Abstract
A common pathological hallmark of protein-conformational brain diseases is the formation of disease-specific protein aggregates. In Alzheimer’s disease, these are comprised of amyloid-β and Tau as opposed to α-synuclein in Parkinson’s disease and N-terminal fragments of mutant huntingtin in Huntington’s disease. Most aggregates also sequester molecular chaperones, a protein family that assists in the folding, refolding, stabilization, and processing of client proteins, including misfolded proteins in brain diseases. Molecular chaperone modulation has achieved remarkable therapeutic effects in some cellular and preclinical animal models of protein-conformational diseases. This has raised hope for chaperone-based strategies to combat these diseases. Here, we review briefly the functional diversity and medical significance of molecular chaperones, their therapeutic potential, and common and specific challenges towards clinical application.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0186-5) contains supplementary material, which is available to authorized users.
Keywords: Molecular chaperones, Protein folding, Aggregation, Neurodegenerative diseases, Protein clearance
Introduction
Pathological accumulations of misfolded proteins in aggregates are a hallmark of many protein-conformational diseases of the central nervous system (CNS). Whether aggregates are toxic remains unclear and highly debated. A more prevailing view is that toxicity is caused by “on- or off-pathway”-soluble oligomers. It is widely accepted that targeting aggregation pathways can lower the concentration of toxic species with the prospect of reversing cellular neuropathology and associated brain pathophysiology. For this reason, molecular chaperones have gained wide interest as potential therapeutic targets to modulate misfolded protein levels in devastating protein-conformational brain diseases. Mounting evidence supports the concept that modulating chaperones can reduce protein misfolding and toxicity in neurons [1]. In most cases, however, the underlying molecular mechanisms are poorly understood and best possible target entry points for therapy remain to be defined.
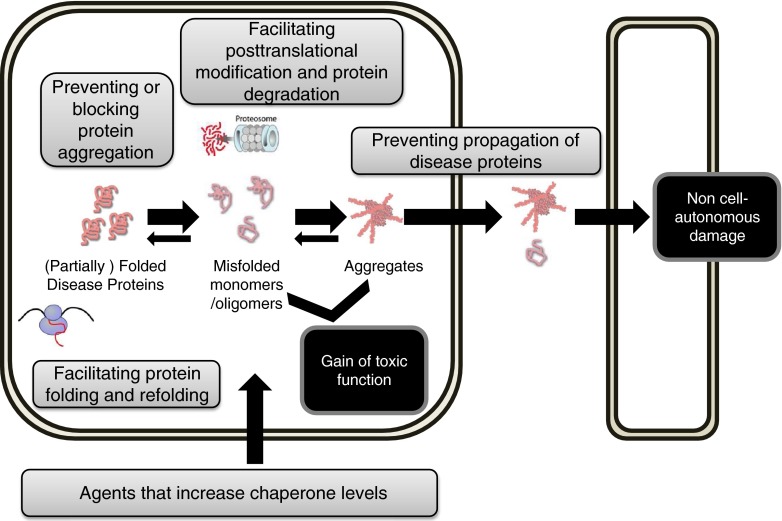
Chaperones form several families of evolutionary highly conserved proteins that are indispensable for cellular function. Chaperones recognize and bind hydrophobic structures that are exposed when proteins misfold to prevent non-native intermolecular interactions. Their wide range of activities include assisting folding of de novo synthesized proteins, help refolding and clearance of misfolded proteins (quality control and proteostasis), assist in the assembly and disassembly of protein complexes, and facilitate protein trafficking [2, 3]. For this reason molecular chaperones, which can modulate protein misfolding at multiple levels (Fig. 1), have gained wide interest as molecular targets, and therapeutic entry points can be envisaged based on chaperone categories with different modes of action. Here, we review briefly their functional diversity and medical significance, and we discuss opportunities and challenges of developing chaperone modulators for therapy.
Fig. 1.
Modulating chaperones offers therapeutic opportunities at multiple levels to combat protein conformational brain diseases. Molecular chaperones can modulate protein misfolding at multiple levels and have gained wide interest as molecular targets. 1. Increasing chaperones, e.g., via the Heat Shock Factor (HSF) mediated cellular stress response. 2. Facilitating protein folding and refolding, e.g., via the Hsp70 (HSPA), Hsp60 (HSPC), or Hsp90 (HSPD) chaperone systems. 3. Preventing or blocking protein aggregation, e.g., via small heat shock protein or Hsp40. 4. Facilitating post-translational modification and protein degradation, e.g., via co-chaperone carboxy terminus of Hsc70-interacting protein. 5. Preventing propagation of disease protein, e.g., via HSPA
Diverse Functions of Molecular Chaperones in Handling Client Proteins
Quality control mechanisms have evolved in organisms to facilitate the proper folding and assembly of proteins, and prevent inappropriate intra- and intermolecular interactions [3]. A superfamily of functionally diverse heat shock proteins (HSPs) plays a major role in these processes. HSPs typically function via iterative binding of client proteins and their release until refolding occurs or the client enters a degradation pathway [4–6]. The Hsp90 (HSPD), Hsp70 (HSPA) and Hsp60 (HSPC) families hydrolyze ATP, with their low intrinsic ATPase activity regulated by co-chaperones. Hsp40 (DNAJ) and the small heat shock protein (HSPB) lack an intrinsic ATPase capability, but interact functionally with ATPase active chaperone systems [3, 4].
HSPB/sHsp Family
Commonly, HSPBs first recognize, bind, and hold unfolded client proteins before transferring their clients to a chaperone (e.g., HSPA members) with intrinsic ATPase activity [4]. HSPBs have a conserved α-crystallin domain that is important for substrate binding [7–10] and the prevailing hypothesis is that they form large, substrate-free oligomeric complexes that dissociate into smaller client-binding multimers when misfolded protein levels in cells increase [4, 9, 10]. In humans, ten HSPBs are known, excluding HSPB11, which shares a number of features with bona fide HSPBs, but lacks the α-crystallin domain. HSPB1 (HSP27) is the most prominent member in this family [4]. Cellular stress induces dissociation of larger complexes of HSPB1 into smaller phophorylated complexes that bind misfolded clients and keep these in a folding-competent non-aggregated state [7, 10]. A phosphorylation-deficient HSPB1 mutant was shown to be insufficient in forming smaller complexes and client binding [10]. When refolding fails, HSPB1 mediates degradation of client proteins via the proteasome [4, 10]. The HSPB family members HSPB6-9 function in a HSPA-independent fashion and do not facilitate client protein refolding, but they all can suppress aggregation of proteins with expanded polyglutamine (polyQ) domains, likely by engaging different mechanisms [4]. For example, HSPB8 was shown to activate eIF2alpha and induce autophagy [11, 12], whereas HSPB7, the most potent inhibitor of polyQ protein aggregation [13, 14], did not increase overall autophagic activity, but facilitated autophagic clearance of polyQ protein aggregates using the autophagy machinery’s existing capacity [14]. It was proposed that HSBP7 might enhance client recognition for capture into autophagosomes, a mechanism that seems to work poorly for mHTT protein [15]. Hence, targeting HSBP7-client interactions might offer unique opportunities for enhancing the clearance of certain misfolded proteins in postmitotic neurons. Specific roles in neurons for HSPB family members are highlighted by mutations in HSPB1, HSPB3, and HSPB8, causing motor neuropathies [16, 17].
DNAJ/Hsp40 Family
Co-chaperones of the DNAJ family (41 genes in humans) are defined by a highly conserved J domain that includes a sequence motif (HPD) for interaction with the nucleotide binding domain of HSPA, stimulating adenosine triphosphate (ATP) hydrolysis and high-affinity client protein binding by HSPA [18]. Functional diversity of DNAJ members and their link to a wide array of pathological conditions likely resides outside their common J domain [4, 18–23]. DNAJB1 is one of the most extensively studied members of this family. DNAJB1 is expressed constitutively in mammalian cells and neurons, and is strongly upregulated by cellular stress via the heat shock response (HSR) [18]. In cooperation with HSPA, DNAJB1 serves important housekeeping functions under basal and stress conditions by recognizing and guiding the fate of unfolded client proteins. DNAJB2 (HSJ-1) is expresssed in neurons and has two isoforms that both contain a ubiquitin interaction motif [24, 25]. DNAJB2 seems to recruit HSPA-dependent clients, including misfolded polyQ protein oligomers to facilitate their proteasomal degradation [24–26]. A loss-of-function mutation in DNAJB2 is the cause of a lower motor neuron disease [27]. DNAJB6 is expressed in a HSR-independent fashion [28]. In neurons, DNAJB6 strongly suppresses the aggregation and toxicity of polyQ proteins and α-synuclein [19, 28, 29]. Oligomerization and acetylation of DNAJB6 appear crucial for exerting its function to inhibit aggregation, but, interestingly, neither its functional J domain nor an interaction with HSPA is required [28]. Missense mutations that prolong the half-life of DNAJB6 reduce its anti-aggregation activity and cause a rare form of muscular dystrophy characterized by abnormal protein accumulations and autophagic pathology [30]. An important role in neurons for other DNAJ proteins is examplified by mutations in the large multidomain protein Sacsin (DNAJC29), which cause an early-onset spastic ataxia with loss of Purkinje cells. Knockdown of wild-type Sacsin enhances toxicity of mutant polyQ ataxin-1 [31].
HSPA/Hsp70 Family
The ATP-dependent HSPA family (13 members) plays a fundamental role in protein homeostasis under normal and stress conditions [2, 32]. Some members in this family (e.g., HSPA1A and B ) are induced strongly in response to stress, whereas others (e.g., HSPA8) are present constitutively in most tissues [2, 4]. Their role in protein quality control depends on an intrinsic ATPase activity, flexible substrate binding and release, and interactions with co-chaperones and other cofactors [32]. Intramolecularly, HSPA functions are coordinated by a crosstalk between the N-terminal nucleotide binding domain and the C-terminal substrate binding domain, which are connected via a short linker region [32, 33]. In the ATP-bound state, HSPAs show low substrate binding affinity and high substrate release rates; the reverse is observed when adenosine diphosphate (ADP) is bound. Communication between the N- and C-terminal domains depends on the conserved hydropobic linker region and on cooperation with cofactors. DNAJs, for example, drive HSPAs into the ADP-bound state favoring substrate interactions [18, 33]. Other cofactors, such as BAG-1, HSPH, or HSPBP1, function as nucleotide exchange factors that facilitate the exchange of ADP for ATP to drive substrate release [32–34]. Multiple rounds of this exchange cycle are often necessary before the final fate (folding or degradation) of a client protein is determined. Some cofactors of HSPAs help to shuttle clients for proteasomal degradation or clearance via autophagy. Increasing HSPA levels has been shown to reduce toxicity and protein aggregation in disease models [35–37].
HSPD/Hsp90 Family
Members of the ATP-dependent HSPD (Hsp90) family also receive help from a host of cofactors. HSPDs play a central role in folding and maintaining the conformation of a variety of signaling proteins, including kinases and steroid hormone receptors [38]. For the most part, HSPDs regulate folding of proteins at advanced, almost native, conformational stages. Hop is a co-factor that transfers substrates from HSPA to HSPD and links these two chaperone mechanisms [38, 39]. HSPDs dimerize via their C-terminal domain, whereas the N-terminal domain binds and hydrolyses ATP [38]. ATP binding drives a conformational dimerization of the N-terminal domains into a closed state such that the substrate is captured in a “molecular clamp” [38]. ATP hydrolysis allows dissociation of the N-terminal domains and substrate release, and the ATPase activity is regulated by co-chaperones, post-transational modifications, and client protein binding. Some co-chaperones inhibit (e.g., Hop and the kinase-specific chaperone Cdc37) [39, 40], whereas others stimulate, the HSPD ATPase function (e.g., Aha1) [41]. Aha1 binds to the middle domain of HSPD showing that this domain also contributes to substrate activation [41]. Increasing HSPD levels has not proven as effective as increasing HSPA levels to counteract toxicity and aggregate formation in disease models. The hypothesis has been raised that HSPD/Hsp90 may have a role as an evolutionary capacitor because HSPD is capable of binding and stabilizing mutated protein variants, avoiding their degradation [3, 42, 43].
HSPC/Hsp60 Family
Finally, the ATP-dependent HSPC or chaperonin members employ a fascinating mechanism. These chaperones encapsulate their unfolded client proteins in a cage that is comprised of large double-ring complexes that can enclose substrates of up to 60 kDa [3]. Mitochondrial HSP60 functionally cooperates with HSP10, which is located at the opening of the cage, and seals it once a substrate has entered [3]. The cytosolic chaperonins (TriC or CCT) usually have two eight-membered rings and a folding-cage that is closed via conformational changes at the apical domain [44, 45]. An ATP-dependent encapsulation cycle drives substrate capture and release [3, 44, 45] and the enclosed cage environment prevents substrates from non-native intermolecular interactions [44, 45]. Increasing HSPC levels has been shown to prevent accumulation of toxic polyQ protein aggregates [46–48]. Mutations in chaperonin and chaperonin-like genes affecting neuronal functions include mutations in mitochondrial HSPD1 that cause hereditary spastic paraplegia [49], and mutations in three chaperonin-like genes that cause Bardet–Biedl ciliopathy syndromes (BBS) by disrupting the assembly of the CCT complex with the BBSome, a complex of seven BBS proteins [50].
Medical Significance of Chaperones in Protein Conformational Brain Diseases
An interesting correlation seems to exist between an age-dependent decline in protein quality control systems, a generally observed late-onset of protein-conformational brain diseases, and the finding that chaperones often colocalize with protein aggregates [3, 51, 52]. Here, we summarize briefly some of the findings in Alzheimer’s disease (AD), Parkinson’s disease (PD), and polyQ disease.
AD
AD is thought to involve the misfolding, aggregation, and deposition of mainly extracellular amyloid-β (Aβ) and intracellular Tau, a microtubule-associated protein that accumulates predominantly in axons [53]. We still lack a comprehensive understanding of how Aβ and/or Tau are detrimental to neurons, but cell autonomous, as well as non-cell autonomous, mechanisms, including inflammatory mediators and transneuronal propagation of pathology, seem to be involved [53–56].
Molecular chaperones are recruited to Aβ and Tau aggregates, and nonphysiological expression levels of chaperones have been detected in post-mortem AD brains [57, 58]. It has been shown that recombinant purified HSPA, DNAJ, and HSPD family members can suppress the formation of large and small oligomeric Aβ aggregates in vitro, suggesting that these chaperones can recognize and bind misfolded Aβ forms at different stages in the aggregation pathway [59]. In an AD-like neuronal model, toxicity of virally-delivered intracellular Aβ was rescued by HSPA overexpression [60]. Furthermore, overexpression of certain HSPBs in cultured cerebrovascular cells and in a Caenorhabditis elegans AD model was shown to reduce intracellular Aβ-induced toxicity [61, 62]. Opposite effects were seen in the same AD model when heat shock factor 1, the major transcription-factor of the HSR, was downregulated, exacerbating intracellular Aβ aggregation and toxicity [63]. However, the physiological relevance of intracellular Aβ aggregates in disease remains speculative and therefore also therapeutic concepts that are based on counteracting this phenomenon. Solid experimental evidence is also still lacking for chaperones being exported from cells where they might counteract extracellular Aβ aggregation. Taken together, various modes of action of chaperones counteracting Aβ aggregation and toxicity may exist, but their physiological relevance to human disease is not known.
ATP-dependent and ATP-independent chaperones also co-localize with Tau aggregates and expression of HSPA, HSPB1, and the co-chaperone carboxy terminus of Hsc70-interacting protein (CHIP) have all been shown to facilitate the degradation of misfolded phosphorylated Tau [64, 65]. It has been demonstrated that chaperones directly recognize and bind to Tau, and that HSPA–HSPD collaboration initiated Tau clearance in cells [64–66]. Furthermore, increased chaperone levels not only blocked Tau aggregation, but also increased the solubility of nonphosphorylated, functional Tau [67]. Finally, HSPD inhibition has been shown to selectively promote the degradation of aberrant P-Tau species [68] and different mechanisms involving the E3 ubiquitin ligase CHIP, prolyl isomerases, and other co-chaperones have been proposed to explain how the HSPD chaperone network regulates Tau biology and pathology [69]. Altogether, these results suggest that chaperone modulators might eventually find a niche in either the Aβ- and/or Tau-directed armamentarium of drugs for AD.
PD
PD is characterized by a progressive degeneration of neurons, primarily dopaminergic neurons in the substania nigra, whose loss accounts for the main clinical manifestations in motor dysfunction. Histopathologic hallmarks are intracellular protein aggregates referred to as Lewy bodies and Lewy neurites. These are composed largely of the small 14-kDa lipid-binding protein α-synuclein (αSN) [70]. Several mutations in the synuclein gene (SNCA), as well as duplications and triplications of the wildtype allele, are a cause of rare familiar forms of PD. Furthermore, Genome Wide Association (GWA) analysis has linked polymorphisms in SNCA to idiopathic PD [71–76]. αSN seems to be involved mainly in synaptic vesicle homeostasis [77–80]. αSN PD mutants show enhanced oligomerization and aggregation properties, but why these or the wildtype counterpart misfold and accumulate in vivo remains speculative. A recent study has demonstrated that, under native conditions, αSN forms a α-helical structured tetramer and that its destabilization is associated with a higher propensity of αSN to aggregate [81]. Hence, targeting αSN misfolding by stabilizing the tetrameric complex or by reducing overall αSN levels could, perhaps, achieve disease-modifying effects in synucleinopathies.
HSPA, HSPB, HSPD, HSPC, and DNAJ chaperone members have been found in Lewy bodies, suggesting that chaperones bind and modulate αSN folding, aggregation, and fate [37, 82–84]. In a fly model for PD, the expression of HSPA has been shown to suppress αSN aggregate formation and increase soluble αSN levels while reducing toxicity [37]. Likewise, the induction of stress-inducible chaperones by geldanamycin was shown to reduce αSN aggregation and toxicity [85]. Nonetheless, it is not fully understood how chaperones modulate αSN aggregation and toxicity. HSPA can inhibit αSN oligomerization and aggregation by recognizing early misfolded αSN conformations in vitro [86]. HSPAs can also bind larger αSN aggregates in the presence of DNAJ and ADP, and can be sequestered into αSN aggregates [87, 88]. Therefore, HSPAs, together with cofactors, seem capable of binding, processing, and guiding the fate of αSN [89]. Studies in live cells suggest that HSPA binds misfolded αSN early in the aggregation process such that oligomerization is inhibited [90–92]. One study reported that HSPA1 overexpression in αSN transgenic mice lowered insoluble αSN aggregate levels [93]. However, another similar study reported that HSPA1 overexpression by itself is not sufficient to counteract αSN pathology [94]. Therefore, HSPA1 may be able to recognize and bind misfolded αSN species, but proper cofactor activity might be required for HSPA to block αSN aggregation and facilitate its degradation. It is interesting, perhaps, to also note that parkin—the product of a gene whose mutations cause a recessive form of familial PD—was shown to ubiquitinate HSPA [95]. HSPDs also bind misfolded αSN. However, in sharp contrast to HSPA, HSPD accelerates aggregate formation in vitro [96]. Chaperone effects of HSPA and HSPD on αSN therefore differ remarkably, and the authors suggested that HSPD may act by scavenging misfolded αSN into large aggregates that are less toxic than αSN oligomers [96]. Other studies showed that CHIP, a cofactor of HSPA and HSPD with an E3-Ligase function, facilitates ubiquitination and proteasomal degradation of αSN [97–99]. HSPD and CHIP also modulates the stability and degradation of another PD gene, namely LRRK2 kinase [100, 101]. These findings could suggest that age-dependent declines in HSPD, HSPA, and CHIP activity might have a role in predisposition to PD. What seems increasingly clear is that the fate of client proteins of HSPA and HSPD can differ remarkably and that much is dictated by the interaction between the main chaperone and its specific co-chaperones. Also, small HSPB chaperones have been reported to accumulate in affected neurons and glial cells and protect against αSN toxicity [102–104]. Therefore, an age-dependent decline in their activity may also lower the threshold for developing protein-conformational diseases. Finally, DNAJ chaperones may play a role in PARK2 patients, who express mutant Parkin proteins that cause autosomal recessive juvenile onset Parkinsonism. DNAJB2a and DNAJB6 have been shown to recover function of Parkin mutants by suppressing aggregation and restoring their capability to locate to damaged mitochondria, and promote their clearance via mitophagy [105].
PolyQ Diseases
PolyQ diseases are inherited disorders caused by CAG/polyglutamine expansions (polyQ) in genes including mHTT and several other proteins that cause spinocerebellar ataxias (SCA). Different chaperones have been shown to interact functionally with polyQ oligomers and aggregates. HSPC was shown to suppress the formation of mHTT inclusion bodies and decrease soluble mHTT oligomer levels [46–48], whereas HSPC inhibition had the opposite effect [46]. HSPA was shown to co-localize with polyQ inclusions and regulate the conformation of misfolded mHTT in a worm Huntington’s disease (HD) model [106]. Mutations in its substrate binding domain reduced its co-localization with inclusion bodies [106]. In SCA animal models, such as a fly model of mutant ataxin-3-induced ocular degeneration, severe neurodegeneration was exacerbated by co-expressing a dominant negative form of HSPA, whereas wildtype HSPA provided rescue [107, 108]. Overexpression of HSPA in the human mutant huntingtin exon-1 R6/2 HD mouse model delayed the loss of body weight compared with R6/2 mice, but otherwise had no effect on inclusions or early death of the mice [109]. Cofactors appear crucial for HSPA function in modulating polyQ protein fate [110]. For example, CHIP binds HSPA and acts as an E3-ligase that transfers polyubiquitin chains to misfolded client proteins and induces their degradation [111–113]. HSPA and DNAJB1 can cooperate, bind, and sequester mHTT monomers and inhibit their conversion into oligomers [114]. In the presence of DNAJB1, HSPA is also able to associate with mHTT oligomers [36]. DNAJB1, DNAJB2, DNAJB6, and DNAJB8 were identified as the most active suppressors of aggregation of a 74-glutamine-containing HTT exon-1 fragment in HEK-293 cells [19]. In vivo, overexpression of DNAJB2a in R6/2 mice reduced mHTT aggregation and improved neurologic performance [25]. In contrast, DNAJA1 overexpression, alone or in combination with HSPA, failed to modulate retinopathy in SCA7 mice [115]. DNAJB6b and DNAJB8 have been identified as the most potent suppressors of polyQ protein aggregation and toxicity [28]. Unlike DNAJB1, both retained activity as aggregation suppressors when mutated in the HSPA interaction J-domain, although this domain is required to facilitate HSPA-dependent degradation of polyQ substrates. Furthermore, HDAC4 was shown to dock on a C-terminal domain of DNAJB6/8 and to be required for their full anti-aggregration activity. The investigators therefore suggested that DNAJB6-type protein pathways might offer interesting targets for therapy in protein misfolding diseases.
Taken together, many findings in preclinical cell and animal models support the notion that chaperone-modulation could become a cornerstone in treatments fighting AD, PD, and/or polyQ protein misfolding diseases. At the same time, our mechanistic understanding is still very incomplete and probably insufficient to pick the right target with the best chances of success for therapy in anyone of the aforementioned diseases.
Targeting Chaperone Pathways for Therapy
Chaperone modulation has been able to achieve powerful therapeutic effects in some cellular and preclinical animal models of protein conformational diseases, which suggests that, eventually, it may become a medically significant strategy to fight neurodegenerative diseases. However, overall success rates of developing CNS drugs are ~8 % from first-in-man to registration, and disappointingly low [116]. Innovation is key to try and improve success rates, and a better understanding of which mechanisms and chaperones to target is pivotal. Generally, CNS protein conformational diseases show slow progression rates and assessing disease-modifying principles in the clinic will be no less of a challenge than targeting, for example, the well-understood amyloid precursor protein pathway in AD [117]. Furthermore, age-dependent changes in protein quality control and disease mechanisms have to be taken into account when selecting patients for proof-of-concept (PoC) studies in humans. When targeting patients suffering from a monogenetic protein conformational disease it will be important to pay attention to age, disease-stage, rate of progression, and best available clinical readouts. Only positive PoC outcomes will encourage the investment needed for clinical development.
Drugs targeting chaperone-based mechanisms in the CNS have many challenges in common with other CNS drugs, most importantly getting across the blood–brain barrier, proof of efficacy, safety, and tolerability. Exposures in brain must be sufficient to trigger pharmacodynamic changes in target activity that translate into meaningful effects on the fate of a client protein and its downstream neuropathophysiologic and clinical effects. It is pivotal to understand the relationship between a drug’s pharmacokinetic properties and its on-target pharmacodynamic effects, and also how the latter translate into driving the molecular process that determines a client’s fate. Ideally, a biomarker for target and process engagement in brain would be available by the time a PoC study starts. Also of key importance to arrive at a clinically meaningful result is to understand the magnitude of effect a treatment needs to achieve, as well as its duration needed. Taking these issues too lightly primes for failure and getting lost in clinical translation. Furthermore, drugs targeting chaperone mechanisms may be prone to on-target safety and toxicology concerns because they likely change a chaperone’s network function beyond the specific role that the chaperone has in quality control of the targeted protein. This can be of advantage in oncology because tumor cells often depend on elevated levels of multiple chaperones [118], but, in the CNS, this could easily turn into a disadvantage, and selective compounds targeting mainly the target-relevant arm of a chaperone’s network seems more promising.
What we can learn from preclinical models also deserves some note of caution. Animal models are great mechanistic pharmacokinetic/pharmacodynamic models, but their predictive power as translational disease models is often very limited. Patient fibroblast-derived induced pluripotent stem cell (iPSC) systems and iPSC-derived neurons, in particular, may offer a better choice to study chaperone-mediated modulation of misfolded proteins. These systems circumvent species-specificity questions and offer advantages with regard to assessing pharmacological effects in genetically heterogeneous human cellular backgrounds, including monogenetic diseases [119–121]. To advance the use of patient cells as disease models, funding agencies support open-access collections of patient-derived fibroblasts with mutations linked to neurologic diseases, including proteinopathies [122]. Encouraging results towards obtaining disease-specific aggregate proteinopathy in iPSC-derived neurons have already been obtained: in iPSC-derived neurons from SCA3 patients treated with L-glutamate, which evoked calcium-dependent proteolysis of the mutant polyQ ataxin-3 protein [123]; in iPSC-derived motor neurons from familial amyloid lateral sclerosis patients with Tar DNA binding protein-43 (TDP-43) mutations [124]; and in cortical neurons generated from iPSC cells from Down’s syndrome patients [125]. Other types of pathologic changes noted in iPSC-derived neurons from patients include a significant increase in lysosomal activity (HD) [126], nuclear aberrations (PD LRRK2) [127], cell morphological changes and defects in autophagic clearance (PD) [128], and increased α-synuclein production in iPSC-derived midbrain dopaminergic neurons generated from a PD patient with a SNCA triplication [129], and a Gaucher patient with a GBA mutation that predisposes to PD [130]. Taken together, the outlook seems bright for finding chaperone-based drugs with potential for disease-modification in patients. Ideally, such drugs would not be disease-specific and could treat a spectrum of protein-conformational diseases. Existing drugs, such as HSPD inhibitors, have, in principle, such features, but, at the same time, perhaps too many liabilities outside use in oncology. For more detailed lists of small molecules acting on molecular chaperones the reader is guided to some excellent reviews [34, 118, 131, 132].
Targeting ATP-Dependent Molecular Chaperone Systems
Molecular chaperones with intrinsic ATPase function such as members of the HSPD, HSPA, and HSPC families, and their co-chaperones assist in the folding and fate determination of a variety of client proteins. These complex machineries provide multiple entry points for pharmacologic intervention. Ideally, the intervention would alter only specific aspects of a chaperone’s protein quality control functions relevant to the targeted client protein without perturbing the rest of that chaperone’s functions. This has been very difficult to achieve with drugs that target core sites, such as the ATP-binding pocket [132].
Targeting HSPD/Hsp90 System
Today, the best characterized drugs that inhibit HSPD function target its ATP-binding domain and were identified as antiproliferative agents in a cancer cell line-based screen. These include fungal antibiotics (radicicol) and the benzoquinone ansamycine (herbimycin A, geldanamycin). All block ATP hydrolysis and cause degradation of client proteins, including specific oncogenic proteins required for tumor cell proliferation [118, 131]. Also, mHTT protein is a HSPD client, and ATP-site inhibitors disrupt the HSPD–mHTT interaction, inducing mHTT clearance via the ubiquitin proteasome system [43]. 17-AAG, a derivative of geldanamycin, has been shown to improve motor deficits in a mouse model of spinal and bulbar muscular atrophy, a disorder caused by a polyQ mutation in the androgen receptor, another HSPD client protein [133]. Whether the action of 17-AAG entailed mainly disruption of the HSPD client protein interaction was questioned because HSPD inhibition also promotes heat shock factor 1-mediated activation of a more general cellular HSR with increased levels of HSPA8, HSPB1, and DNAJB1 [131, 134]. Irrespective, the results demonstrated that inhibiting the HSPD function alleviated excessive accumulation of mutant androgen receptors, which translated into some functional recovery in vivo [133]. Geldanamycin has also been shown to protect against αSN toxicity in a fly PD model [85]. However, despite remarkable activities in preclinical models, stability and safety issues of geldanamycin and radicicol derivatives in the clinic has precluded approval [131]. Furthermore, it was shown that the capacity of HSPD inhibitors to mount a HSR is reduced in R6/2 and HdhQ150 knock-in mouse brains [135], and similar observations have been made in cells expressing full-length polyQ-expanded mHTT [136]. Also note that all compounds with this mechanism of action will increase stress-inducible chaperone levels in a nonspecific fashion, which could cause undesirable side effects, narrow the therapeutic window, and limit translation to non-oncology indications in the clinic. A new generation of chemically distinct, more selective, safer, and potent HSPD inhibitors has been developed and has entered clinical trials in cancer. Some of these compounds seem well tolerated and achieved clinical benefit [131]. As an additional benefit, decreased chaperone levels and reduced HSR activity have been noted in patients with protein conformational brain diseases [135, 136], and HSPD inhibitors might be able to overcome these molecular deficits.
There is some evidence that targeting the linker region or the C-terminal domain of HSPD might avoid mechanistic side effects associated with targeting the ATP-binding pocket. At nanomolar concentrations, novobiocin analogues bind to the C-terminus of HSPD and induce HSPD and HSPA expression [137, 138]. Compounds of this class were shown to protect neurons from Aß-induced cellular toxicity, but the underlying mechanism remains to be elucidated [139]. Finally, targeting cofactors, such as aha1, which stimulates the ATPase function of HSPD, or mechanisms, such as post-translational acetylation of HSPD [140, 141], might have fewer liabilities.
Targeting The HSPA/Hsp70 System
Members of the HSPA family also provide multiple entry points for pharmacologic intervention. However, little progress has been made in bringing HSPA inhibitors to the clinic. In addition to inhibiting the enzymatic activity of HSPA, its chaperone network provides multiple in-roads for modulating more selectively aspects of these chaperone-mediated protein quality control mechanisms. HSPA functional diversity is guided largely by a large network of co-chaperones that includes J proteins, nucleotide exchange factors (NEFs) and tetratricopeptide repeat (TPR) domain-containing proteins. DNAJB1 and DNAJB6, for example, inhibit the aggregation and toxicity of mHTT, and DNAJA1 overexpression increases mHTT aggregation [19, 36, 142]. DNAJA1 overexpression facilitates the degradation of Tau and DNAJB1 inhibits Tau aggregation [143–145]. Therefore, targeting specifically the interaction between HSPA and specific J proteins seems like a promising approach to intervene therapeutically in protein conformational disorders. Two more specific entry points for targeting DNAJs might be worth considering: their oligomerization mechanism and specific post-translational modifications. For example, DNAJB6 functionally oligomerizes [18, 19], and its acteylation is necessary to exert its anti-aggregation effects [19].
Although modulating interactions of DNAJ proteins with misfolded disease proteins seems the more specific route to therapy, most efforts thus far were directed towards modulating DNAJ–HSPA cooperativity. [132]. An interesting group of compounds with this mechanism of action are the dihydropyrimidines, which specifically modulate DNAJ effects on HSPA ATPase activity. One of these compounds, MAL3-101, inhibits DNAJ-stimulated ATPase activity [145], whereas 115-7c stimulates ATP turnover [146]. This shows that modulating HSPA activity is possible in both directions. MAL3-101 was shown to have antimyeloma effects [147], whereas other dihydopydimines appeared to control the stability of HSPA substrates, including Tau and polyQ proteins [148, 149]. Importantly, compounds in this class are free of cytotoxicity and do not induce HSR [132, 148, 149], suggesting a greatly reduced side effect potential than compounds targeting the ATP binding pocket of HSPA.
There is also evidence that targeting members of the three main subclasses of NEFs (Hsp110, HspBP1 and BAGs) may facilitate the fate decision of HSPA-bound substrates. NEFs influence HSPA function by regulating nucleotide exchange and substrate release [3, 4, 34, 150, 151]. For example, HSPA–BAG2 facilitates proteosomal clearance of Tau, while HSPA–BAG1 stabilizes Tau [151–153]. These findings indicate that two cofactors that use a similar domain for interacting with HSPA determine the fate of the same HSPA client in a fundamentally different fashion. Which one offers a better target for therapy to reduce, for example, tauopathy remains an open question. Another example provides the NEF Hsp110. Hsp110 remodels the HSPA–DNAJ system to efficiently disaggregate and refold protein aggregates [150].
It is also early in the assessment of modulators targeting HSPA–TPR complexes [132]. The TPR co-chaperone CHIP directs ubiquitination and proteasomal degradation of HSPA-bound substrates [154, 155], whereas the TPR Hop coordinates the transfer of substrates between HSPA and HSPD and the folding of non-native proteins [156]. Hop and CHIP compete for binding to HSPA and, accordingly, determine the fate of clients.
Finally, evidence is growing that several misfolded protein pathologies, including synucleinopathy, can spread and cause non-cell autonomous damage [54, 55, 157]. HSPA was reported to chaperone αSN and reduce extracellular oligomer formation and related toxicity [158], suggesting that modulation of certain chaperones can also reduce non-cell autonomous neuronal damage caused by propagating aggregate pathologies. Altogether, many findings support the notion that small molecule modulators of HSPA–co-chaperone interactions may find their way to combat brain protein conformational disorders. For example, cystamine and its Food and Drugs Administraion-approved reduced form cysteamine (currently in a clinical trial for HD) upregulate DNAJ protein levels, enhance brain BDNF levels, and exert neuroprotection in HD models [159]. Compounds with HDAC inhibitor activity, such as valproate [160], and activation of the deacetylase SIRT1 have been shown to induce HSPA [161]. Furthermore, HDAC inhibitors can alter chaperone function also by changing their acetylation state [162], a mechanism proposed for HDAC4 enhancing DNAJB6 activity in suppressing polyQ protein aggregation [28]. HDAC inhibitors have been shown to counteract polyQ disease in fly, mouse, and cellular models, but reducing the expression of single HDACs in the R6/2 HD mouse model has not yet met with great success in terms of pointing specifically at therapeutic potential for a specific HDAC subtype [163]. Therapeutic success of any of these avenues will depend clearly on a more in-depth understanding of target and mechanism, especially as therapeutic effects of HDAC inhibitors are often limited by toxicity.
Targeting the HSPC/Hsp60 System
HSPC family members have been shown to prevent the accumulation of polyQ mHTT aggregates and cellular toxicity, whereas their RNAi-mediated knock-down was shown to result in opposite effects [46–48, 164, 165]. HSPC (TriC) binds a sequence N-terminal to the polyQ tract in mHTT, based on the specificity of this mHTT–CCT1 interaction, Tam et al. [164] proposed that the CCT1 substrate binding domain might be a particularly suitable site for drugs to counteract mHTT aggregation and toxicity. Accordingly, targeting specific family members of the HSPC family seems another promising therapeutic approach—perhaps particularly suited for HD. What makes this mechanism particularly interesting is that an interaction between a short hydrophobic N-terminal sequence element and not the polar polyQ stretch that accounts for the specific mHTT–CCT1 interaction and a suppression of mHTT protein aggregation. It shows that an aggregation-promoting element of a specific polyQ protein is recognized by a specific chaperone subunit—perhaps a kind of “ideal case scenario” for developing highly selective drugs to modulate HSPC–mHTT interactions without perturbing TriC’s essential roles in folding [165]. Smart assay designs to screen for such modulators seem, therefore, a promising way forward.
Prospects
Modulating molecular chaperone mechanisms seems to have great potential for treating protein conformational brain disorders. Of the small number of compounds that target molecular chaperones today, only few have entered the clinic; the majority act on HSR through HSPD, and these are tested in cancer patients. Significant improvements are still needed both at the level of compounds, targets, and specific mechanisms before good PoC studies can be conducted in patients with protein conformational brain diseases. The complex cascade of chaperone folding pathways offers many more additional entry points than those that have been targeted today. Also, our in-depth understanding of chaperone biology in relation to specific protein targets is progressing and the prospects for actually finding new drugs seem good. Translating any of these principles successfully to the clinic will depend on having good assays to monitor drug target and process engagement, and, ideally, also assays that can measure the misfolded culprit protein and its fate over time. Tremendous progress is being made in the biomarker field both with respect to markers in cerebrospinal fluid (CSF) and the use of noninvasive imaging technologies. This should encourage us to take mechanistically well-founded target projects in the chaperone arena forward to the clinic for testing new therapies in devastating diseases. Unfortunately, today, we have nothing more than a handful of drugs treating some symptoms.
Electronic supplementary material
(PDF 890 kb)
Acknowledgments
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Contributor Information
Herman van der Putten, Email: hermanvdp87@gmail.com.
Gregor P. Lotz, Email: glotz54@gmail.com
References
- 1.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 2.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 3.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 4.Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- 5.Feldman DE, Frydman J. Protein folding in vivo: the importance of molecular chaperones. Curr Opin Struct Biol. 2000;10:26–33. doi: 10.1016/s0959-440x(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 6.Ellis RJ, Hartl FU. Principles of protein folding in the cellular environment. Curr Opin Struct Biol. 1999;9:102–110. doi: 10.1016/s0959-440x(99)80013-x. [DOI] [PubMed] [Google Scholar]
- 7.Shashidharamurthy R, Koteiche HA, Dong J, McHaourab HS. Mechanism of chaperone function in small heat shock proteins: Dissociation of the HSP27 oligomer is required for recognition and binding of destabilized T4 lysozyme. J Biol Chem. 2005;280:5281–5289. doi: 10.1074/jbc.M407236200. [DOI] [PubMed] [Google Scholar]
- 8.Ito H, Kamei K, Iwamoto I, Inaguma Y, Nohara D, Kato K. Phosphorylation-induced change of the oligomerization state of RB-crystallin. J Biol Chem. 2001;276:5346–5352. doi: 10.1074/jbc.M009004200. [DOI] [PubMed] [Google Scholar]
- 9.van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 10.Bryantsev AL, Kurchashova SY, Golyshev SA, et al. Regulation of stress-induced intracellular sorting and chaperone function of Hsp27 (HspB1) in mammalian cells. Biochem J. 2007;407:407–417. doi: 10.1042/BJ20070195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 12.Carra S. The stress-inducible HspB8-Bag3 complex induces the eIF2alpha kinase pathway: implications for protein quality control and viral factory degradation? Autophagy. 2009;5:428–429. doi: 10.4161/auto.5.3.7894. [DOI] [PubMed] [Google Scholar]
- 13.Vos MJ, Zijlstra MP, Kanon B, et al. HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum Mol Genet. 2010;19:4677–4693. doi: 10.1093/hmg/ddq398. [DOI] [PubMed] [Google Scholar]
- 14.Vos MJ, Zijlstra MP, Carra S, Sibon OC, Kampinga HH. Small heat shock proteins, protein degradation and protein aggregation diseases. Autophagy. 2011;7:101–103. doi: 10.4161/auto.7.1.13935. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carra S, Crippa V, Rusmini P, et al. Alteration of protein folding and degradation in motor neuron diseases: Implications and protective functions of small heat shock proteins. Prog Neurobiol. 2012;97:83–100. doi: 10.1016/j.pneurobio.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Boncoraglio A, Minoia M, Carra S. The family of mammalian small heat shock proteins (HSPBs): implications in protein deposit diseases and motor neuropathies. Int J Biochem Cell Biol. 2012;44:1657–1669. doi: 10.1016/j.biocel.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hageman J, van Waarde MA, Zylicz A, Walerych D, Kampinga HH. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem J. 2011;435:127–142. doi: 10.1042/BJ20101247. [DOI] [PubMed] [Google Scholar]
- 20.Sterrenberg JN, Blatch GL, Edkins AL. Human DNAJ in cancer and stem cells. Cancer Lett. 2011;312:129–142. doi: 10.1016/j.canlet.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Kalia SK, Kalia LV, McLean PJ. Molecular chaperones as rational drug targets for Parkinson's disease therapeutics. CNS Neurol Disord Drug Targets. 2010;9:741–753. doi: 10.2174/187152710793237386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra A, Shevde LA, Samant RS. Multi-faceted role of HSP40 in cancer. Clin Exp Metastasis. 2009;26:559–567. doi: 10.1007/s10585-009-9255-x. [DOI] [PubMed] [Google Scholar]
- 23.Harms MB, Sommerville RB, Allred P, et al. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann Neurol. 2012;71:407–416. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westhoff B, Chapple JP, van der Spuy J, Hohfeld J, Cheetham ME. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr Biol. 2005;15:1058–1064. doi: 10.1016/j.cub.2005.04.058. [DOI] [PubMed] [Google Scholar]
- 25.Labbadia J, Novoselov SS, Bett JS, et al. Suppression of protein aggregation by chaperone modification of high molecular weight complexes. Brain. 2012;135:1180–1196. doi: 10.1093/brain/aws022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howarth JL, Kelly S, Keasey MP, et al. Hsp40 molecules that target to the ubiquitin-proteasome system decrease inclusion formation in models of polyglutamine disease. Mol Ther. 2007;15:1100–1105. doi: 10.1038/sj.mt.6300163. [DOI] [PubMed] [Google Scholar]
- 27.Blumen SC, Astord S, Robin V, et al. A rare recessive distal hereditary motor neuropathy with HSJ1 chaperone mutation. Ann Neurol. 2012;71:509–519. doi: 10.1002/ana.22684. [DOI] [PubMed] [Google Scholar]
- 28.Hageman J, Rujano MA, van Waarde MA, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell. 2010;37:355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Durrenberger PF, Filiou MD, Moran LB, et al. DnaJB6 is present in the core of Lewy bodies and is highly up-regulated in parkinsonian astrocytes. J Neurosci Res. 2009;87:238–245. doi: 10.1002/jnr.21819. [DOI] [PubMed] [Google Scholar]
- 30.Sarparanta J, Jonson PH, Golzio C, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 2012;44:450–455. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parfitt DA, Michael GJ, Vermeulen EG, et al. The ataxia protein sacsin is a functional co-chaperone that protects against polyglutamine-expanded ataxin-1. Hum Mol Genet. 2009;18:1556–1565. doi: 10.1093/hmg/ddp067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans CG, Chang L, Gestwicki JE. Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaffar G, Breuer P, Boteva R, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 36.Lotz GP, Legleiter J, Aron R, et al. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J Biol Chem. 2010;285:38183–38193. doi: 10.1074/jbc.M110.160218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auluck PK, Chan HYE, Trojanowski JQ, Lee VMY, Bonini NM. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 38.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 39.Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the atpase cycle. J Biol Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- 40.Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 42.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 43.Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J Biol Chem. 2012;287:1406–1414. doi: 10.1074/jbc.M111.294801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz IG, Yébenes H, Zhou M, et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol. 2011;18:14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- 45.Douglas NR, Reissmann S, Zhang J, et al. Dual action of ATP hydrolysis couples lid closure to substrate release into the Group II chaperonin chamber. Cell. 2011;144:240–252. doi: 10.1016/j.cell.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitamura A, Kubota H, Pack CG, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 47.Behrends C, Langer CA, Boteva R, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 48.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bross P, Naundrup S, Hansen J, et al. The Hsp60-(p.V98I) mutation associated with hereditary spastic paraplegia SPG13 compromises chaperonin function both in vitro and in vivo. J Biol Chem. 2008;283:15694–15700. doi: 10.1074/jbc.M800548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo S, Baye LM, Schulz NP, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A. 2010;107:1488–1489. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev. 2011;10:205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calderwood SK, Murshid A, Prince T. The shock of aging: molecular chaperones and the heat shock response in longevity and aging—a mini-review. Gerontology. 2009;55:550–558. doi: 10.1159/000225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kfoury N, Holmes BB, Jiang H, Holtzman DM, Diamond MI. Trans-cellular propagation of Tau aggregation by fibrillar species. J Biol Chem. 2012;287:19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo BC, Seidl R, Cairns N, Lubec G. Heat-shock protein 70 levels in brain of patients with Down syndrome and Alzheimer's disease. J Neural Transmission. 1999;57(Suppl):315–322. doi: 10.1007/978-3-7091-6380-1_22. [DOI] [PubMed] [Google Scholar]
- 58.Renkawek K, Bosman GJ, de Jong WW. Expression of small heat-shock protein hsp 27 in reactive gliosis in Alzheimer disease and other types of dementia. Acta Neuropathol. 1994;87:511–519. doi: 10.1007/BF00294178. [DOI] [PubMed] [Google Scholar]
- 59.Evans CG, Wisén S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1–42) aggregation in vitro. J Biol Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 60.Magrané J, Smith RC, Walsh K, Querfurth HW. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed beta-amyloid in neurons. J Neurosci. 2004;24:1700–1706. doi: 10.1523/JNEUROSCI.4330-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fonte V, Kipp DR, Yerg J, 3rd, et al. Suppression of in vivo beta-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J Biol Chem. 2008;283:784–791. doi: 10.1074/jbc.M703339200. [DOI] [PubMed] [Google Scholar]
- 62.Wilhelmus MM, Boelens WC, Otte-Höller I, Kamps B, de Waal RM, Verbeek MM. Small heat shock proteins inhibit amyloid-beta protein aggregation and cerebrovascular amyloid-beta protein toxicity. Brain Res. 2006;17:67–78. doi: 10.1016/j.brainres.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 63.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 64.Petrucelli L, Dickson D, Kehoe K, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 65.Shimura H, Miura-Shimura Y, Kosik KS. Binding of tau to heat shock protein 27 leads to decreased concentration of hyperphosphorylated tau and enhanced cell survival. J Biol Chem. 2004;279:17957–17962. doi: 10.1074/jbc.M400351200. [DOI] [PubMed] [Google Scholar]
- 66.Thompson AD, Scaglione KM, Prensner J, et al. Analysis of the tau-associated proteome reveals that exchange of hsp70 for hsp90 is involved in tau degradation. ACS Chem Biol. 2012;7:1677–1686. doi: 10.1021/cb3002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dou F, Netzer WJ, Tanemura K, et al. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci U S A. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dickey CA, Kamal A, Lundgren K, et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salminen A, Ojala J, Kaarniranta K, Hiltunen M, Soininen H. Hsp90 regulates tau pathology through co-chaperone complexes in Alzheimer's disease. Prog Neurobiol. 2011;93:99–110. doi: 10.1016/j.pneurobio.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 70.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 71.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 72.Chartier-Harlin MC, Kachergus J, Roumier C, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 73.Cronin KD, Ge D, Manninger P, et al. Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet. 2009;18:3274–3285. doi: 10.1093/hmg/ddp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 75.Kruger R, Kuhn W, Muller T, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 76.Zarranz JJ, Alegre J, Gomez-Esteban JC, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 77.Cooper AA, Gitler AD, Cashikar A, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nemani VM, Lu W, Berge V, et al. Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65:66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thayanidhi N, Helm JR, Nycz DC, Bentley M, Liang Y, Hay JC. Alpha-synuclein delays endoplasmic reticulum (ER)-to-Golgi transport in mammalian cells by antagonizing ER/Golgi SNAREs. Mol Biol Cell. 2010;21:1850–1863. doi: 10.1091/mbc.E09-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakamura K, Nemani VM, Azarbal F, et al. Direct membrane association drives mitochondrial fission by the Parkinson’s disease-associated protein α-synuclein. J Biol Chem. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bartels T, Choi JG, Selkoe DJ. alpha-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McLean PJ, Kawamata H, Shariff S, et al. TorsinA and heat shock proteins act as molecular chaperones: suppression of alpha-synuclein aggregation. J Neurochem. 2002;83:846–854. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- 83.Uryu K, Richter-Landsberg C, Welch W, et al. Convergence of heat shock protein 90 with ubiquitin in filamentous alpha-synuclein inclusions of alphasynucleinopathies. Am J Pathol. 2006;168:947–961. doi: 10.2353/ajpath.2006.050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leverenz JB, Umar I, Wang Q, et al. Proteomic identification of novel proteins in cortical lewy bodies. Brain Pathol. 2007;17:139–145. doi: 10.1111/j.1750-3639.2007.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Auluck PK, Meulener MC, Bonini NM. Mechanisms of Suppression of {alpha}-Synuclein Neurotoxicity by Geldanamycin in Drosophila. J Biol Chem. 2005;280:2873–2878. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 86.Dedmon MM, Christodoulou J, Wilson MR, Dobson CM. Heat shock protein 70 inhibits alpha-synuclein fibril formation via preferential binding to prefibrillar species. J Biol Chem. 2005;280:14733–14740. doi: 10.1074/jbc.M413024200. [DOI] [PubMed] [Google Scholar]
- 87.Lindersson E, Beedholm R, Hojrup P, et al. Proteasomal inhibition by alpha-synuclein filaments and oligomers. J Biol Chem. 2004;279:12924–12934. doi: 10.1074/jbc.M306390200. [DOI] [PubMed] [Google Scholar]
- 88.Pemberton S, Madiona K, Pieri L, Kabani M, Bousset L, Melki R. Hsc70 Protein interaction with soluble and fibrillar {alpha}-Synuclein. J Biol Chem. 2011;286:34690–34699. doi: 10.1074/jbc.M111.261321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roodveldt C, Bertoncini CW, Andersson A, et al. Chaperone proteostasis in Parkinson's disease: stabilization of the Hsp70/alpha-synuclein complex by Hip. EMBO J. 2009;28:3758–3770. doi: 10.1038/emboj.2009.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luk KC, Mills IP, Trojanowski JQ, Lee VM. Interactions between Hsp70 and the hydrophobic core of alpha-synuclein inhibit fibril assembly. Biochemistry. 2008;47:12614–12625. doi: 10.1021/bi801475r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang C, Cheng H, Hao S, et al. Heat shock protein 70 inhibits alpha-synuclein fibril formation via interactions with diverse intermediates. J Mol Biol. 2006;364:323–336. doi: 10.1016/j.jmb.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 92.Klucken J, Outeiro TF, Nguyen P, McLean PJ, Hyman BT. Detection of novel intracellular alphasynuclein oligomeric species by fluorescence lifetime imaging. FASEB J. 2006;20:2050–2057. doi: 10.1096/fj.05-5422com. [DOI] [PubMed] [Google Scholar]
- 93.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 Reduces alpha-synuclein aggregation and toxicity. J Biol Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 94.Shimshek DR, Mueller M, Wiessner C, Schweizer T, van der Putten PH. The HSP70 molecular chaperone is not beneficial in a mouse model of alpha-synucleinopathy. PLoS One. 2010;5:e10014. doi: 10.1371/journal.pone.0010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 96.Falsone SF, Kungl AJ, Rek A, Cappai R, Zangger K. The molecular chaperone Hsp90 modulates intermediate steps of amyloid assembly of the Parkinson-related protein alpha-synuclein. J Biol Chem. 2009;284:31190–31199. doi: 10.1074/jbc.M109.057240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kalia LV, Kalia SK, Chau H, Lozano AM, Hyman BT, McLean PJ. Ubiquitinylation of alpha-synuclein by carboxyl terminus Hsp70-interacting protein (CHIP) is regulated by Bcl-2- associated athanogene 5 (BAG5) PLoS One. 2011;6:e14695. doi: 10.1371/journal.pone.0014695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 99.Tetzlaff JE, Putcha P, Outeiro TF, et al. CHIP targets toxic alpha-Synuclein oligomers for degradation. J Biol Chem. 2008;283:17962–17968. doi: 10.1074/jbc.M802283200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ding X, Goldberg MS. Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PloS One. 2009;4:e5949. doi: 10.1371/journal.pone.0005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ko HS, Bailey R, Smith WW, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci U S A. 2009;106:2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zourlidou A, Payne Smith MD, Latchman DS. HSP27 but not HSP70 has a potent protective effect against alpha-synuclein-induced cell death in mammalian neuronal cells. J Neurochem. 2004;88:1439–1448. doi: 10.1046/j.1471-4159.2003.02273.x. [DOI] [PubMed] [Google Scholar]
- 103.Iwaki T, Wisniewski T, Iwaki A, et al. Accumulation of alpha B-crystallin in central nervous system glia and neurons in pathologic conditions. Am J Pathol. 1992;140:345–356. [PMC free article] [PubMed] [Google Scholar]
- 104.Braak H, Del Tredici K, Sandmann-Kiel D, Rub U, Schultz C. Nerve cells expressing heat-shock proteins in Parkinson’s disease. Acta Neuropathol. 2001;102:449–454. doi: 10.1007/s004010100395. [DOI] [PubMed] [Google Scholar]
- 105.Rose JM, Novoselov SS, Robinson PA, Cheetham ME. Molecular chaperone-mediated rescue of mitophagy by a Parkin RING1 domain mutant. Hum Mol Genet. 2011;20:16–27. doi: 10.1093/hmg/ddq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 107.Warrick JM, Paulson HL, Gray-Board GL, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 108.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 109.Hansson O, Nylandsted J, Castilho RF, Leist M, Jäättelä M, Brundin P. Overexpression of heat shock protein 70 in R6/2 Huntington's disease mice has only modest effects on disease progression. Brain Res. 2003;970:47–57. doi: 10.1016/s0006-8993(02)04275-0. [DOI] [PubMed] [Google Scholar]
- 110.Rujano MA, Kampinga HH, Salomons FA. Modulation of polyglutamine inclusion formation by the Hsp70 chaperone machine. Exp Cell Res. 2007;313:3568–3578. doi: 10.1016/j.yexcr.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 111.Miller VM, Nelson RF, Gouvion CM, et al. CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci. 2005;25:9152–9161. doi: 10.1523/JNEUROSCI.3001-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jana NR, Dikshit P, Goswami A, et al. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J Biol Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 113.Al-Ramahi I, Lam YC, Chen HK, et al. CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J Biol Chem. 2006;281:26714–26724. doi: 10.1074/jbc.M601603200. [DOI] [PubMed] [Google Scholar]
- 114.Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol. 2004;11:1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- 115.Helmlinger D, Bonnet J, Mandel JL, Trottier Y, Devys D. Hsp70 and Hsp40 chaperones do not modulate retinal phenotype in SCA7 mice. J Biol Chem. 2004;279:55969–55977. doi: 10.1074/jbc.M409062200. [DOI] [PubMed] [Google Scholar]
- 116.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 117.Selkoe DJ. Preventing Alzheimer's disease. Science. 2012;337:1488–1492. doi: 10.1126/science.1228541. [DOI] [PubMed] [Google Scholar]
- 118.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.The HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wernig M, Zhao JP, Pruszak J, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of Parkinson’s disease. Proc Natl Acad Sci U S A. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tomaskovic-Crook E, Crook JM. Human embryonic stem cell therapies for neurodegenerative diseases. CNS Neurol Disord Drug Targets. 2011;10:440–448. doi: 10.2174/187152711795564001. [DOI] [PubMed] [Google Scholar]
- 122.Wray S, Self M, NINDS Parkinson's Disease iPSC Consortium, NINDS Huntington's Disease iPSC Consortium, et al. Creation of an Open-access, mutation-defined fibroblast resource for neurological disease research. PLoS One 2012;7(8):e43099. [DOI] [PMC free article] [PubMed]
- 123.Koch P, Breuer P, Peitz M, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 124.Bilican B, Serio A, Barmada SJ, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci U S A. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shi Y, Kirwan P, Smith J, MacLean G, Orkin SH, Livesey FJ. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Sci Transl Med. 2012;4:124–129. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Camnasio S, Delli Carri A, Lombardo A, et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 127.Liu GH, Qu J, Suzuki K, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sánchez-Danés A, Richaud-Patin Y, Carballo-Carbajal I, et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mol Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Devine MJ, Ryten M, Vodicka P, et al. Parkinson's disease induced pluripotent stem cells with triplication of the α-synuclein locus. Nat Commun. 2011;2:440. doi: 10.1038/ncomms1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mazzulli JR, Xu YH, Sun Y, et al. Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Soga S, Akinaga S, Shiotsu Y. Hsp90 Inhibitors as anti-cancer agents, from basic discoveries to clinical development. Curr Pharm Des. 2013;19:366–376. doi: 10.2174/138161213804143617. [DOI] [PubMed] [Google Scholar]
- 132.Assimon VA, Gillies AT, Rauch JN, Gestwicki JE. Hsp70 protein complexes as drug targets. Curr Pharm Des. 2013;19:404–417. doi: 10.2174/138161213804143699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Waza M, Adachi H, Katsuno M, et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 134.Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280:33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 135.Labbadia J, Cunliffe H, Weiss A, et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Invest. 2011;121:3306–3319. doi: 10.1172/JCI57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chafekar SM, Duennwald ML. Impaired heat shock response in cells expressing full-length polyglutamine-expanded huntingtin. PLoS One. 2012;7:e37929. doi: 10.1371/journal.pone.0037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Donnelly A, Blagg BS. Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr Med Chem. 2008;15:2702–2717. doi: 10.2174/092986708786242895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yi F, Regan L. A novel class of small molecule inhibitors of Hsp90. ACS Chem Biol. 2008;3:645–654. doi: 10.1021/cb800162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ansar S, Burlison JA, Hadden MK, et al. A non-toxic Hsp90 inhibitor protects neurons from Abeta-induced toxicity. Bioorg Med Chem Lett. 2007;17:1984–1990. doi: 10.1016/j.bmcl.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 140.Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 141.Koulov AV, Lapointe P, Lu B, et al. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell. 2010;21:871–884. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Miyata Y, Koren J, Kiray J, Dickey CA, Gestwicki JE. Molecular chaperones and regulation of tau quality control: strategies for drug discovery in tauopathies. Future Med Chem. 2011;3:1523–1537. doi: 10.4155/fmc.11.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sahara N, Maeda S, Yoshiike Y, et al. Molecular chaperone mediated tau protein metabolism counteracts the formation of granular tau oligomers in human brain. J Neurosci Res. 2007;85:3098–3108. doi: 10.1002/jnr.21417. [DOI] [PubMed] [Google Scholar]
- 144.Abisambra JF, Jinwal UK, Suntharalingam A, et al. DnaJA1 Antagonizes Constitutive Hsp70-Mediated Stabilization of Tau. J Mol Biol. 2012;421:653–661. doi: 10.1016/j.jmb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fewell SW, Smith CM, Lyon MA, et al. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J Biol Chem. 2004;279:51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 146.Wisen S, Bertelsen EB, Thompson AD, et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem Biol. 2010;5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Braunstein MJ, Scott SS, Scott CM, et al. Antimyeloma effects of the heat shock protein 70 molecular chaperone inhibitor MAL3–101. J Oncol. 2011;2011:232037. doi: 10.1155/2011/232037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Walter GM, Smith MC, Wisen S, et al. Ordered assembly of heat shock proteins, Hsp26, Hsp70, Hsp90, and Hsp104, on expanded polyglutamine fragments revealed by chemical probes. J Biol Chem. 2011;286:40486–40493. doi: 10.1074/jbc.M111.284448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jinwal UK, Miyata Y, Koren J, 3rd, et al. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J Neurosci. 2009;29:12079–12088. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rampelt H, Kirstein-Miles J, Nillegoda NB, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kabbage M, Dickman MB. The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol Life Sci. 2008;65:1390–1402. doi: 10.1007/s00018-008-7535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Carrettiero DC, Hernandez I, Neveu P, Papagiannakopoulos T, Kosik KS. The cochaperone BAG2 sweeps paired helical filamentinsoluble tau from the microtubule. J Neurosci. 2009;29:2151–2161. doi: 10.1523/JNEUROSCI.4660-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Elliott E, Tsvetkov P, Ginzburg I. BAG-1 associates with Hsc70Tau complex and regulates the proteasomal degradation of Tau protein. J Biol Chem. 2007;282:37276–37284. doi: 10.1074/jbc.M706379200. [DOI] [PubMed] [Google Scholar]
- 154.Connell P, Ballinger CA, Jiang J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 155.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hutchison KA, Dittmar KD, Czar MJ, Pratt WB. Proof that hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with hsp90. J Biol Chem. 1994;269:5043–5049. [PubMed] [Google Scholar]
- 157.Danzer KM, Kranich LR, Ruf WP, et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Danzer KM, Ruf WP, Putcha P, et al. Heatshock protein 70 modulates toxic extracellular alphα-synuclein oligomers and rescues transsynaptic toxicity. FASEB J. 2011;25:326–336. doi: 10.1096/fj.10-164624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Borrell-Pagès M, Canals JM, Cordelières FP, et al. Cystamine and cysteamine increase brain levels of BDNF in Huntington disease via HSJ1b and transglutaminase. J Clin Invest. 2006;116:1410–1424. doi: 10.1172/JCI27607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marinova Z, Ren M, Wendland JR, et al. Valproic acid induces functional heat-shock protein 70 via Class I histone deacetylase inhibition in cortical neurons: a potential role of Sp1 acetylation. J Neurochem. 2009;111:976–987. doi: 10.1111/j.1471-4159.2009.06385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rao R, Fiskus W, Ganguly S, Kambhampati S, Bhalla KN. HDAC inhibitors and chaperone function. Adv Cancer Res. 2012;116:239–262. doi: 10.1016/B978-0-12-394387-3.00007-0. [DOI] [PubMed] [Google Scholar]
- 163.Moumné L, Campbell K, Howland D, Ouyang Y, Bates GP. Genetic knock-down of HDAC3 does not modify disease-related phenotypes in a mouse model of Huntington's disease. PLoS One. 2012;7(2):e31080. doi: 10.1371/journal.pone.0031080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tam S, Spiess C, Auyeung W, et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 890 kb)