 mg supplement of folic acid. If participants developed toxicity to MTX or SSZ, the drug was discontinued or the dosage was decreased at the discretion of the treating physician. If the treatment changes resolved the toxicity in 2 weeks, the drug was continued at that dose.
Eligibility criteria were reported in detail in prior publications.12 Briefly, entry criteria included age of 18 years or older; duration of disease <3 years; RA diagnosis by American College of Rheumatology (ACR) criteria;15 have active disease at the time of screening, defined as at least 4 swollen and 4 tender joints due to RA (using the 28 joint count) and either the presence of erosions or positive rheumatoid factor; if taking corticosteroids, receiving stable doses (≤10 mg/day of prednisone) at least 2 weeks prior to screening; if taking non-steroidal anti-inflammatory drugs, receiving stable doses for at least 1 week prior to screening. Participants were excluded if they were pregnant or lactating; had contraindications to study medications; received corticosteroid injections during the 4 weeks prior to screening; had a diagnosis of serious infection; or used biologic therapy. Mean disease duration ranged from 2.9 to 4.5 months across study arms and RF seropositivity approached 90%. Prior use of leflunomide, HCQ, and SSZ was allowable if for no more than 2 months, as was a total dose of ≤ 40 mg of MTX. The TEAR Trial was registered with clinicaltrials.gov (NCT00259610) and approved by the appropriate IRB committees.
The primary outcome of the TEAR Trial was defined as the mean Disease Activity Score on 28 joints (DAS28) from weeks 48 to 102, with secondary endpoints defined by ACR criteria for improvement (ACR20, ACR50, and ACR70) as assessed by a questionnaire and radiographic evidence of joint damage. DAS28 has been previously validated as an outcome measure for RA disease activity.16 Participants were evaluated during clinic visits at 6-week intervals for the first 48 weeks and at 12-week intervals thereafter. Drug toxicity was assessed at 6-week intervals via laboratory measures and a self-report of adverse events at each scheduled visit. Because participants were given a folic acid supplement, the risk of adverse events was reduced, limiting our statistical power to consider different classes of toxicity outcomes. Thus, we have grouped together all instances of adverse events, including infections (i.e. upper respiratory, urinary tract, or general infection), as well as gastrointestinal, oral, and skin conditions.
Of the 755 participants, 630 consented to genotyping and provided DNA. Of those, 471 had complete information on adverse events and covariates, and were included in the analysis.
Genotyping
Genomic DNA was extracted from a whole blood sample using the PureGene system (Gentra Systems, Minneapolis, MN, USA). The DMET platform (Affymetrix, Santa Clara, CA, USA) enables multiplexed genotyping of 1,936 markers in 225 genes previously determined to have functional significance in phase I and phase II drug metabolism enzymes and drug transporters. Genotyping was carried out using methods as described by the manufacturer.17
Genotype calls were made using the DMET Command Console. Samples were considered passed or in bounds if they had genotyping calls of >95%. Samples that were out of bounds or had genotyping calls of ≤95% were re-run. Of the original 1936 markers, 1931 were successfully genotyped; of those, 1068 were found to be monomorphic in the study population and removed, leaving 863 markers in 224 genes in the analysis.
Statistical analysis
We tested the hypothesis that genes represented by DMET chip SNPs are associated with MTX efficacy and toxicity using two different approaches. The first approach was a conventional multiple regression model of the association between genetic markers from the DMET chip and DAS28 at 24 weeks, adjusted for the baseline DAS28, treatment arm, race (European American, African American, or other), sex, age (as linear and quadratic terms), and smoking status. The genetic marker effects were assumed to be co-dominant with the genotypes coded as {0, 1, 2} according to the number of minor alleles. Additionally, we conducted sensitivity analyses modeling genotype effects using recessive and dominant modes of inheritance, as well as restricting the analysis to Caucasian participants (data not shown). The 863 markers were tested one at a time. The freely available software for genetic data analysis, PLINK v1.07,18 was used to fit each of the models. We were primarily interested in the P-value from the test of the additional variation explained by the marker vs. a null model, which included all covariates except the marker effect. The same variables were used to test the association of the individual markers on the toxicity outcome except that multiple logistic regression analysis was used as implemented by PLINK.
The issue of adjusting the type 1 error rate to account for multiple testing is not as simple as performing a Bonferroni correction because of the linkage disequilibrium (LD) between markers on the DMET chip. Assuming complete pairwise LD between markers within a gene, we implemented a gene-wise correction for multiple testing, estimating the significance level at α= 0.05/224 genes= 2.2 × 10−4.
The second approach to testing the hypothesis that DMET markers were associated with variation in efficacy and toxicity was to regard the gene discovery analysis as a model selection problem. Because the number of explanatory variables to consider far exceeds the number of genotyped participants, we used a form of penalized regression that shrinks small marker associations while allowing for greater probability of detecting markers with larger associations. In this framework, we tested the hypothesis that the covariates, genes and markers were jointly associated with toxicity or the change in DAS28. To test this hypothesis, we used a regression model that penalizes both individual markers as well as groups of markers, e.g. all markers within a specific gene.19, 20 This approach is a natural application to the DMET chip data because each marker can be grouped into a specific gene. The approach differs from standard penalized regression approaches (e.g. LASSO, ridge), which penalize only the individual markers. The tuning constants, λL and λE, both control the penalty on the marker effects, but λL (lasso penalty) applies to individual markers, and the λE (Euclidian or ridge penalty) applies to groups of markers, i.e., reflecting the combined influence of markers on the outcome across an entire gene.
The key component of the penalized regression model fitting process is choosing the total tuning constant (penalty), λ = λL+λE. The model optimization procedure developed by Wu et al.21 and Zhou et al.22 is controlled by the number of explanatory variables, which in the present case are DMET markers and/or covariates. In general, λ is a decreasing function of the number of explanatory variables, i.e. as the dimension of the model grows the penalty constant is relaxed.21 Given a preset number of explanatory variables and ratio of individual and group penalties λL/λL+λE, the algorithm uses a “bracketing and bisection” strategy that finds a solution for the total tuning penalty. In order to observe the model optimization as the penalty varied, we extracted shrinkage estimates of regression parameters as the number of variables changed from 1 to 20. The order of entry of markers/covariates into the model provides a rubric for selecting an appropriately sized model, which a priori is expected to contain markers from a small proportion of the 224 analyzed genes from the DMET array. The models were fit using the software Mendel, following the methods outlined by Zhou et al.22
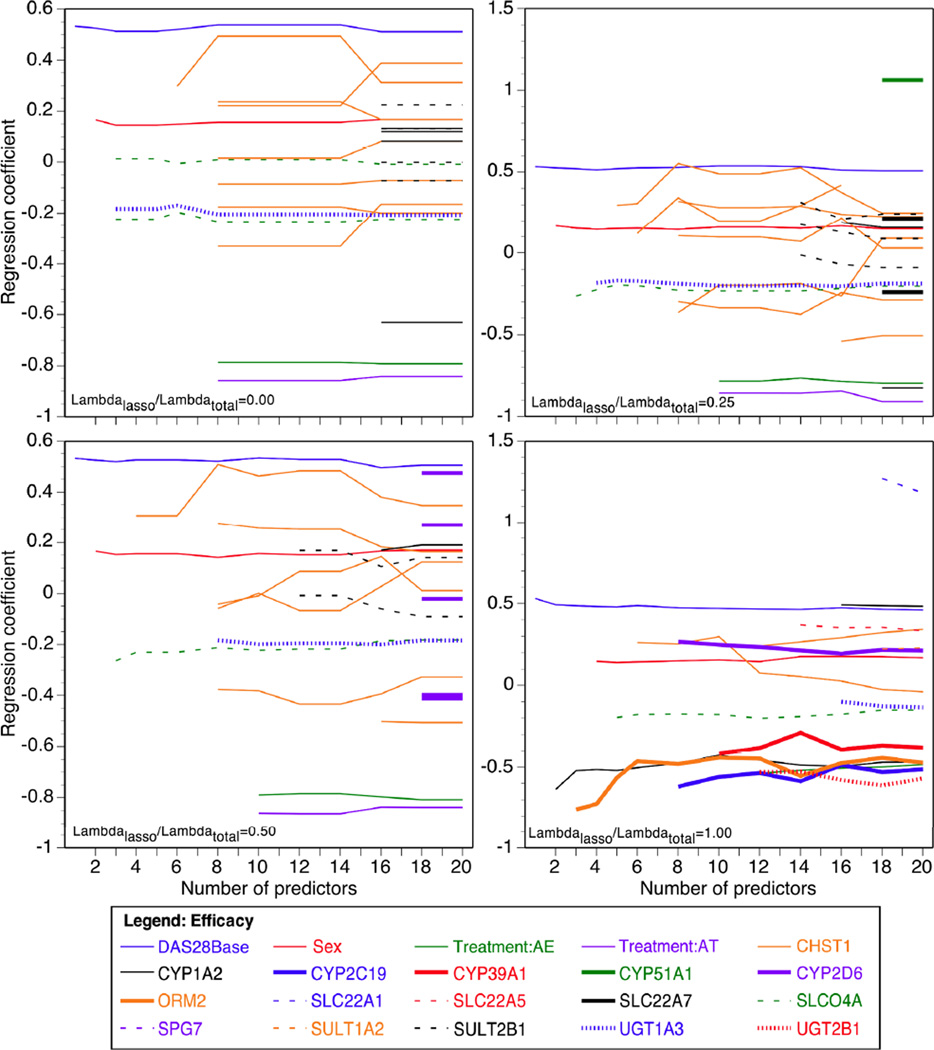
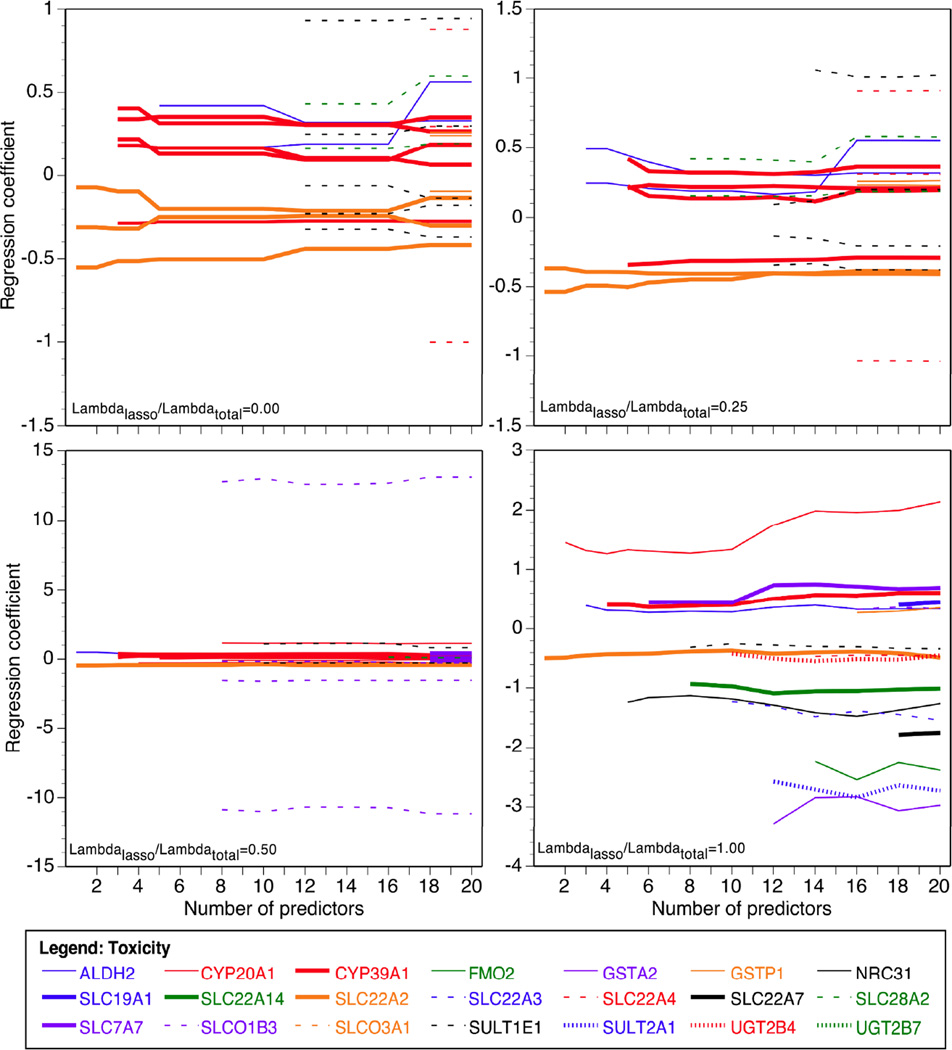
The purpose of the penalties λL, λE is to limit the actual number of variants, because clearly a small minority of the 863 markers is expected to be important. Here we do not consider models with more than 20 variables, including markers and covariates. Because simulation studies suggest that penalizing both is the optimal choice,22 we examined the robustness of our findings by fitting models with the ratio of individual penalty to total penalty λL/ (λL+λE) of 1 (pure lasso penalty), 0.5, 0.25, and 0 (pure group penalty) (Figures 1 and 2). The models were fit using the software Mendel.23
Figure 1.
Genetic variants selected by group lasso penalized regression for entrance into the additive model of methotrexate efficacy in TEAR participants. Panels show the following proportions of lasso to total penalty: λL/λ= 0 (pure group penalty), λL/λ=0.25, λL/λ=0.5, and λL/λ=1 (pure lasso penalty).
Figure 2.
Genetic variants selected by group lasso penalized regression for entrance into the additive model of methotrexate-associated adverse events in TEAR participants. Panels show the following proportions of lasso to total penalty: λL/λ= 0 (pure group penalty), λL/λ=0.25, λL/λ=0.5, and λL/λ=1 (pure lasso penalty).
Finally, multiple regression was used to compare the proportion of variance in efficacy and toxicity explained by the markers selected from the penalized regression analysis with 5 allowed variables and penalties of λL/ (λL+λE) = 1 or 0.5. These models were fit using R software.24
Results
Demographic and clinical characteristics of the study population are listed in Table 1. In this analysis, the proportion of patients in each treatment arm did not differ from that in the parent study.12 The majority of participants were female and of self-reported European ancestry, with a mean age of 50 years and body mass index of 30 (obese). The average decrease in DAS28 from baseline to week 24 across treatment arms was 1.9 units.
Table 1.
General characteristics of the study population (n=471).
| Variable | ||
|---|---|---|
| Age, years | 49.6 ± 12.6 | |
| Female, n (%) | 340 (72) | |
| Treatment | ||
| Early intensive therapy—ETN, n (%) | 157 (33) | |
| Early intensive therapy—SSZ + HCQ, n (%) | 81 (17) | |
| Step-up therapy, n (%) | 233 (49) | |
| Race | ||
| Caucasian, n (%) | 374 (79) | |
| African American, n (%) | 51 (11) | |
| Other, n (%) | 46 (10) | |
| Body mass index, kg/m2 | 30.2 ± 7.5 | |
| DAS28 at baseline, points | 5.8 ± 1.0 | |
| DAS28 at 24 weeks, points | 3.9 ± 1.5 | |
| Experienced any side effects or infection, n (%) | 168 (36) | |
In single marker analyses of MTX efficacy, the DAS28 score at 24 weeks was associated with 42 out of 863, or 4.8%, of the markers at a P<0.05. However, no marker was statistically significant after Bonferroni correction at the marker or gene levels. Five of the top six associated with DAS28 were in CHST11, which encodes carbohydrate (chondroitin 4) sulfotransferase 11. For all these markers, the minor allele was associated with greater MTX efficacy, as indicated by the change in DAS28 from baseline to 24 weeks. Modeling the genotype as dominant vs. additive or restricting the data set to Caucasian participants only did not appreciably change the estimates of association (data not shown).
Table 2 shows the marker groups and genes selected by the penalized regression model listed by order of entrance into the model under varying proportions of lasso to overall penalty. In all four penalized regression models, the baseline DAS28 score was selected first, followed by sex in all models except with the pure lasso (i.e. marker only, λL/λ=1) penalty. The percentage of overall variance in efficacy explained by DMET chip markers when 5 variables were allowed in the model was 2% and 1% with λL/λ = 0.5 and 1.0, respectively.
Table 2.
Top groups of variables selected under varying ratios of lasso to total penalties and an additive model of methotrexate efficacy in TEAR participants.
| λL/λ | First | Second | Third | Fourth |
|---|---|---|---|---|
| 1 (pure lasso penalty) | DAS28 at baseline |
CYP1A21 rs2069514 |
ORM2 rs2636889 |
Sex |
| 0.5 | DAS28 at baseline |
Sex |
SLCO4A1 rs2236553 |
CHST11 rs903247 rs2463437 rs2463018 rs2468110 rs7847 |
| 0.25 | DAS28 at baseline |
Sex |
SLCO4A1 rs2236553 |
UGT1A3 rs6706232 |
| 0 (pure group penalty) | DAS28 at baseline |
Sex | SLCO4A1 | UGT1A3 |
Genes listed in the table encode the following proteins: ORM2—orosomucoid 2; CYP1A2—cytochrome p450 family 1 subfamily A polypeptide 2; SLCO4A1— solute carrier organic anion transporter family member 4A1; UGT1A3—UDP glucuronosyltransferase 1 family polypeptide A3; CHST11— carbohydrate (chondroitin 4) sulfotransferase 11.
In single marker analyses adjusted for multiple testing, toxicity was associated with 43 out of 863, or 5.0%, of the markers with P< 0.05. No single marker was associated after the conservative multiple testing correction. Similarly to the efficacy analyses, the top hits did not vary depending on the assumed mode of inheritance or by racial group (Caucasians vs. the total study population). In contrast to efficacy, non-genetic covariates such as age, race, and treatment were not among the top variables included in the penalized regression model of MTX toxicity. Notably, one of the top hits for toxicity, rs671 in the ALDH2 gene, was significantly out of Hardy-Weinberg equilibrium in our population (P-value= 8×10−35), while other SNPs in the same gene were not. The percentage of overall variance in toxicity explained by DMET chip markers when 5 variables were allowed in the model was more than threefold higher than for efficacy when λL/λ=0.5 (5% vs. 2%) and twice as high under the pure lasso penalty (9% vs. 1%), suggesting that functional variants such as those included on the DMET array may play a larger role in explaining the heterogeneity in MTX treatment toxicity rather than efficacy.
Discussion
Using the DMET genotyping array, we have evaluated associations between a set of known pharmacogenetic variants and MTX response in patients with early RA enrolled in the TEAR Trial. Of these, the ALDH2 finding is of particular interest because evidence suggests that in vitro human aldehyde dehydrogenase activity is highly variable, and that genetic variation in enzymes that catalyze the conversion of MTX to 7-OH-MTX is associated with two phenotypes of MTX metabolism among RA patients.25, 26 In single marker analysis, the presence of one copy of the minor allele at the rs886205 locus was associated with a 42% reduction in the relative odds of adverse events. One possible explanation is that the variant genotype is associated with higher enzymatic activity in the formation of 7-OH-MTX from MTX, reduced amount of cellular MTX, and therefore reduced MTX toxicity. Under that hypothesis, a positive relation would also be expected between the dosage of the minor allele at the rs886205 locus and MTX efficacy; however, the association with the DAS28 at 24 weeks observed in our study population was negative and did not reach statistical significance in the single marker analysis.
Associations with solute carrier genes were observed for both efficacy and toxicity outcomes. These results are notable as SLC19A1, which encodes solute carrier family 19 member A1 and belongs to the same family of genes as SLCO4A1, SLC22A2 and SLC28A2, plays a crucial role in the pharmacokinetics of both MTX and 7-OH-MTX by transporting them into the cell.27 Accordingly, several studies including a genome-wide scan have linked polymorphisms in SLC19A1 and SLCO1B3 to MTX treatment outcomes in childhood cancers.28, 29 Our study is the first to report associations between SLCO4A1, SLC22A2, SLC28A2, and MTX response; future investigations are warranted to consider associations between other solute carrier genes and response to anti-folate drugs.
One of the strengths of the penalized regression analysis lies in its ability to identify subsets of features, the number of which on the DMET chip far exceeds the available sample size, that explain the most amount of variability in the outcomes. For example, we established that non-genetic covariates were consistently selected first in the models of MTX efficacy, in accordance with other published algorithms.30, 31 Furthermore, our findings suggest that the polymorphisms contained on the DMET chip explain at least twice the amount of variance in the toxicity outcome when compared to the efficacy outcome. This is consistent with prior studies32, 33 of RA patients that reported statistically significant associations between several genetic polymorphisms in the folate metabolic pathway and MTX treatment toxicity but not efficacy. Possible explanations for this discrepancy include: 1) differential distribution of genetic effects by outcome, e.g. many small effects for efficacy vs. larger effects for toxicity and 2) lower heritability of MTX treatment efficacy, which could be due to higher phenotypic variance for a given amount of additive genetic variance.34 Finally, the variants associated with MTX treatment efficacy may simply be not as well represented on the DMET array as those that are important for toxicity. Future studies, including genome-wide scans, are warranted to explore the differential genetic architecture of MTX efficacy and toxicity.
Other strengths of our study lie in its uniquely comprehensive ascertainment of genetic variation typed in participants of the TEAR Trial, novelty of the statistical approach, and consistency between the results of single marker and penalized regression analyses. However, several limitations are germane to interpreting our findings. First, the folic acid supplement that was provided to all participants reduces the risk of MTX toxicity, thus lowering the number of adverse events and limiting the power of the toxicity analyses.35 Second, the DMET chip does not include markers in several important genes (e.g. those in the folate pathway) that are likely to explain additional variability in MTX response. Third, although RF status has been shown to be significantly associated with treatment response, we were not able to evaluate it as a modifier of the association between genetic markers and the outcomes because almost 90% of the TEAR participants were RF-positive. Fourth, as the majority of TEAR participants were of self-reported European ancestry, the results may not be generalizable to other populations given the evidence of effect modification by racial group.36 Finally, although we were able to confirm that our findings were robust to modeling the genotype effects as either dominant or additive, we could not evaluate them against the recessive mode of inheritance because the low minor allele frequency (<0.05) of almost half of the DMET chip markers resulted in the extremely low number of homozygote individuals, and thus inadequate statistical power. In conclusion, we have confirmed associations with known pharmacogenetic markers in the context of early RA MTX therapy. Future investigations can build on our findings to develop personalized therapeutic approaches for early aggressive RA.
Table 3.
Top groups of DMET platform SNPs selected under varying ratios of lasso to total penalties and an additive model of methotrexate toxicity in TEAR participants.
| λL/λ | First | Second | Third | Fourth |
|---|---|---|---|---|
| 1 (pure lasso penalty) |
SLC22A22 rs624249 |
CYP20A1 rs2043449 |
ALDH2 rs886205 |
CYP39A1 rs7761731 |
| 0.5 |
SLC22A2 rs316019 rs624249 |
ALDH2 rs886205 |
CYP39A1 rs9369629 rs2277119 rs953062 rs7761731 |
CYP20A1 rs1048013 rs2043449 |
| 0.25 |
SLC22A2 rs316019 rs624249 |
ALDH2 rs886205 rs671 |
CYP39A1 rs9369629 rs2277119 rs953062 rs7761731 |
SLC28A2 rs10519020 rs1060896 |
| 0 (pure group penalty) | SLC22A2 | CYP39A1 | ALDH2 | SLC28A2 |
Genes listed in the table encode the following proteins: SLC22A2—solute carrier family 22 member 2; CYP20A1—cytochrome p450 family 20 subfamily A polypeptide 1; ALDH2—aldehyde dehudrogenase 2; CYP39A1—cytochrome p450 family 39 subfamily A polypeptide 1; SLC28A2—solute carrier family 28 member 2.
Acknowledgments
We are grateful to the Heflin Genomics Core Laboratory at the University of Alabama at Birmingham for conducting the genotyping of the TEAR Trial participants using the DMET array. This work was funded by the following grants: NIH P60 AR048095 (R.P. Kimberly, PI: D.K. Arnett, Project PI) and ARRA supplement 3P60AR048095-07S1; NIH R01 AR052658 (S. Louis Bridges, Jr., PI); University of Alabama at Birmingham Center for Clinical and Translational Science through the NIH National Center for Research Resources as part of its Clinical and Translational Science Award Program (5UL1RR025777-03, 5KL2RR025776-03, 5TL1RR025775-03); University of Alabama at Birmingham Comprehensive Cancer Center Core Grant 5P30-CA13148-38 (Genotyping), K01 AR060848 (Richard J. Reynolds, PI), and Amgen (the parent TEAR trial).
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
References
- 1.Saag KG, Teng GG, Patkar NM, Anuntiyo J, Finney C, Curtis JR, et al. American College of Rheumatology 2008 recommendations for the use of nonbiologic and biologic disease-modifying antirheumatic drugs in rheumatoid arthritis. Arthritis Rheum. 2008;59:762–784. doi: 10.1002/art.23721. [DOI] [PubMed] [Google Scholar]
- 2.Smolen JS, Landewe R, Breedveld FC, Dougados M, Emery P, Gaujoux-Viala C, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs. Ann Rheum Dis. 2010;69:964–975. doi: 10.1136/ard.2009.126532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drouin J, Haraoui B 3e Initiative Group. Predictors of clinical response and radiographic progression in patients with rheumatoid arthritis treated with methotrexate monotherapy. J Rheumatol. 2010;37:1405–1410. doi: 10.3899/jrheum.090838. [DOI] [PubMed] [Google Scholar]
- 4.Saevarsdottir S, Wallin H, Seddighzadeh M, Ernestam S, Geborek P, Petersson IF, et al. Predictors of response to methotrexate in early DMARD naïve rheumatoid arthritis: results from the initial open-label phase of the SWEFOT trial. Ann Rheum Dis. 2011;70:469–475. doi: 10.1136/ard.2010.139212. [DOI] [PubMed] [Google Scholar]
- 5.Kremer JM. Methotrexate pharmacogenomics. Ann Rheum Dis. 2006;65:1121–1123. doi: 10.1136/ard.2006.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dervieux T, Greenstein N, Kremer J. Pharmacogenomic and metabolic biomarkers in the folate pathway and their association with methotrexate effects during dosage escalation in rheumatoid arthritis. Nat Clin Pract Rheumatol. 2007;3:256–257. doi: 10.1002/art.22129. [DOI] [PubMed] [Google Scholar]
- 7.Criswell LA, Lum RF, Turner KN, Woehl B, Zhu Y, Wang J, et al. The influence of genetic variation in the HLA–DRB1 and LTA–TNF regions on the response to treatment of early rheumatoid arthritis with methotrexate or etanercept. Arthritis Rheum. 2004;50:2750–2756. doi: 10.1002/art.20469. [DOI] [PubMed] [Google Scholar]
- 8.Sharma S, Das M, Kumar A, Marwaha V, Shankar S, Singh P, et al. Purine biosynthetic pathway genes and methotrexate response in rheumatoid arthritis patients among north Indians. Pharmacogenet Genomics. 2009;19:823–828. doi: 10.1097/fpc.0b013e328331b53e. [DOI] [PubMed] [Google Scholar]
- 9.Wessels JA, de Vries-Bouwstra JK, Heijmans BT, Slagboom PE, Goekoop-Ruiterman YP, Allaart CF, et al. Efficacy and toxicity of methotrexate in early rheumatoid arthritis are associated with single-nucleotide polymorphisms in genes coding for folate pathway enzymes. Arthritis Rheum. 2006;54:1087–1095. doi: 10.1002/art.21726. [DOI] [PubMed] [Google Scholar]
- 10.Wessels JA, Kooloos WM, De Jonge R, De Vries-Bouwstra JK, Allaart CF, Linssen A, et al. Relationship between genetic variants in the adenosine pathway and outcome of methotrexate treatment in patients with recent-onset rheumatoid arthritis. Arthritis Rheum. 2006;54:2830–2839. doi: 10.1002/art.22032. [DOI] [PubMed] [Google Scholar]
- 11.Breedveld FC, Weisman MH, Kavanaugh AF, Cohen SB, Pavelka K, van Vollenhoven R, et al. The PREMIER study: A multicenter, randomized, double-blind clinical trial of combination therapy with adalimumab plus methotrexate versus methotrexate alone or adalimumab alone in patients with early, aggressive rheumatoid arthritis who had not had previous methotrexate treatment. Arthritis Rheum. 2006;54:26–37. doi: 10.1002/art.21519. [DOI] [PubMed] [Google Scholar]
- 12.Moreland LW, O'Dell JR, Paulus HE, Curtis JR, Bathon JM, St Clair EW, et al. A randomized comparative effectiveness study of oral triple therapy versus etanercept plus methotrexate in early, aggressive rheumatoid arthritis. Arthritis Rheum. 2012;64:2824–2835. doi: 10.1002/art.34498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreland LW, O'Dell JR, Paulus HE, Curtis JR, Bridges SL, Zhang X, et al. TEAR - Treatment of early aggressive rheumatoid arthritis: a randomized, double-blind 2 year trial comparing immediate triple DMARD vs methotrexate and etanercept to step-up from initial methotrexate monotherapy. Arthritis Rheum. 2009;60:S707. [Google Scholar]
- 14.Moreland LW, O'Dell JR, Paulus HE, Curtis JR, Bathon JM, St Clair EW, et al. Two-year radiographic results from the TEAR trial. Arthritis Rheum. 2010;62:S1368. [Google Scholar]
- 15.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 16.Prevoo ML, van 't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–48. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 17.Burmeister JK, Sedova M, Shapero MH, Mansfield E. DMET microarray technology for pharmacogenomics-based personalized medicine. Methods Mol Biol. 2010;632:99–124. doi: 10.1007/978-1-60761-663-4_7. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draper N, Smith H. Applied regression analysis. 3rd ed. New York: Wiley; 1998. [Google Scholar]
- 20.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;58:267–288. [Google Scholar]
- 21.Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25:714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H, Sehl ME, Sinsheimer JS, Lange K. Association screening of common and rare genetic variants by penalized regression. Bioinformatics. 2010;26:2375–2382. doi: 10.1093/bioinformatics/btq448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, et al. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69(Supplement):504. [Google Scholar]
- 24.The R Core Group. R (Version 2.13.0) Available at http://CRAN.R-project.org/
- 25.Baggott JE, Bridges SL, Morgan SL. Evidence for two phenotypes in the metabolism of methotrexate to 7-hydroxymethotrexate in patients with rheumatoid arthritis. Arthritis Rheum. 2005;52:356–358. doi: 10.1002/art.20742. [DOI] [PubMed] [Google Scholar]
- 26.Baggott JE, Morgan SL. Methotrexate catabolism to 7-hydroxymethotrexate in rheumatoid arthritis alters drug efficacy and retention and is reduced by folic acid supplementation. Arthritis Rheum. 2009;60:2257–2261. doi: 10.1002/art.24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscow JA, Gong M, He R, Sgagias MK, Dixon KH, Anzick SL, et al. Isolation of a gene encoding a human reduced folate carrier (RFC1) and analysis of its expression in transport-deficient, methotrexate-resistant human breast cancer cells. Cancer Res. 1995;55:3790–3794. [PubMed] [Google Scholar]
- 28.Trevino LR, Shimasaki N, Yang W, Panetta JC, Cheng C, Pei D, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faganel Kotnik B, Grabnar I, Bohanec Grabar P, Dolzan V, Jazbec J. Association of genetic polymorphism in the folate metabolic pathway with methotrexate pharmacokinetics and toxicity in childhood acute lymphoblastic leukaemia and malignant lymphoma. Eur J Clin Pharmacol. 2011;67:993–1006. doi: 10.1007/s00228-011-1046-z. [DOI] [PubMed] [Google Scholar]
- 30.Dervieux T, Wessels JA, Kremer JM, Padyukov L, Seddighzadeh M, Saevarsdottir S, et al. Patterns of interaction between genetic and nongenetic attributes and methotrexate efficacy in rheumatoid arthritis. Pharmacogenet Genomics. 2012;22:1–9. doi: 10.1097/FPC.0b013e32834d3e0b. [DOI] [PubMed] [Google Scholar]
- 31.Wessels JA, van der Kooij SM, le Cessie S, Kievit W, Barerra P, Allaart CF, et al. A clinical pharmacogenetic model to predict the efficacy of methotrexate monotherapy in recent-onset rheumatoid arthritis. Arthritis Rheum. 2007;56:1765–1775. doi: 10.1002/art.22640. [DOI] [PubMed] [Google Scholar]
- 32.Grabar PB, Logar D, Lestan B, Dolzan V. Genetic determinants of methotrexate toxicity in rheumatoid artritis patients: a study of polymorphisms affecting methotrexate transport and folate metabolism. Eur J Clin Pharmacol. 2008;64:1057–1068. doi: 10.1007/s00228-008-0521-7. [DOI] [PubMed] [Google Scholar]
- 33.Berkun Y, Levartovsky D, Rubinow A, Orbach H, Aamar S, Grenader T, et al. Methotrexate related adverse effects in patients with rheumatoid arthritis are associated with the A1298C polymorphism of the MTHFR gene. Ann Rheum Dis. 2004;63:227–231. doi: 10.1136/ard.2003.016337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falconer DS, Mackay TFC. Introduction to quantitative genetics. 4th edition. Essex: Pearson Education Limited; 1996. [Google Scholar]
- 35.Morgan SL, Baggott JE. Folate supplementation during methotrexate therapy for rheumatoid arthritis. Clin Exp Rheumatol. 2010;28:S102–S109. [PubMed] [Google Scholar]
- 36.Hughes LB, Beasley TM, Patel H, Tiwari HK, Morgan SL, Baggott JE, et al. Racial or ethnic differences in allele frequencies of single-nucleotide polymorphisms in the methylenetetrahydrofolate reductase gene and their influence on response to methotrexate in rheumatoid arthritis. Ann Rheum Dis. 2006;65:1213–1218. doi: 10.1136/ard.2005.046797. [DOI] [PMC free article] [PubMed] [Google Scholar]