Abstract
Huntington’s disease (HD) typifies a class of inherited neurodegenerative disorders in which a CAG expansion in a single gene leads to an extended polyglutamine tract and misfolding of the expressed protein, driving cumulative neural dysfunction and degeneration. HD is invariably fatal with symptoms that include progressive neuropsychiatric and cognitive impairments, and eventual motor disability. No curative therapies yet exist for HD and related polyglutamine diseases; therefore, substantial efforts have been made in the drug discovery field to identify potential drug and drug target candidates for disease-modifying treatment. In this context, we review here a range of early-stage screening approaches based in in vitro, cellular, and invertebrate models to identify pharmacological and genetic modifiers of polyglutamine aggregation and induced neurodegeneration. In addition, emerging technologies, including high-content analysis, three-dimensional culture models, and induced pluripotent stem cells are increasingly being incorporated into drug discovery screening pipelines for protein misfolding disorders. Together, these diverse screening strategies are generating novel and exciting new probes for understanding the disease process and for furthering development of therapeutic candidates for eventual testing in the clinical setting.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0195-4) contains supplementary material, which is available to authorized users.
Keywords: Polyglutamine diseases, High-throughput screening, Drug discovery, Model organism
Introduction
At least nine dominantly inherited human neurodegenerative diseases, including Huntington’s disease (HD), spinal-bulbar muscular atrophy, dentatorubral-pallidoluysian atrophy, and spinocerebellar ataxias (SCAs) are caused by CAG-repeat expansions in the coding region of the respective disease gene [1]. The huntingtin gene (HTT) mutation was identified in 1993 [2] as a trinucleotide CAG expansion encoding polyglutamine (polyQ) near the amino terminus (N-terminus) of the large Huntingtin (HTT) protein. Similarly, spinal-bulbar muscular atrophy, dentatorubral-pallidoluysian atrophy, and ataxias were found to be caused by CAG-repeat expansions in the genes encoding the androgen receptor, atrophin, and ataxin proteins, respectively. In each case, an abnormally expanded polyQ tract causes misfolding and accumulation of the mutant protein in cellular inclusions, which are associated with progressive neuronal degeneration and cell loss in different regions of the brain. These neurodegenerative diseases are characterized by progressive motor, cognitive, and neuropsychiatric deficits, with only modest symptomatic management achievable with medication and physical and occupational therapy [3, 4]. As no curative therapy exists for any of these polyQ-driven diseases, there is an urgent need to develop and improve drug discovery screening technologies targeting these devastating disorders.
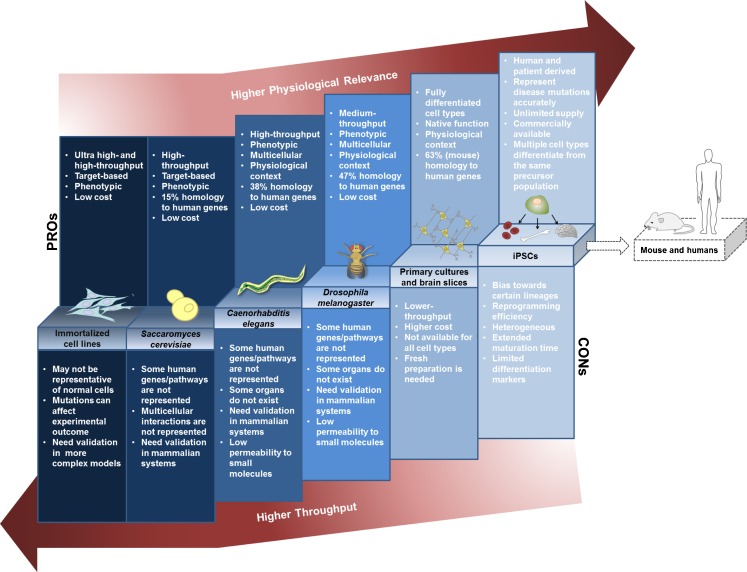
The discovery that the proximal causes of these diseases emanate from CAG expansions within single genes has enabled the generation of genetic models with high relevance and specificity to each disease. For example, expression of all, or part of the HTT gene containing the CAG expansion in mice recapitulates key aspects of HD, including the development of cellular inclusions, motor and cognitive symptoms, and neuronal pathology [5, 6]. Analogous genetic models for HD and other polyQ diseases have also been developed in the budding yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans, and the fruit fly Drosophila melanogaster, as well as in neuronal cell lines and most recently in induced pluripotent stem cells (iPSCs) derived from patients (Fig. 1). While any such model will have intrinsic advantages, as well as limitations, in the context of drug discovery—generally weighing physiological/clinical relevance against throughput and cost—collectively these screening models provide a powerful critical path for the discovery, optimization, and validation of new candidate drugs and drug targets.
Fig. 1.
Model systems for drug and target discovery in polyQ disorders balance physiological relevance against cost and throughput. The simplest models have high throughput, but limited biological complexity, whereas tissue and organismal models retain high biological complexity, but have more modest throughput capacities. Note that the categories shown are not intended to represent hard distinctions, but rather a continuum of screening solutions balancing throughput against biological complexity
Here we will review some recently developed models of polyQ disease and how these models have been implemented in high-throughput screening (HTS) campaigns to identify new therapeutic targets and compounds for disease modification. Rather than describing each type of molecular or cell/organism-based model in turn, we will discuss how these models have been employed to achieve different goals in the drug discovery development process, including compound screening with disease gene-specific and non-specific assay readouts; genetic screening using RNA interference (RNAi), chemical mutagenesis, or transposon insertions; and proteomic screening. The latter two approaches have been applied primarily for target identification and validation, while compound screens have been done in the full range of such disease models, from yeast to cell- and tissue-based models to whole animal model organisms. While we will provide examples of new drug and drug target candidates emerging from such studies, we will regrettably not be able to review the field in a comprehensive manner owing to space limitations. We thus refer the reader to recent and excellent reviews focusing on other aspects of polyQ drug discovery and disease models [7–14].
Molecular Screening to Target PolyQ Protein Levels and Aggregation
The monogenic nature of HD, together with the likelihood that a major portion of disease pathogenesis arises from gain-of-function mechanisms [15, 16], suggest that molecular screening assays based on mutant HTT (mHTT) proteins could be used to identify proximal events in disease initiation. In fact, one of the first observable molecular phenotypes of polyQ diseases was the presence of large visible protein inclusions, comprised of the mutant misfolded protein and associated cellular proteins [17–19]. Thus, molecular screening assays to inhibit mutant polyQ protein aggregation were among the first high-throughput screens developed for these diseases (see [8] and [20] for review). A number of such campaigns done in vitro and in cellular models identified small molecules and peptides that reduced aggregation (Table 1) [21–25].
Table 1.
Chemical modifier screens
| Phenotype | Expressed protein | Assay system | Hits | Secondary assays | Ref. |
|---|---|---|---|---|---|
| HSF-1 activation | ~1 × 106 compound screen in HeLa cells | A1, A3, C1, D1, F1 | Mammalian cellular and Caenorhabditis elegans models of HD | [51] | |
| HSF-1 activation | 10,000 compound library screen in yeast | HSF1A (TRiC/CCT complex) | Mammalian cellular and Drosophila models of HD | [52] | |
| Fly motor defect | HTT Q128 | 521 quinazoline-derived compound screen in Drosophila neurons | EVP4593 | Medium spiny neurons | [81] |
| Toxicity | HTT480 Q68 | 40,000 compounds in primary striatal neuron culture | [56] | ||
| Toxicity | HTTN90 Q8 and Q73 | ~400 selected small molecule screen in primary cultures of cortical and striatal neurons | Inhibitors of Rho kinase, phosphodiesterase, adenosine 2A receptor, and IKKβ | [57] | |
| Toxicity | HTT588 Q138 and Q15 | ~2600 small molecule screen in Drosophila primary neuronal culture | Camptothecin, OH-camptothecin, 18β-glycyrrhetinic acid, and carbenoxolone | Drosophila HD model | [66] |
| Toxicity | HTT exon 1 Q25- and Q103-EGFP | 68,887 compound screen in PC12 cells | 16F16, thiomuscimol, cystamine | Cortico-striatal brain slice models of HD and AD | [61] |
| Toxicity | HTTN90 Q73 | 74 drug-like compound screen in rat brain slices | Inhibitors of IKK complex, CXCR3 chemokine receptor, c-Jun N-terminal kinase, and adenosine 2A receptor | [60] | |
| Toxicity | HTT exon 1 Q150 | 9 selected compound screen in C. elegans ASH neurons | LiCl, TSA, SAHA, and mithramycin | [80] | |
| Aggregation and toxicity | HTT Q103-EGFP | 16,000 compound library screen in yeast | C2-8 | Mammalian cells, brain-slice, and Drosophila models of HD; R6/2 transgenic mouse | [41] |
| Aggregation | GST-HTT exon 1 Q51 | In vitro screen of ~184,000 small molecule library screen | Benzothiazoles | 293 Tet-Off cells | [21] |
| Aggregation | HTT171 Q58 | In vitro screen of 1040 FDA-approved drugs and bioactive compounds | Gossypol, gambogic acid, juglone, celastrol, sanguinarine, and anthralin | HdhQ111/Q111 striatal cells | [22] |
| Aggregation | GST-Q62 | In vitro screen of peptide phage display library | Six tryptophan-rich peptides (polyQ-binding peptide 1) | COS-7 cell model of HD | [23] |
| Aggregation | GST-HTT exon 1 Q51 | In vitro screen of ~5000 natural substances | EGCG and related polyphenols | Yeast and Drosophila models of HD | [24] |
| Aggregation | AR127 Q65 | In vitro screen of 2800 biologically active compounds | Y-27632 | Drosophila model of HD | [25] |
| Aggregation | GST-HTT exon 1 Q51 | In vitro screen of 11 small molecules | Congo red, thioflavine S, chrysamine G, and Direct fast yellow | COS-7 cells transfected with HTT 52Q | [26] |
| Aggregation | HTTNT fragment | In vitro screen of 15 peptides | HTTNT-based peptides | SH-SY5Y cells | [27] |
| Aggregation | HTTQ72-Luciferase | 2687 small molecule screen in HEK-293 cells | Leflunomide and teriflunomide | [40] | |
| Degradation (clearance) | HTT573 Q72 | 10,000 natural compound library screen in HN10 neuronal cell line | TSA analogue, staurosporine, anisomycin, cycloheximide, borrelidin, BAY 61-3606 | [46] | |
| Degradation (clearance) | HTT573 Q72 and Q25 | ~2 × 106 compound screen in HN10 neuronal cell line | Heat shock protein 90 inhibitors | HdhQ150 embryonic stem cells and in embryonic stem cell-derived neurons | [48] |
Blank entry indicates not included in the study.
HSF = heat shock factor; HTT = huntingtin; TriC/CCT = chaperonin containing TCP-1; EGFP = enhanced green fluorescent protein; GST = glutathione-S-transferase; FDA = Food and Drug Administration; HEK = human embryonic kidney; IKKβ = IκB kinase; CXCR3 = chemokine (C-X-C motif) receptor 3; LiCl = lithium chloride; TSA = trichostatin A; SAHA = suberoylanilide hydroxamic acid; EGCG = (−)-epigallocatechin-3-gallate; AR = androgen receptor; HD = Huntington’s disease; AD = Alzheimer’s disease.
Molecular assays directed against polyQ disease proteins have been designed in diverse ways. Examples of pure in vitro assays for aggregation of recombinant proteins include quantification of aggregate formation using the filter-trap aggregation assay [21, 24, 26] and the sedimentation assay [27]. Both assays have been used for the identification of compounds with inhibitory properties against mHTT aggregation. More recently, in vitro assays have been used to define more clearly the toxic protein species [28, 29]. For example, methylene blue (MB) reduced aggregation as measured by the filter trap method when added to monomers or preformed fibrils in vitro [30]. MB was then shown to diminish different mHTT species isolated from primary cortical neurons using an agarose gel method that distinguishes aggregates and oligomers [31]. MB is currently in late-stage clinical trials as a tau aggregation inhibitor for mild-to-moderate Alzheimer’s disease (AD) [32].
To incorporate cellular context, a number of laboratories have designed assay platforms in which aggregation can be monitored within living cells. For example, aggregation-prone proteins fused to fluorescent tags that emit a Förster/fluorescence resonance energy transfer (FRET) signal were used to screen a ~2,800 compound library and identified the Rho kinase (ROCK) inhibitor Y-27632 as a hit compound [25]. Y-27632 reduced the FRET signal by nearly half and reduced the loss of photoreceptor neurons expressing mHTT exon 1 in a Drosophila model. Although Y-27632 has limited potency [33], the discovery of ROCK inhibitors as aggregation inhibitors enabled the characterization of a ROCK–profilin signaling pathway [34, 35]. Profilin binding to mHTT appears to maintain mHTT solubility, and ROCK phosphorylation of profilin reduces the protective profilin–mHTT interaction. Recently, administration of the clinically-approved ROCK inhibitor HA-1077 (Fasudil) was shown to rescue retinal degeneration in the R6/2 mouse model expressing human HTT exon 1 with >150 CAGs [36, 37]. Thus, ROCK inhibition has been strongly implicated as a mechanistic pathway for intervention in HD.
Automated time-lapse microscopy has been used to analyze live primary neuronal cultures for aggregation of GFP-tagged polyQ proteins [38, 39]. By following individual cells over time, visible aggregate morphology could be anticorrelated with the risk of cell death. Another aggregation model that used restoration of luciferase activity from a polyQ-luciferase reporter identified MB and several other compounds as aggregation inhibitors [40]. Terflunomide reduced incorporation of polyQ-containing proteins into aggregates resulting in smaller aggregates, but did not disaggregate polyQ protein or reduce polyQ-associated toxicity in human embryonic kidney (HEK) 293 cells [40]. Such complex screening methodologies highlight the value of distinguishing mechanisms of action for aggregation inhibitors, even at the primary screening stage.
Aggregation-based screens can also be done in the context of genetic model organisms, such as a yeast-based primary screen of 16,000 small molecules for suppressors of HTT103Q-mediated aggregation [41]. A subsequent secondary screen of a focused compound library in mammalian cell models of polyQ disease identified compounds that increased soluble HTT103Q levels. The potency of these compounds in inhibiting mHTT aggregation was then tested in organotypic hippocampal slices obtained from R6/2 mice. Among the 7 compounds tested in this model, only the sulfobenzoic acid derivative C2-8 showed a marked effect in reducing mHTT aggregate load in neurons, despite having no significant effect in reducing the total number of aggregates. Compound C2-8 was also shown to have neuroprotective activity in vivo by rescuing photoreceptor degeneration in a Drosophila eye model of HD. Although the precise mechanism by which C2-8 reduced aggregation remains unknown, the authors provided evidence that the compound could be targeting the polymerization step of the polyQ aggregation process. Of interest, compound C2-8 was recently shown to cross the blood–brain barrier, improve motor function, and reduce striatal neuronal atrophy in R6/2 mice, although overall survival was not extended [42]. The discovery of C2-8 and other sulfobenzoic acid derivatives also uncovered a hitherto unknown connection between SIRT2 inhibition, cholesterol homeostasis, and mHTT aggregation [43, 44]. Compounds with SIRT2 inhibitory activity were also neuroprotective against mutated alpha-synuclein in cell culture models [45].
Finally, methods that employ time-resolved FRET (TR-FRET) have also been designed to quantify soluble and aggregated mHTT levels from samples derived from screening campaigns, such as those described above, from whole-animal models, and from patient samples as potential progression and drug response biomarkers in the clinical context [46, 47]. Detection of mHTT in this system depends on the use of two labeled antibodies, including one that recognizes the polyQ expansion; thus, the TR-FRET signal is specific for mHTT, with little or no crossover to the wild type (WT) protein. Importantly, this method has the benefit of being homogeneous, robust and scalable for HTS [46]. Although, to date, TR-FRET screens have primarily identified previously known target classes [46, 48], they have strong potential to identify novel targets as well. A quantification method for aggregated mHTT TR-FRET has also been developed based on dual labeling of a single antibody that recognizes mHTT [49].
Target-Based and Phenotypic Screening Using High-Content Analysis
In addition to reducing soluble and/or aggregated mHTT, screening assays can also be based on defined molecular targets (target-based) or specific cellular states, and/or features believed to be associated with the disease state (phenotypic screening; including high-content analysis). Such assay endpoints are often linked within a screening campaign to associate target engagement directly with a desired phenotypic outcome, such as protection against cell death. A limitation in the field is that no truly validated molecular targets for HD are known in the strictest sense, as there are, currently no approved drugs for HD with disease-modifying ability. However, numerous candidate molecular targets have emerged from basic science studies, as well as target screening campaigns (see below), and many of these have been employed in screening assays to generate tool compounds and/or biologics for subsequent hypothesis testing and potential clinical development. Alternatively, many laboratories have generated phenotypic screening assays that are hypothesis-neutral with respect to molecular target(s), and instead focus on rescue of particular cellular phenotypic/pathogenic features induced by the introduction of mutant polyQ proteins.
HTT is intrinsically the most relevant molecular target in HD, and itself can be the focus of a targeted molecular screen. For example, a high-throughput Western blot-based screen done in cell-based models demonstrated the involvement of matrix metalloproteinases in proteolytic cleavage of mHTT—a key event in the pathogenesis of HD—and in associated cellular toxicity [50]. Another example of a target-based screen was one in which the primary endpoint was the activation of a heat shock protein HSP70 promoter-luciferase reporter construct responsive to the activity of heat shock factor-1 (HSF-1), the master regulator of the heat shock response [51]. Large-scale screening was designed to identify compounds that could increase HSF-1 activity and thus drive the luciferase reporter. Hit compounds were then advanced to show suppression of aggregate formation and reduction of toxicity in a C. elegans polyQ model (see following section on model organisms). Importantly, the hit compounds did not seem to act by inducing protein misfolding, or by inhibiting HSP90 or the proteasome.
A similar approach was taken in a yeast strain that depended on human HSF-1 activation for growth in which a library of 10,000 non-biased small molecules was screened for activators of HSF-1 [52]. Two benzyl-pyrazole compounds (HSF1A and HSF1C) were identified as effective HSF-1 activators and subsequently shown to reduce polyQ aggregation and cell death in a cell culture model; they were also shown to ameliorate disease-like phenotype in a Drosophila model of SCA3. The compound HSF1A was shown to bind to the TRiC/CCT cytosolic chaperone complex, which had been previously identified as a mHTT binding partner in a proteomic screen [53], and had also been shown to reduce polyQ aggregation/toxicity in mammalian and yeast models [54, 55]. Thus, CCT might play an essential role in modulating the folding of polyQ-expanded proteins and could be a prime target for further therapeutic development.
Cell-based phenotypic screening for polyQ disorders has frequently used cell degeneration and/or death as primary endpoints. For example, expressing a 480-amino acid mHTT fragment with 68Q in primary striatal neuronal cultures led to 50 % cell death in a 6-day assay window [56]. This assay was used to screen 40,000 compounds and generated candidate compounds undergoing further characterization. Similarly, a high-content, composite cortical neuron, striatal neuron, and astroglial co-culture screening platform was described [57]. Striatal and cortical cell populations could be visualized separately through electroporation of different fluorescent proteins before co-culturing, with loss of fluorescence as a quantitative readout for cell survival. In this platform, expression of mHTT exon 1 fragment (73 CAG) reduced striatal and cortical neuron survival compared with normal Q-length controls. Screening in 96-well plate format led to identification of several compound classes with known and novel neuroprotective actions, including compounds that had been previously identified in screens using different endpoints (e.g., the ROCK inhibitor Y-27632) [25] or acted on gene targets implicated in other HD models (e.g., inhibitor of kappa B (IκB) kinase and the adenosine 2A receptor) [58–60].
Induction of apoptosis by a mHTT fragment in the pheochromocytoma cell line PC12 was used to screen nearly 70,000 compounds [61]—several hits from which appeared to bind to a common protein target. The cellular target was subsequently identified as the molecular chaperone protein disulfide isomerase by using a novel adaptation of the "click" chemistry approach [62]. These compound hits were subsequently shown to be neuroprotective in ex vivo HD and AD brain slice models, supporting the predictive value of phenotypic screens based in PC12 cells. Moreover, the authors presented evidence that protein disulfide isomerase may also be a molecular target for cystamine, which had previously been shown to provide benefit in a whole-animal HD model [63, 64].
A large-scale screen using nearly 8,000 small interfering RNA pools representing the "druggable genome" was carried out in a HEK293 cell line that underwent caspase activation when transfected with an N-terminal mHTT fragment (558 amino acids with 141Q) [65]. Interestingly, although this initial screen was done in a non-neuronal cell line, mHTT modifier gene hits were enriched in neurologic disease functions. Moreover, connecting nodes in a suppressor gene network included genes such as nuclear factor-kappa B [58, 60] with established roles in HD pathology. A novel pathway was also identified, as small interfering RNAs to multiple components of the related rat sarcoma (RRAS) viral oncogene homolog pathway emerged as mHTT toxicity suppressors. Subsequently, aberrant RRAS signaling was confirmed in the striatum of R6/2 mice, and RRAS inhibition was protective in a knock-in cell line (HdhQ111), as well as a Drosophila HD model [65].
For phenotypic screening in neurons, the neuronal dendritic arbor can be a very sensitive and quantitative indicator of cell state and vitality. An interesting example of such a screen was done in primary neuronal cultures generated from Drosophila transgenic lines expressing a 588-amino acid fragment of HTT with either 15Q or 138Q, and tagged with monomeric red fluorescent protein (mRFP) to monitor aggregation [66]. The strain was also transgenic for a membrane-targeted green fluorescent protein (GFP), which allowed for visualization of dendrites in the primary neurons. mHTT-expressing primary neurons showed altered dendritic and axonal morphologies that could be distinguished from the normal Q-length control. Using these assay endpoints for mHTT aggregation, neuromere size, and dendritic length, 2,600 compounds and a 468-gene kinase/phosphatase RNAi sub-library were screened in 384-well plates with the mammalian target of rapamycin/insulin pathway negative regulator lkb1, as well as camptothecins, emerging as hits.
Similarly, synapses and synaptic function can be very sensitive surrogate endpoints for neuronal phenotype and function, and for states of neural circuits. In this context, a HTS was established to measure synaptic activity using a pH-sensitive GFP fused to synaptophysin [67]. Fluorescence develops only upon synaptic vesicle fusion and exposure of the GFP tag to the normal pH of the synaptic cleft. Using a fully automated 96-well plate based system, neurons could be stimulated and the rate of exocytosis of the synaptic vesicle label measured. Adenosine 1 receptor agonists were found to suppress synaptic vesicle movement using this technique. Alternatively, synaptic connections can be directed through microchannels (“synapse microarray”) in a more stereotyped fashion to facilitate automated analysis [68]. Using this system, 22 compounds were evaluated at 3 doses and several histone deacetylase (HDAC) inhibitors that enhanced the synaptic signal were identified.
High-content analysis methods have also been used in primary cultured cortical neurons to detect compounds that specifically influenced synaptogenesis and dendritic outgrowth [69]. Using immunocytochemistry against microtubule-associated protein 2 to label dendrites and against synapsin to label synapses, several compounds were tested and found to reduce dendritic lengths and/or synapse numbers in a concentration-dependent manner. Interestingly, the compound mevastatin appeared to suppress synapse number specifically without shortening dendrites.
Three-dimensional (3D) culturing systems represent a way to build biological complexity while maintaining the scalability of cell culture. For example, 3D hydrogel tissue constructs formed through extracellular matrix remodeling can measure contractile force, which does not occur in two-dimensional culture [70]. Similarly, 3D spheroid cultures derived from SH-SY5Y neuroblastoma cells expressing tau variants have been used as a model of tauopathy [71].
To mimic the native 3D tissue context of neurons even more closely, brain slice explants can be used as a screening platform [72, 73]. For example, biolistic transfection has been used to introduce mHTT constructs together with fluorescent visual reporters to develop phenotypic assays for mHTT-induced neuronal degeneration [74]. The brain slice preparation allows a variety of neuronal features to be assayed, including dendritic arborization, and allows direct quantitative readouts of neuronal loss. Moreover, intact brain tissue also retains cell-based neuroinflammatory responses, and a number of neuroprotective compounds identified in brain slice screens were likely to have neuroinflammatory pathways as their primary targets [60]. The application of novel methodologies such as these to HD cell- and tissue-based models are supporting the development of valuable new screening platforms for phenotypic features that may not be expressed in conventional monolayer cell cultures (Fig. 1).
Drug and Drug Target Screening in Invertebrate Animal Models
Combining biological complexity with amenability to high-throughput technologies, invertebrate model organisms, such as yeast, flies, and worms, can help to bridge the gap between in vitro and mammalian in vivo assays (Fig. 1). Such model organisms can provide powerful genetic screening platforms for drug and especially drug target discovery in polyQ diseases. In the following sections we will describe examples of small molecule, genetic, and RNAi screens that have been performed in model organisms, both at the small and large scale (Table 2). We will also discuss recent proteomic studies and associated follow-on validation studies in model organisms. As more data emerge from such screens and can be harmonized and analyzed, key concepts and driving pathways for disease pathogenesis should increasingly emerge.
Table 2.
Genetic, RNA interference (RNAi) and proteomic modifier screens
| Phenotype | Expressed protein | Tissue | Assay system | Hits | Secondary assays | Ref. |
|---|---|---|---|---|---|---|
| Toxicity | HTT exon 1 Q20 and Q53 | Genome-wide loss-of-function enhancer screen in yeast | Genes involved in stress responses, protein folding, and ubiquitin-dependent protein degradation | [84] | ||
| Toxicity | HTT exon 1 Q103 | Genome-wide loss-of-function suppressor screen in yeast | Genes involved in vacuolar transport, transcriptional regulation/maintenance of chromatin structure, other processes (bna4) and prion genes | [85] | ||
| Aggregation | Ade-2 Q25 and Q97 | Transposon insertion mutants in yeast | Genes involved in vacuolar transport, transcriptional regulation (Spt4) | Murine striatal neuron cell models of HD | [86] | |
| Toxicity | HTT exon 1 Q2 and Q150 | ASH sensory neurons | Mutation screen in Caenorhabditis elegans | polyQ enhancer-1 (pqe-1) gene | [95] | |
| Toxicity | HTT exon 1 Q128 | Touch receptor neurons | 6034 candidate RNAi screen in C. elegans | Genes involved in cell death, protein folding, intracellular transport, metabolic processes, response to stress, stress-activated pathways | Striatum of HD mouse models | [101] |
| Aggregation | Q35-YFP | BWM | Genome-wide RNAi in C. elegans | Genes involved in RNA synthesis/processing, protein synthesis, transport, folding, and degradation | [102] | |
| Aggregation | Q35-YFP | BWM | Genome-wide RNAi in C. elegans | Genes involved in cell cycle, DNA and RNA synthesis/processing, energy and metabolism, and protein synthesis, transport and folding | C. elegans expressing Q37-YFP, SODG93A, and temperature sensitive proteins | [105] |
| Toxicity | Q127 | Retina | 7000 P-element insertions in Drosophila | DNAJ domain-containing proteins dHDJ1 and dTPR2 | Transgenic Drosophila lines | [90] |
| Toxicity and aggregation | Ataxin-1 Q30 and Q82 | Retina, central nervous system | Transposon insertion in Drosophila | Genes involved in protein folding, degradation, transcriptional regulation, RNA binding and stress response | [91] | |
| Toxicity | Ataxin-1 Q82, ataxin-3 Q78 and Q127 | Retina | 55 modifier strains in Drosophila | Genes involved in protein folding and apoptosis | Drosophila brain | [145] |
| Toxicity and aggregation | Ataxin-3tr Q78 | Retina | 2300 enhancer/promoter element insertion library in Drosophila | Chaperones and ubiquitin-pathway components, and genes involved in transcription, translation, and RNA binding | Drosophila model of tau-induced degeneration | [92] |
| Toxicity and aggregation | HTT588 Q138 | Neuronal and non-neuronal tissues | Autosomal deficiency (Df) kit for chromosome II and III (160 Df lines) | Small C-terminal domain phosphatases and Antennapedia complex | Drosophila HD model | [93] |
| Toxicity | HTT588 Q138 and Q15 | Whole genome (468 genes) kinase/phosphatase RNAi sub-library in Drosophila neuronal culture | lkb1 | Drosophila HD model | [66] | |
| Toxicity | HTT exon 1 Q97 | Mouse brain, mouse muscle | Y2H, MS | Proteins involved in folding, synaptic transmission, cytoskeletal organization, signal transduction, and transcription | Drosophila | [53] |
| HTT Q144 | Mouse brain | Affinity purification/MS | Proteins involved in translation | [113] | ||
| Toxicity | HTT Q97 | Mouse brain | Affinity purification/MS | Proteins involved in protein folding, 14-3-3 signaling, microtubule-based intracellular transport, and mitochondrial function | Drosophila | [114] |
| Toxicity | HTT Q72 | iPSC lines | 2-DE/MS | Genes involved in oxidative stress and apoptosis | [136] |
Blank entry indicates not included in the study.
HTT = huntingtin; HD = Huntington’s disease; YFP = yellow fluorescent protein; BWM = body wall muscle; SOD = superoxide dismutase; TRP2 = tetratricopeptide repeat protein 2; Y2H = yeast 2-hybrid; MS = mass spectrometry; iPSC = inducible pluripotent stem cell; 2-DE = two-dimensional electrophoresis.
Model organisms commonly used in polyQ HTS drug and target discovery include S. cerevisiae, C. elegans, and D. melanogaster. Despite being a simple unicellular organism S. cerevisiae has made fundamental contributions to the elucidation of essential eukaryotic pathways and cellular mechanisms (reviewed in [75]). In fact, about one third of the yeast genome has direct human orthologs; in addition, nearly 500 human disease genes are orthologous to yeast genes, making this organism an apt tool for the study of human diseases [76]. While yeast cannot model the complexity of mammalian neural networks, the presence of conserved eukaryotic pathways allows extrapolation of findings obtained in this organism to humans. For example, conservation of protein-control quality machinery between yeast and higher eukaryotes makes S. cerevisiae an extremely useful model organism to study neurodegenerative diseases associated with protein misfolding and aggregation [75]. Finally, yeast is highly amenable to HTS, allowing for the identification of compounds with inhibitory properties against disease protein-mediated cellular phenotypes.
C. elegans also offers an effective model in which to study neurodegenerative diseases, including diseases associated with polyQ expansions (reviewed in [77]). Although like yeast C. elegans does not contain its own HTT ortholog, expression of expanded CAG constructs induces cellular and behavioral phenotypes reminiscent of cellular dysfunction and degeneration in mammalian systems. Worm polyQ models have thus been used to test the effects of known drugs or to identify novel drugs and gene candidates that modify aggregation/toxicity of polyQ-disease proteins.
Finally, D. melanogaster has proven to be a powerful tool for studying the pathology of human diseases both at the molecular and cellular levels (reviewed in [78]). Drosophila genes share about 50 % homology with the human genome, and important biological pathways are highly conserved. In addition, Drosophila has a relatively complex nervous system and a brain, providing relevance for the study of neurodegenerative diseases. Together with the extensive genetic tools available, these features have enabled Drosophila to be incorporated very effectively into large-scale genetic and pharmacologic screens for the identification of human disease genes and potential therapeutic leads [78].
Small Molecule Screening With Model Organisms
Several groups have taken advantage of the amenability of model organisms to high-throughput technologies to screen libraries of chemical compounds for the identification of inhibitors of polyQ-induced phenotypes (Table 2). For example, a C. elegans model of HD in which neuronal expression of the N-terminal fragment of human HTT carrying a tract of 150Q (HTT exon 1 Q150) induced progressive neurodegeneration of ASH sensory neurons was used to screen a collection of compounds previously reported to ameliorate death in cellular and animal models of polyQ diseases [79]. A subset of the compounds tested was able to decrease polyQ-mediated death of ASH sensory neurons [80].
Analogously, a Drosophila HD model in which pan-neuronal expression of mHTT resulted in a motor phenotypic impairment (limb tremors and climbing) was used in a screen of 521 quinazoline-derived compounds that identified compound EVP4593 as a potent suppressor of the climbing defect [81]. In secondary assays, EVP4593 was also shown able to protect cultured primary medium spiny neurons (MSNs) isolated from transgenic HD mice (YAC128) from glutamate-induced toxicity. Follow-up studies suggested that one or more subunits of the transient receptor potential (TRP) channels could be the molecular targets of the compound, adding to other reports supporting a role of TRP channel family members [82] in neurodegeneration [83].
Genetic and RNAi Screens
In addition to allowing the identification of lead compounds, model organisms provide an excellent tool for the discovery of genes (this section) and proteins (see following section on proteomic screening approaches) as potential drug target candidates. Genome-wide genetic screens for modifiers of mHTT-mediated toxicity in yeast have identified several genetic pathways and molecular mechanisms underlying neurodegeneration in HD that could be exploited for therapeutic purposes. For example, genes that enhance mHTT-mediated toxicity have been found to be involved in the response to cellular stress, protein folding, and degradation [84], whereas genes with a predicted role in vesicle transport, vacuolar degradation, transcriptional regulation, and prion-like aggregation have been shown to be suppressors of mHTT toxicity [85].
A recent phenotypic yeast-based screen was used to assay for transposon insertion mutants that could restore the activity of the Ade-2 protein containing 97Q [86]. An interesting hit from this screen was SPT4, a transcription elongation factor that reduces RNA polymerase II dissociation from its template, thereby increasing polymerase processing [86]. Accordingly, a deletion in SPT4 led to a general decrease in transcription elongation, which, surprisingly, was more pronounced in genes encoding long stretches of CAG or other repetitive sequences (e.g., CAA and AAA repeats). In turn, reduced transcription of expanded polyCAG stretches was shown to reduce aggregation of the mutant Ade-2 protein, and also to diminish its co-aggregation with Ade-2-containing short polyQ stretches. The beneficial effects of deleting SPT4 in yeast were reproduced in a mouse model of HD, where knockdown of the ortholog Supt4h led to reduction of HTTQ111 aggregation, with no effect on HTTQ7 expression.
Transgenic Drosophila models of human polyQ-mediated diseases have been also used in high-throughput genetic screens for modifiers of mutant protein-induced aggregate/toxicity (reviewed in [87–89]). Using the fly eye as a model to study neurodegeneration, two groups ran genetic screens to identify modifiers of either pure polyQ tracts (127Q repeats) or ataxin-1 82Q-mediated toxicity [90, 91]. A screen for modifiers of polyQ-induced toxicity identified two suppressor genes encoding the dHDJ1 and dTPR2 proteins, which contain a J domain known to stimulate HSP70 activity and prevent protein aggregation [90]. Similarly, a screen for modifiers of mutant ataxin-1 toxicity identified components of the protein folding (dHDJ1) and clearance pathways (ubiquitin conjugases), as well as novel modifiers involved in RNA processing, transcriptional regulation, and cellular detoxification [91]. A genetic screen for modifiers of Drosophila eye degeneration induced by mutant ataxin-3 (ataxin-3tr Q78) identified 14 suppressors and one enhancer [92]. Analogous to the two studies described above [90, 91], some suppressors belonged to the chaperone and ubiquitin-pathway component classes in addition to suppressor genes with roles in transcription, translation, and nuclear export. Interestingly, some of the suppressor genes, such as the co-chaperone TPR2 and polyubiquitin, were also able to reduce tau-mediated degeneration, indicating that chaperones and the ubiquitin-proteasome system may be general modifiers for different protein conformation diseases [92].
More recently, a haplo-insufficiency screen for suppressors of aggregation and toxicity mediated by expression of a 588-amino acid N-terminal fragment of HTT with a 138Q repeats (HTT588 138Q) was done in Drosophila [93]. Although these toxicity and aggregation screens were done in neuronal and non-neuronal tissues, respectively, they both identified the same chromosomal regions as hits. Two major classes of suppressors were identified: one class that rescued fly viability by decreasing HTT expression and aggregation, and a second class that rescued viability without reducing HTT aggregation, suggesting that aggregation and toxicity are separable, and that mutant HTT-associated toxicity depends on both the soluble and aggregation-prone forms of the mutant protein. Two single genes were mapped: one was a piggyBac insertion that reduced the expression of CG5830, which shares homology with small nuclear protein phosphatases known to silence neuronal gene expression. As disruption of transcriptional regulation (e.g., of brain-derived neurotrophic factor) has been implicated in HD [14, 94], it is possible that loss of CG5830 rescues mutant HTT-induced toxicity by restoring transcription. The other mapped gene was Labial, a member of the Antennapedia complex that has been linked to neural stem cell survival in Drosophila; the rescue in Drosophila viability elicited by this mutation was attributed to its ability to reduce mutant HTT levels. Interestingly, other members of the Antennapedia complex were also able to suppress HTTQ138-induced toxicity.
Caenorhabditis elegans models of HD and other polyQ-associated diseases have also been widely used to screen for genetic modifiers of polyQ aggregation and toxicity. The C. elegans model in which mHTT exon 1 Q150 expression caused degeneration of ASH neurons (see above) was also used in a genetic screen for mutations that increase mHTT neurotoxicity, leading to the identification of the polyQ enhancer-1 (pqe-1) gene [95]. A loss of function mutation in pqe-1 was shown to increase mHTT toxicity, whereas its overexpression rescued the toxic phenotype. The authors suggested that pqe-1 provides protection against polyQ toxicity in C. elegans neurons through its glutamine/proline-rich domain by competing with other proteins for binding to mHTT.
This mHTT exon 1 Q150 model has also been used to investigate the roles of specific histone deacetylases in regulating mHTT toxicity [96]. As several studies indicated that mHTT can sequester cyclic adenosine monophosphate response element-binding protein-binding protein (CBP), reducing its histone acetyltransferase activity [97, 98]; inhibition of histone deacetylase activity could counteract this effect and subsequently reduce mHTT toxicity. In fact, using RNAi, Bates et al. [96] reported that lowering hda-3 in C. elegans reduced polyQ toxicity. Analogously, chemical manipulation of HDAC function by histone deacetylase inhibitors such as trichostatin A also led to a decrease in polyQ-mediated neurodegeneration in this study.
C. elegans expressing an N-terminal fragment of the HTT exon 1 protein containing expanded (Q88 and Q128) or unexpanded (Q19) polyQ stretches in touch receptor neurons showed polyQ-dependent neuronal dysfunction [99]. This system was used to investigate the effect of increased sir2.1 deacetylase activity in rescuing nematode neurons from polyQ-mediated toxicity [100]. The Q128 nematode model was also used in a large-scale RNAi screen to identify genes that would either reduce or exacerbate the loss of response to light touch [101]. An intriguing observation was that some of the RNAi hits from this C. elegans screen overlapped with striatal gene expression data derived from the CHL2 (Q150/Q150 knock-in) and the transgenic R6/2 mouse models of HD, underscoring the relevance of using invertebrate models for the identification of potential therapeutic targets for human neurodegenerative diseases. In particular, the druggable pha gene, which encodes the enzyme phenylalanine-4-hydroxylase, was of interest because of its connection to the tyrosine–dopamine biosynthesis pathway, which is known to be associated with early HD phenotypes [101]
In a broader approach, a genome-wide RNAi screen was done in a C. elegans transgenic strain expressing stretches of 35 Q (polyQ35) fused with yellow fluorescent protein specifically in muscle cells [102]. This study yielded 186 genes that, when knocked down, caused premature aggregation of polyQ35 aggregates, including the TCP-1 chaperonin ortholog later confirmed to have a role in aggregation of mHTT in mammalian cells [54, 55, 103]. In addition, a subsequent RNAi screen of human orthologs of these C. elegans modifiers showed a subset to also be suppressors of mHTT aggregation in human cells (HEK293) expressing HTT-Q74–GFP [104].
A complementary approach was undertaken recently [105] in which the polyQ35 C. elegans muscle model was used to screen for genes that, when down-regulated, increased cellular protein folding capacity and thereby suppressed polyQ35-mediated toxicity [105]. Gene hits were subsequently tested for suppression of aggregation in a mutant superoxide dismutase 1 (SOD1G93A) model of amyotrophic lateral sclerosis. This strategy led to the identification of 9 genes that work as core proteostasis network modulators and not only reduce protein aggregation but also enhance the protein-folding environment [106, 107].
An interesting observation from these C. elegans genetic screens was the minimal overlap between the hits derived from aggregation screens [102, 105] and those from the toxicity screen described previously [101]: only 15 modifiers were identified in common. This emphasizes the critical roles of assay readouts (e.g., aggregation vs toxicity), and of molecular and cellular context in such genetic screens. For example, whereas the two aggregation screens used a polyQ-only expansion expressed in muscle cells, the toxicity screen used HTT exon 1 Q128 expressed in neurons.
Proteomic Screening
A parallel and complementary systems biology approach to the identification of genetic modifiers is the direct identification of mHTT interacting proteins that may play mechanistic roles in polyQ-related disease pathogenesis. Initial yeast two-hybrid screens identified interactors of mHTT with widespread roles in protein trafficking, turnover, mRNA biogenesis and metabolism, and transcription and signaling pathways [108–111], underscoring the broad range of proteins and core cellular pathways that interact with mHTT.
More recently, an extensive proteomic study was done using a combination of yeast two-hybrid and pull-down assay/mass spectrometry [53]. A total of 234 novel HTT-fragment-interacting proteins were identified that belonged to different functional classes, such as protein folding and degradation, signal transduction, synaptic transmission, and metabolism. Sixty of these protein hits were subsequently tested in a Drosophila model of HD, with 48 of the 60 shown to be either enhancers or suppressors of mHTT-induced neurodegeneration. An interesting observation was that some of the modifiers were involved in synaptic vesicle fusion, suggesting that WT HTT could have a role in modulating neurotransmitter secretion, which could be impaired by mHTT. In addition, two components of the cytosolic CCT (TriC) were identified, and these proteins were also confirmed to alter the course of polyQ-induced toxicity in mammalian cells [54, 55].
Recently, proteomic studies to identify HTT-associated proteins have also been done in the context of full-length HTT expressed in mouse brain, using affinity purification-mass spectrometry [112, 113]. Using subcellular fractionation of brain homogenates, one study found molecular chaperones as mutant HTT interactors, in support of previous findings and highlighting the central role of these folding machineries as HTT modifiers [112]. Interestingly, components of the translational machinery were also identified as mutant HTT interacting proteins, providing a novel molecular pathway for HTT-mediated toxicity. In another study distinct regions of WT and BACHD mouse brains of different ages were used to identify 747 HTT interacting proteins [113], of which several had previously been found as HTT interactors or as modifiers of mHTT toxicity [53, 114]. A subset of these protein hits was also shown to act as genetic modifiers of mHTT-induced neurodegeneration in a Drosophila model. Of importance, several of these proteins are targets of compounds that have been effective in mouse HD models, such as geldanamycin targeting HSP90. Interestingly, although the majority of HTT interactors were relatively conserved across different brain tissue and age, brain region- and developmental stage-specific interactors were also found, suggesting that some of these proteins might contribute to the selective neuronal susceptibility and age-dependent degeneration observed in HD patients. A comparison to other proteomic datasets reported previously [53, 111, 115] showed greater overlap among HTT datasets than between HTT and ataxia datasets, suggesting that unique protein–protein interactions may contribute to specific pathogenic mechanisms in different, but related, polyQ-mediated diseases [113].
Chemical, Genetic and Proteomic Screens: What Have We Learned?
With an increasing number of studies completed we can begin to compare the hits emerging from a range of diverse screening strategies (Tables 1 and 2). Perhaps not surprisingly, there have been relatively few common genes [89] and compounds found across screens. Numerous factors are likely to contribute to such low hit overlap. For example, different technical methodologies may intrinsically bias towards or limit against certain subsets of targets and mechanisms: proteomic interaction methods tend to identify abundant proteins and may miss rare interacting partners, while genetic screens may fail to detect essential genes owing to lethality or gene families with redundant functions. In addition, not all models share all genetic pathways. Differences in constructs used to drive disease models (e.g., fragment vs full-length HD models) may also emphasize different hit pathways and categories.
However, such high-throughput genetic and proteomic screens in the context of different polyQ-driven neurodegenerative diseases are also identifying functional gene classes represented across disease states, suggesting conserved roles for protein classes in common and, perhaps, core disease mechanisms. For example, molecular chaperones and components of the proteolytic machineries involved in refolding and degradation of misfolded proteins, respectively, have commonly been found as hits in screens for suppression of polyQ aggregation and toxicity [106, 107]. Overexpression of selected or multiple chaperones by genetic and pharmacologic strategies has, indeed, proven effective in rescuing aggregation-mediated toxicity of mHTT and other polyQ-related proteins in multiple models. Analogously, induction of autophagy by rapamycin, an inhibitor of mammalian target of rapamycin, was shown to ameliorate neurodegenerative phenotypes both in fly and mouse models of HD [116] in support of these screening results, as described in previous sections.
In addition many genes involved in transcriptional regulation and RNA processing are consistently found as modifiers of polyQ-mediated aggregation and toxicity in yeast, Drosophila, and C. elegans [2]. Indeed, accumulating evidence suggests that chromatin structure and epigenetic regulation are involved in polyQ-mediated pathology. It has been shown in several models that mutant polyQ proteins localize in the nucleus, often as inclusions, and cause the redistribution of key transcription factors, such as the nuclear receptor co-repressor, CBP, and p300/CBP-associated factor [117]. Sequestration of such factors could compromise the acetylation level of the chromatin and lead to altered transcription [110].
Remarkably, in both flies and mice loss in histone deacetylation and subsequent HD-induced neurodegeneration could be rescued not only by CBP overexpression, but also by pharmacological means, using HDAC inhibitors [117]. HDAC inhibitors show a therapeutic potential for the treatment of several disorders, including HD [118]. For example, the HDAC inhibitors suberoylanilide hydroxamic acid [119], sodium butyrate [120], phenylbutyrate [121], and 4b [122] have all been shown to ameliorate mutant HTT-mediated degeneration in different HD mouse models, and some (e.g., sodium butyrate) have reached clinical trials for the treatment of HD. These encouraging studies suggest that clinical development of HDAC inhibitors for HD therapy may be possible.
That certain targets and mechanisms could be common to all polyQ diseases, such as the involvement of molecular chaperones and the ubiquitin-proteasome system [123], may not be surprising. It might be expected, however, that some pathways involved in polyQ-mediated disease progression will be disease-specific. In fact, a comparative study of known ataxin-1 Q82 modifiers in Drosophila found that only certain genes were able to modify mutant ataxin-1- and HTT-induced phenotypes in a similar manner, whereas other genes were disease-specific or even modified the two disease models in opposite ways [124]. In addition, post-transcriptional/translational modifications outside of the polyQ tract have been found to have profound effects on the pathogenicity and mechanisms of action of polyQ proteins, including HTT [125, 126], and potentially driving disease-specific or "disease-emphasized" pathways.
The Promise of iPSC
Recent advances in iPSC technology represent a tremendous opportunity to apply drug discovery assays and approaches such as those described in the previous sections to patient-derived cells that can be differentiated into neurons. Monogenic diseases such as HD were among the first to take advantage of iPSC technology to generate neurons and other cells types from patient fibroblasts (reviewed in [127–131]). The benefits of using bona fide human and patient-derived cells that can be differentiated into relevant neuronal and/or glial species are inestimable for new drug discovery and development. Moreover, the pluripotency of iPSCs can theoretically provide a nearly unlimited and consistent supply of such cellular reagents. In addition, embryonic and neural stem cells from HD mouse models have also been characterized and could serve as critical bridging tools for supporting preclinical translational studies [132, 133]. Initial studies showed no differences in the ability of HD iPSCs to differentiate into neurons [123, 134, 135].
The first iPSC-based proteomic analysis for HD has recently been reported. This study compared normal with HD patient-derived iPSC lines expressing HTT Q25 and identified 26 differentially regulated genes involved in a range of processes [136]. In particular, genes involved in programmed cell death and oxidative stress were enriched in HD iPSC lines compared with normal iPSC lines, indicating that these cell lines could be highly vulnerable to oxidative stress and apoptotic stimuli. In fact, levels of antioxidant enzymes, such as SOD1 and glutathione-S-transferase, were found to be lower in HD iPSC lines, which were also more susceptible to apoptotic cell death than control lines. Critically, it has been recently shown that correction of the polyQ expansion in HD patient iPSCs (i.e., in the identical genetic backgrounds) normalizes pathogenic HD signaling pathways and phenotypes such as susceptibility to cell death and altered mitochondrial bioenergetics [137].
For use in phenotypic studies and screening, a set of HD iPSCs consisting of normal (28- and 33-CAG) and long CAG repeats characteristic of juvenile onset HD (60-, 109-, and 180-CAG) was recently generated, with HD neurons found to have increased caspase activity and susceptibility to stressors such as glutamate and brain-derived neurotrophic factor withdrawal [138]. Interestingly, iPSC-derived neurons with shorter polyQ repeat lengths may require more extended times in culture to mature and develop HD-associated cellular phenotypes. For example, iPSC derived neurons with adult onset CAG length (42, 44) showed little phenotype in culture [139], whereas the longest (109, 180) CAG repeat length had more pronounced deficits [138]. However, significant challenges remain in implementing iPSC in HTS protocols [129], including definition, consistency, and yields of cellular differentiation and phenotypes.
Conclusions and Perspectives
While current clinical options remain limited for HD and other polyQ diseases, rapid progress in developing focused preclinical models and drug screening technologies for these disorders promises that new generations of drug candidates will be entering clinical evaluation in coming years, including biologic molecules targeting the mutant genes and proteins themselves [10, 140–142]. Moreover, emerging robotic and automated imaging technologies, together with powerful new analytic tools, are offering new possibilities for the higher throughput use of complex models in drug discovery, ranging from iPSCs to brain slices to C. elegans and D. melanogaster. For example, the Complex Object Parametric Analyzer and Sorter Biosort [143], which is able to automatically sort and dispense an accurate number of worms and Drosophila eggs and embryos into multiwell plates, and the WormToolbox [144], an automated image analysis tool for HTS of C. elegans phenotypes, are contributing considerably to increase the throughput of screens using model organisms. Together with recent and rapid progress in novel therapeutic modalities; disease and response biomarker development, including image-based biomarkers and target engagement assays; blood barrier penetrant antibodies; and inhibitory RNAs, peptides, and antisense oligonucleotides for protein misfolding and aggregation, there is much optimism that the polyQ diseases may soon make the transition from "fatal" to manageable.
Electronic supplementary material
(PDF 510 kb)
Acknowledgments
Supported in part by the CHDI Foundation, Inc., and NIH grants NS074379 and NS080514.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Nance MA. Therapy in Huntington's disease: where are we? Curr Neurol Neurosci Rep. 2012;12:359–366. doi: 10.1007/s11910-012-0277-4. [DOI] [PubMed] [Google Scholar]
- 4.Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 5.Ingram MA, Orr HT, Clark HB. Genetically engineered mouse models of the trinucleotide-repeat spinocerebellar ataxias. Brain Res Bull. 2012;88:33–42. doi: 10.1016/j.brainresbull.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heng MY, Detloff PJ, Albin RL. Rodent genetic models of Huntington disease. Neurobiol Dis. 2008;32:1–9. doi: 10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Weydt P, La Spada AR. Targeting protein aggregation in neurodegeneration—lessons from polyglutamine disorders. Expert Opin Ther Targets. 2006;10:505–513. doi: 10.1517/14728222.10.4.505. [DOI] [PubMed] [Google Scholar]
- 8.Fecke W, Gianfriddo M, Gaviraghi G, Terstappen GC, Heitz F. Small molecule drug discovery for Huntington's Disease. Drug Discov Today. 2009;14:453–464. doi: 10.1016/j.drudis.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Watson LM, Wood MJ. RNA therapy for polyglutamine neurodegenerative diseases. Expert Rev Mol Med. 2012;14:e3. doi: 10.1017/erm.2011.1. [DOI] [PubMed] [Google Scholar]
- 10.Butler DC, McLear JA, Messer A. Engineered antibody therapies to counteract mutant huntingtin and related toxic intracellular proteins. Prog Neurobiol. 2012;97:190–204. doi: 10.1016/j.pneurobio.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steffan JS, Thompson LM. Targeting aggregation in the development of therapeutics for the treatment of Huntington's disease and other polyglutamine repeat diseases. Expert Opin Ther Targets. 2003;7:201–213. doi: 10.1517/14728222.7.2.201. [DOI] [PubMed] [Google Scholar]
- 12.Katsuno M, Banno H, Suzuki K, et al. Molecular genetics and biomarkers of polyglutamine diseases. Curr Mol Med. 2008;8:221–234. doi: 10.2174/156652408784221298. [DOI] [PubMed] [Google Scholar]
- 13.Weir DW, Sturrock A, Leavitt BR. Development of biomarkers for Huntington's disease. Lancet Neurol. 2011;10:573–590. doi: 10.1016/S1474-4422(11)70070-9. [DOI] [PubMed] [Google Scholar]
- 14.Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 15.Jacobsen JC, Gregory GC, Woda JM, et al. HD CAG-correlated gene expression changes support a simple dominant gain of function. Hum Mol Genet. 2011;20:2846–2860. doi: 10.1093/hmg/ddr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 17.Davies SW, Turmaine M, Cozens BA, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 18.Scherzinger E, Lurz R, Turmaine M, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 19.DiFiglia M, Sapp E, Chase KO, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 20.Varma H, Lo DC, Stockwell BR. High-throughput and high-content screening for Huntington's disease therapeutics. In: Lo DC, Hughes RE (eds) Neurobiology of Huntington's Disease: applications to drug discovery. Frontiers in Neuroscience, Boca Raton, FL; 2011.
- 21.Heiser V, Engemann S, Brocker W, et al. Identification of benzothiazoles as potential polyglutamine aggregation inhibitors of Huntington's disease by using an automated filter retardation assay. Proc Natl Acad Sci U S A. 2002;99(Suppl. 4):16400–16406. doi: 10.1073/pnas.182426599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Gines S, MacDonald ME, Gusella JF. Reversal of a full-length mutant huntingtin neuronal cell phenotype by chemical inhibitors of polyglutamine-mediated aggregation. BMC Neurosci. 2005;6:1. doi: 10.1186/1471-2202-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai Y, Tucker T, Ren H, et al. Inhibition of polyglutamine protein aggregation and cell death by novel peptides identified by phage display screening. J Biol Chem. 2000;275:10437–10442. doi: 10.1074/jbc.275.14.10437. [DOI] [PubMed] [Google Scholar]
- 24.Ehrnhoefer DE, Duennwald M, Markovic P, et al. Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models. Hum Mol Genet. 2006;15:2743–2751. doi: 10.1093/hmg/ddl210. [DOI] [PubMed] [Google Scholar]
- 25.Pollitt SK, Pallos J, Shao J, et al. A rapid cellular FRET assay of polyglutamine aggregation identifies a novel inhibitor. Neuron. 2003;40:685–694. doi: 10.1016/s0896-6273(03)00697-4. [DOI] [PubMed] [Google Scholar]
- 26.Heiser V, Scherzinger E, Boeddrich A, et al. Inhibition of huntingtin fibrillogenesis by specific antibodies and small molecules: implications for Huntington's disease therapy. Proc Natl Acad Sci U S A. 2000;97:6739–6744. doi: 10.1073/pnas.110138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra R, Jayaraman M, Roland BP, et al. Inhibiting the nucleation of amyloid structure in a huntingtin fragment by targeting alpha-helix-rich oligomeric intermediates. J Mol Biol. 2012;415:900–917. doi: 10.1016/j.jmb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nucifora LG, Burke KA, Feng X, et al. Identification of novel potentially toxic oligomers formed in vitro from mammalian-derived expanded huntingtin exon-1 protein. J Biol Chem. 2012;287:16017–16028. doi: 10.1074/jbc.M111.252577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beam M, Silva MC, Morimoto RI. Dynamic imaging by fluorescence correlation spectroscopy identifies diverse populations of polyglutamine oligomers formed in vivo. J Biol Chem. 2012;287:26136–26145. doi: 10.1074/jbc.M112.362764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sontag EM, Lotz GP, Agrawal N, et al. Methylene blue modulates huntingtin aggregation intermediates and is protective in Huntington's disease models. J Neurosci. 2012;32:11109–11119. doi: 10.1523/JNEUROSCI.0895-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss A, Klein C, Woodman B, et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington's disease. J Neurochem. 2008;104:846–858. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 32.Schirmer RH, Adler H, Pickhardt M, Mandelkow E. Lest we forget you--methylene blue…. Neurobiol Aging. 2011;32:2325. doi: 10.1016/j.neurobiolaging.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Li M, Huang Y, Ma AA, Lin E, Diamond MI. Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol Dis. 2009;36:413–420. doi: 10.1016/j.nbd.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Shao J, Welch WJ, Diamond MI. ROCK and PRK-2 mediate the inhibitory effect of Y-27632 on polyglutamine aggregation. FEBS Lett. 2008;582:1637–1642. doi: 10.1016/j.febslet.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao J, Welch WJ, Diprospero NA, Diamond MI. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol. 2008;28:5196–5208. doi: 10.1128/MCB.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Yasumura D, Ma AA, et al. Intravitreal administration of HA-1077, a ROCK inhibitor, improves retinal function in a mouse model of huntington disease. PLoS One. 2013;8:e56026. doi: 10.1371/journal.pone.0056026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangiarini L, Sathasivam K, Seller M, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 38.Sharma P, Ando DM, Daub A, Kaye JA, Finkbeiner S. High-throughput screening in primary neurons. Methods Enzymol. 2012;506:331–360. doi: 10.1016/B978-0-12-391856-7.00041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 40.Fuentealba RA, Marasa J, Diamond MI, Piwnica-Worms D, Weihl CC. An aggregation sensing reporter identifies leflunomide and teriflunomide as polyglutamine aggregate inhibitors. Hum Mol Genet. 2012;21:664–680. doi: 10.1093/hmg/ddr500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Smith DL, Meriin AB, et al. A potent small molecule inhibits polyglutamine aggregation in Huntington's disease neurons and suppresses neurodegeneration in vivo. Proc Natl Acad Sci U S A. 2005;102:892–897. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chopra V, Fox JH, Lieberman G, et al. A small-molecule therapeutic lead for Huntington's disease: preclinical pharmacology and efficacy of C2-8 in the R6/2 transgenic mouse. Proc Natl Acad Sci U S A. 2007;104:16685–16689. doi: 10.1073/pnas.0707842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor DM, Balabadra U, Xiang Z, et al. A brain-permeable small molecule reduces neuronal cholesterol by inhibiting activity of sirtuin 2 deacetylase. ACS Chem Biol. 2011;6:540–546. doi: 10.1021/cb100376q. [DOI] [PubMed] [Google Scholar]
- 44.Luthi-Carter R, Taylor DM, Pallos J, et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci U S A. 2010;107:7927–7932. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 46.Paganetti P, Weiss A, Trapp M, et al. Development of a method for the high-throughput quantification of cellular proteins. Chembiochem. 2009;10:1678–1688. doi: 10.1002/cbic.200900131. [DOI] [PubMed] [Google Scholar]
- 47.Weiss A, Grueninger S, Abramowski D, et al. Microtiter plate quantification of mutant and wild-type huntingtin normalized to cell count. Anal Biochem. 2011;410:304–306. doi: 10.1016/j.ab.2010.11.044. [DOI] [PubMed] [Google Scholar]
- 48.Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J Biol Chem. 2012;287:1406–1414. doi: 10.1074/jbc.M111.294801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldo B, Paganetti P, Grueninger S, et al. TR-FRET-based duplex immunoassay reveals an inverse correlation of soluble and aggregated mutant huntingtin in huntington's disease. Chem Biol. 2012;19:264–275. doi: 10.1016/j.chembiol.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 50.Miller JP, Holcomb J, Al-Ramahi I, et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron. 2010;67:199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calamini B, Silva MC, Madoux F, et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2012;8:185–196. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaltenbach LS, Romero E, Becklin RR, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitamura A, Kubota H, Pack CG, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 56.Pruss RM. Phenotypic screening strategies for neurodegenerative diseases: a pathway to discover novel drug candidates and potential disease targets or mechanisms. CNS Neurol Disord Drug Targets. 2010;9:693–700. doi: 10.2174/187152710793237377. [DOI] [PubMed] [Google Scholar]
- 57.Kaltenbach LS, Bolton MM, Shah B, et al. Composite primary neuronal high-content screening assay for Huntington's disease incorporating non-cell-autonomous interactions. J Biomol Screen. 2010;15:806–819. doi: 10.1177/1087057110373392. [DOI] [PubMed] [Google Scholar]
- 58.Khoshnan A, Patterson PH. The role of IkappaB kinase complex in the neurobiology of Huntington's disease. Neurobiol Dis. 2011;43:305–311. doi: 10.1016/j.nbd.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Popoli P, Blum D, Domenici MR, Burnouf S, Chern Y. A critical evaluation of adenosine A2A receptors as potentially "druggable" targets in Huntington's disease. Curr Pharm Des. 2008;14:1500–1511. doi: 10.2174/138161208784480117. [DOI] [PubMed] [Google Scholar]
- 60.Reinhart PH, Kaltenbach LS, Essrich C, et al. Identification of anti-inflammatory targets for Huntington's disease using a brain slice-based screening assay. Neurobiol Dis. 2011;43:248–256. doi: 10.1016/j.nbd.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffstrom BG, Kaplan A, Letso R, et al. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2010;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 63.Karpuj MV, Becher MW, Springer JE, et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med. 2002;8:143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 64.Dedeoglu A, Kubilus JK, Jeitner TM, et al. Therapeutic effects of cystamine in a murine model of Huntington's disease. J Neurosci. 2002;22:8942–8950. doi: 10.1523/JNEUROSCI.22-20-08942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JP, Yates BE, Al-Ramahi I, et al. A genome-scale RNA-interference screen identifies RRAS signaling as a pathologic feature of Huntington's disease. PLoS Genet. 2012;8:e1003042. doi: 10.1371/journal.pgen.1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schulte J, Sepp KJ, Wu C, Hong P, Littleton JT. High-content chemical and RNAi screens for suppressors of neurotoxicity in a Huntington's disease model. PLoS One. 2011;6:e23841. doi: 10.1371/journal.pone.0023841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hempel CM, Sivula M, Levenson JM, et al. A system for performing high throughput assays of synaptic function. PLoS One. 2011;6:e25999. doi: 10.1371/journal.pone.0025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi P, Scott MA, Ghosh B, et al. Synapse microarray identification of small molecules that enhance synaptogenesis. Nat Commun. 2011;2:510. doi: 10.1038/ncomms1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harrill JA, Robinette BL, Mundy WR. Use of high content image analysis to detect chemical-induced changes in synaptogenesis in vitro. Toxicol In Vitro. 2011;25:368–387. doi: 10.1016/j.tiv.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 70.Lam V, Wakatsuki T. Hydrogel tissue construct-based high-content compound screening. J Biomol Screen. 2011;16:120–128. doi: 10.1177/1087057110388269. [DOI] [PubMed] [Google Scholar]
- 71.Seidel D, Krinke D, Jahnke HG, et al. Induced tauopathy in a novel 3D-Culture model mediates neurodegenerative processes: a real-time study on biochips. PLoS One. 2012;7:e49150. doi: 10.1371/journal.pone.0049150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho S, Wood A, Bowlby MR. Brain slices as models for neurodegenerative disease and screening platforms to identify novel therapeutics. Curr Neuropharmacol. 2007;5:19–33. doi: 10.2174/157015907780077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murphy RC, Messer A. Gene transfer methods for CNS organotypic cultures: a comparison of three nonviral methods. Mol Ther. 2001;3:113–121. doi: 10.1006/mthe.2000.0235. [DOI] [PubMed] [Google Scholar]
- 74.Lo DC. Neuronal transfection using particle-mediated gene transfer. Curr Protoc Neurosci 2001;Chapter 3:Unit 3 15. [DOI] [PubMed]
- 75.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker's yeast? Nat Rev Neurosci. 2010;11:436–449. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 76.Sherman MY, Muchowski PJ. Making yeast tremble: yeast models as tools to study neurodegenerative disorders. Neuromolecular Med. 2003;4:133–146. doi: 10.1385/NMM:4:1-2:133. [DOI] [PubMed] [Google Scholar]
- 77.Brignull HR, Morley JF, Morimoto RI. The stress of misfolded proteins: C. elegans models for neurodegenerative disease and aging. Adv Exp Med Biol. 2007;594:167–189. doi: 10.1007/978-0-387-39975-1_15. [DOI] [PubMed] [Google Scholar]
- 78.Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 79.Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci U S A. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Voisine C, Varma H, Walker N, Bates EA, Stockwell BR, Hart AC. Identification of potential therapeutic drugs for huntington's disease using Caenorhabditis elegans. PLoS One. 2007;2:e504. doi: 10.1371/journal.pone.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu J, Shih HP, Vigont V, et al. Neuronal store-operated calcium entry pathway as a novel therapeutic target for Huntington's disease treatment. Chem Biol. 2011;18:777–793. doi: 10.1016/j.chembiol.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patapoutian A, Tate S, Woolf CJ. Transient receptor potential channels: targeting pain at the source. Nat Rev Drug Discov. 2009;8:55–68. doi: 10.1038/nrd2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol Rev. 2007;87:165–217. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- 84.Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 85.Giorgini F, Guidetti P, Nguyen Q, Bennett SC, Muchowski PJ. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet. 2005;37:526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu CR, Chang CR, Chern Y, et al. Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell. 2012;148:690–701. doi: 10.1016/j.cell.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 87.Marsh JL, Pallos J, Thompson LM. Fly models of Huntington's disease. Hum Mol Genet 2003;12 Spec No 2:R187-193. [DOI] [PubMed]
- 88.Rincon-Limas DE, Jensen K, Fernandez-Funez P. Drosophila models of proteinopathies: the little fly that could. Curr Pharm Des. 2012;18:1108–1122. doi: 10.2174/138161212799315894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Ham TJ, Breitling R, Swertz MA, Nollen EA. Neurodegenerative diseases: Lessons from genome-wide screens in small model organisms. EMBO Mol Med. 2009;1:360–370. doi: 10.1002/emmm.200900051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 91.Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408:101–6. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 92.Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weiss KR, Kimura Y, Lee WC, Littleton JT. Huntingtin aggregation kinetics and their pathological role in a Drosophila Huntington's disease model. Genetics. 2012;190:581–600. doi: 10.1534/genetics.111.133710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- 95.Faber PW, Voisine C, King DC, Bates EA, Hart AC. Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity. Proc Natl Acad Sci U S A. 2002;99:17131–17136. doi: 10.1073/pnas.262544899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCampbell A, Taylor JP, Taye AA, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 98.Jiang H, Nucifora FC, Jr, Ross CA, DeFranco DB. Cell death triggered by polyglutamine-expanded huntingtin in a neuronal cell line is associated with degradation of CREB-binding protein. Hum Mol Genet. 2003;12:1–12. doi: 10.1093/hmg/ddg002. [DOI] [PubMed] [Google Scholar]
- 99.Parker JA, Connolly JB, Wellington C, Hayden M, Dausset J, Neri C. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc Natl Acad Sci U S A. 2001;98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parker JA, Arango M, Abderrahmane S, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 101.Lejeune FX, Mesrob L, Parmentier F, et al. Large-scale functional RNAi screen in C. elegans identifies genes that regulate the dysfunction of mutant polyglutamine neurons. BMC Genomics. 2012;13:91. doi: 10.1186/1471-2164-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nollen EA, Garcia SM, van Haaften G, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Behrends C, Langer CA, Boteva R, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 104.Teuling E, Bourgonje A, Veenje S, et al. Modifiers of mutant huntingtin aggregation: functional conservation of C. elegans-modifiers of polyglutamine aggregation. PLoS Curr. 2011;3 doi: 10.1371/currents.RRN1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Silva MC, Fox S, Beam M, Thakkar H, Amaral MD, Morimoto RI. A genetic screening strategy identifies novel regulators of the proteostasis network. PLoS Genet. 2011;7:e1002438. doi: 10.1371/journal.pgen.1002438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 107.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 108.Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 109.Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 110.Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 111.Goehler H, Lalowski M, Stelzl U, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 112.Culver BP, Savas JN, Park SK, et al. Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J Biol Chem. 2012;287:21599–21614. doi: 10.1074/jbc.M112.359307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shirasaki DI, Greiner ER, Al-Ramahi I, et al. Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron. 2012;75:41–57. doi: 10.1016/j.neuron.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Omi K, Hachiya NS, Tanaka M, Tokunaga K, Kaneko K. 14-3-3zeta is indispensable for aggregate formation of polyglutamine-expanded huntingtin protein. Neurosci Lett. 2008;431:45–50. doi: 10.1016/j.neulet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 115.Lim J, Hao T, Shaw C, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 116.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 117.Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 118.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 119.Hockly E, Richon VM, Woodman B, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Orlando ER, Caroli E, Ferrante L. Management of the cervical esophagus and hypofarinx perforations complicating anterior cervical spine surgery. Spine (Phila Pa 1976) 2003;28:E290–295. doi: 10.1097/01.BRS.0000087093.89889.0A. [DOI] [PubMed] [Google Scholar]
- 121.Gardian G, Browne SE, Choi DK, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 122.Thomas EA, Coppola G, Desplats PA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Voisine C, Pedersen JS, Morimoto RI. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis. 2010;40:12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Branco J, Al-Ramahi I, Ukani L, et al. Comparative analysis of genetic modifiers in Drosophila points to common and distinct mechanisms of pathogenesis among polyglutamine diseases. Hum Mol Genet. 2008;17:376–390. doi: 10.1093/hmg/ddm315. [DOI] [PubMed] [Google Scholar]
- 125.Appl T, Kaltenbach L, Lo DC, Terstappen GC. Targeting mutant huntingtin for the development of disease-modifying therapy. Drug Discov Today. 2012;17:1217–1223. doi: 10.1016/j.drudis.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 126.Ehrnhoefer DE, Sutton L, Hayden MR. Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist. 2011;17:475–492. doi: 10.1177/1073858410390378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Trounson A, Shepard KA, DeWitt ND. Human disease modeling with induced pluripotent stem cells. Curr Opin Genet Dev. 2012;22:509–516. doi: 10.1016/j.gde.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 128.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells–opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 130.Gunaseeli I, Doss MX, Antzelevitch C, Hescheler J, Sachinidis A. Induced pluripotent stem cells as a model for accelerated patient- and disease-specific drug discovery. Curr Med Chem. 2010;17:759–766. doi: 10.2174/092986710790514480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Benraiss A, Goldman SA. Cellular therapy and induced neuronal replacement for Huntington's disease. Neurotherapeutics. 2011;8:577–590. doi: 10.1007/s13311-011-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ritch JJ, Valencia A, Alexander J, et al. Multiple phenotypes in Huntington disease mouse neural stem cells. Mol Cell Neurosci. 2012;50:70–81. doi: 10.1016/j.mcn.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Conforti P, Camnasio S, Mutti C, et al. Lack of huntingtin promotes neural stem cells differentiation into glial cells while neurons expressing huntingtin with expanded polyglutamine tracts undergo cell death. Neurobiol Dis. 2012;50C:160–170. doi: 10.1016/j.nbd.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 134.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of human Huntington's disease cell model from induced pluripotent stem cells. PLoS Curr. 2010;2 doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Juopperi TA, Kim WR, Chiang CH, et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington's disease patient cells. Mol Brain. 2012;5:17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chae JI, Kim DW, Lee N, et al. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington's disease patient. Biochem J. 2012;446:359–371. doi: 10.1042/BJ20111495. [DOI] [PubMed] [Google Scholar]
- 137.An Mahru C, Zhang N, Scott G, et al. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Camnasio S, Carri AD, Lombardo A, et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 140.Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121:500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Denovan-Wright EM, Davidson BL. RNAi: a potential therapy for the dominantly inherited nucleotide repeat diseases. Gene Ther. 2006;13:525–531. doi: 10.1038/sj.gt.3302664. [DOI] [PubMed] [Google Scholar]
- 142.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 143.Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol. 2006;351:275–286. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- 144.Wahlby C, Kamentsky L, Liu ZH, et al. An image analysis toolbox for high-throughput C. elegans assays. Nat Methods. 2012;9:714–716. doi: 10.1038/nmeth.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ghosh S, Feany MB. Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum Mol Genet. 2004;13:2011–2018. doi: 10.1093/hmg/ddh214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 510 kb)