Abstract
Viral fusion proteins undergo dramatic conformational transitions during membrane fusion. For viruses that enter through the endosome, these conformational rearrangements are typically pH sensitive. Here we provide a comprehensive review of the molecular interactions that govern pH-dependent rearrangements and introduce a novel paradigm for electrostatic residue pairings that regulate progress through the viral fusion coordinate. Analysis of structural data demonstrates a significant role for side chain protonation in triggering conformational change. To characterize this behavior we identify two distinct residue pairings, which we define as Histidine-Cation (HisCat) and Anion-Anion (AniAni) interactions. These side chain pairings destabilize a particular conformation via electrostatic repulsion through side chain protonation. Furthermore, two energetic control mechanisms, thermodynamic and kinetic, regulate these structural transitions. This review expands on the current literature by identification of these residue clusters, discussion of data demonstrating their function, and speculation of how these residue pairings contribute to the energetic controls.
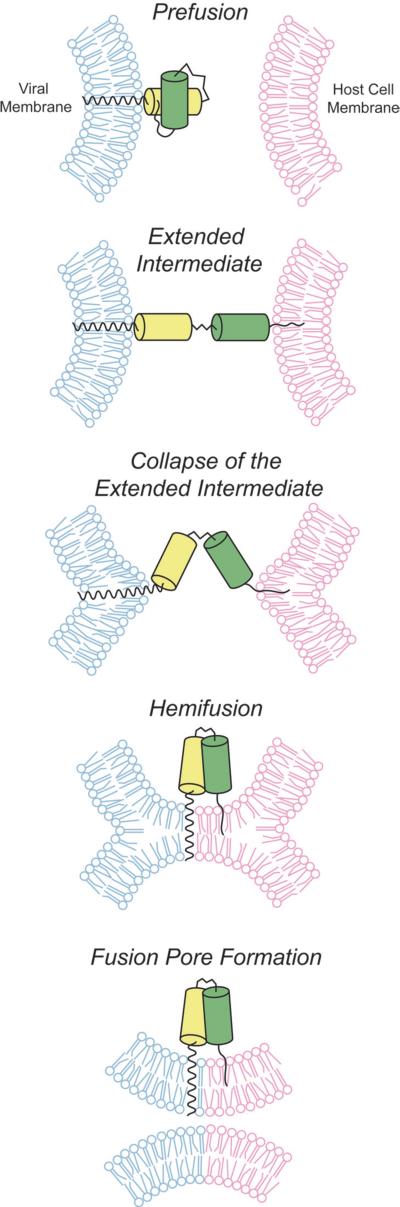
Both cells and enveloped viruses are surrounded by phospholipid bilayers that act as physical barriers between the cellular and viral genomes. Viruses have evolved efficient mechanisms to circumvent this barrier by fusing their membrane with that of the host (Kielian and Rey, 2006; Weissenhorn et al., 2007; Harrison, 2008; White and Delos, 2008). This process results in the formation of a pore in the membrane, allowing the virus to deliver its genetic material to the cell (Figure 1) (Chernomordik and Kozlov, 2008). Although, fusion of two lipid membranes is an energetically favorable process, there is a large activation barrier due to electrostatic repulsion between the polar head groups of the phospholipids (Chernomordik et al., 2006; Kozlov et al., 2010). This energetic barrier is overcome by glycoproteins embedded in the viral envelope. These proteins generally adopt at least three distinct conformational states during the membrane fusion process; (i) the prefusion state; (ii) the extended intermediate state; and (iii) the post-fusion state (Figure 1) (Chernomordik and Kozlov, 2008; Harrison, 2008; Kielian and Rey, 2006; White and Delos, 2008). The pre- and post-fusion states of many envelope glycoproteins have been characterized with high resolution by X-ray crystallography and with low resolution by various methods. However, evidence of the extended intermediate has been largely indirect (Jiang et al., 1993; Miller et al., 2011; Pessi et al., 2012). Two recent reports have provided direct information about the extended intermediates in paramyxoviruses (Kim et al., 2011) and Avian Sarcoma Leukosis Virus (ASLV) (Cardone et al., 2012; Matsuyama et al., 2004). At present, no extended intermediate conformation from any virus has been characterized in high resolution.
Figure 1. Schematic of viral fusion reaction coordinate.
In the prefusion state the fusion proteins exist in a stable conformation on the surface of the virus. After a triggering step, which varies among viruses, the extended intermediate is formed. This transient conformation collapses to form the final post-fusion structure where lipid mixing can occur. First, in hemifusion, only the outer membranes have mixed. Finally, when the fusion pore is formed, the inner membranes can mix, and the virus can enter the cell.
Many viruses enter the cell through the endocytic pathway where vesicle acidification triggers progression through the viral fusion cascade (Lozach et al., 2011; Mercer et al., 2010). Thus, the viral envelope proteins function as pH sensors, sensing the pH decrease, to ~pH 5, which is encountered as the endocytic vesicles mature (Huotari and Helenius, 2011). From a chemical perspective, differential side chain protonation likely triggers the conformational rearrangements. Of the functional groups in canonical proteins, only three amino acid side chains (Asp, Glu, and His) titrate in the necessary pH range to function as candidate sensors. Indeed, numerous structural studies have demonstrated the critical role of histidines in these conformational changes, (Boo et al., 2012; Carneiro et al., 2003, 2006; Chanel-Vos and Kielian, 2004; Huang et al., 2002; Kampmann et al., 2006; Liu and Kielian, 2009; Mueller et al., 2008; Qin et al., 2009; Schowalter et al., 2009; Stauffer et al., 2007) while the role of anionic side chains has only recently been elucidated (Chang et al., 2012; Harrison et al., 2011, 2012; Liu and Kielian, 2009). Therefore, electrostatic changes provide some of the forces behind these conformational changes. Note that in contrast, the hydrophobic effect — not electrostatics — is the dominant stabilizing force for protein folding (Baldwin, 2007; Dill, 1990).
pH-dependent residue pairs
Mutational data from many pH-dependent viral fusion systems have implicated two discrete residue pairings as potential pH sensitive elements. These residue pairs function by destabilizing a conformation at a particular pH through electrostatic repulsion. Undoubtedly other intermolecular interactions can contribute to pH sensitive conformational rearrangements, such as hydrogen bonds and salt bridges, but neither of these forces can provide a destabilizing force like repulsion. Moreover, the concept of repulsion influencing pH-dependent rearrangements provides a plausible explanation for free energy changes between the prefusion and post- fusion conformation, discussed later.
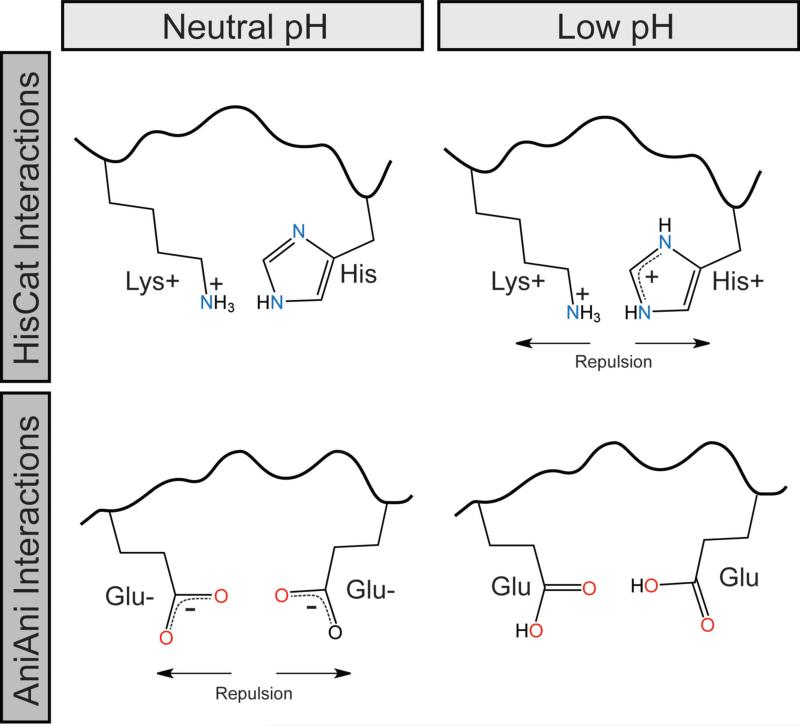
The first pair, HisCat, consists of an interaction between a histidine residue and another cationic residue, Lys, Arg, or His (Figure 2). The pKa of free histidine is 6.5, however in the context of a folded protein's microenvironment this value is commonly altered. As the pH decreases the imidazole ring accepts a proton rendering this residue cationic (Figure 2). In HisCat pairs, histidine residues are found in close proximity, usually less then 7Å, to another His or a basic residue (Arg or Lys). When the His is protonated, these clusters acquire cationic charge and repel, destabilizing the prefusion state, contributing to the formation of the extended intermediate.
Figure 2.
Schematic of HisCat and AniAni interactions at neutral and endosomal pH
Recently, a HisCat interaction that is critical for pH triggering in human metapneumovirus was dissected with mutagenesis (Chang et al., 2012). His435 is found clustered with basic residues, Lys295, Arg396, and Lys438 and variation of this histidine to an arginine resulted in a hyperfusogenic glycoprotein (Chang et al., 2012). Intriguingly, many HisCat pairs cluster at the interfaces between domains or subunits that undergo large spatial rearrangements during the fusion coordinate. For example, mutation of His3 from the Semliki Forest Virus glycoprotein, which is found at the trimer interface and contacts the cognate residue in the other subunits, markedly decreased the pH requirement for membrane fusion (Qin et al., 2009). The rapid change in electrostatic potential upon protonation may be a contributing force to spatial reorganization in these proteins.
The second pH-dependent interaction occurs between the side chains of two anionic residues (AniAni), Asp or Glu, in close proximity, often below 4Å and as close as 2.5Å (Figure 2). Asp and Glu side chains have pKa values of 3.9 and 4.2 respectively, but the pKa of these residues are often elevated in the context of a folded protein's microenvironment (Harms et al., 2009). At neutral pH, Asp and Glu are negatively charged; as the pH decreases they are protonated forming the conjugate acid. The proximity of these anionic side chains in the post-fusion conformation disfavors its formation at neutral pH (Figure 2). However, as the pH decreases this repulsion is relieved, thereby increasing the stability of the post-fusion conformation. For example, variation of Asp188, which is buried in the trimer core of Semliki Forest Virus, has profound effects on the pH-dependence of membrane fusion (Liu and Kielian, 2009). AniAni interactions may provide a stabilizing force in the post-fusion conformation because they fulfill the theoretical definition of low barrier hydrogen bond partners (Cleland, 2000). This type of interaction has been observed between Asp residues in aspartic acid proteases (Northrop, 2001). This phenomenon may contribute to the variation of the side chain pKa, although more extensive studies are necessary to demonstrate this effect.
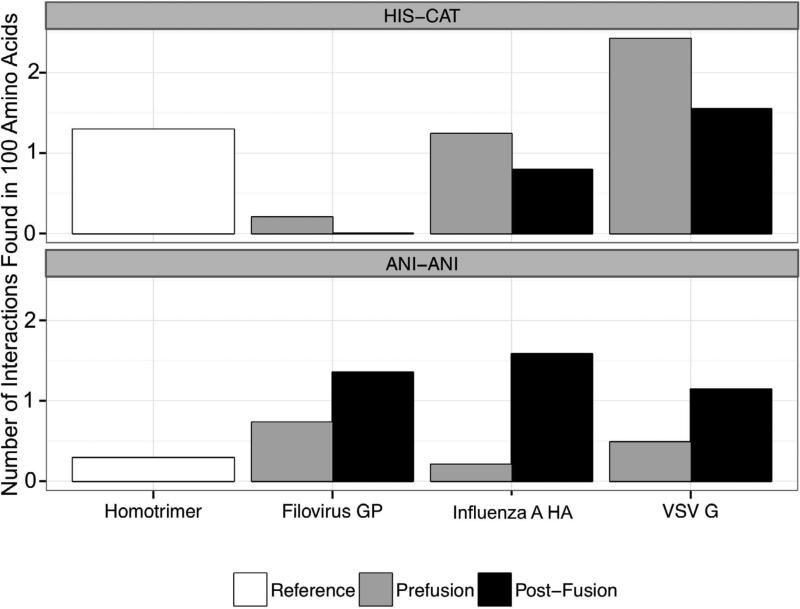
To analyze the distribution of HisCat and AniAni interactions in different conformations of viral fusion proteins, we used the Rosetta3 Scientific Benchmarking ChargeCharge feature reporter (Leaver-Fay et al., 2013) to measure interatomic distances between chemical moieties on charged residues’ side chains in both the pre- and post- fusion conformations of three model viral fusion systems discussed in detail herein: hemaggluttinin from influenza A (HA), protein G from vesicular stomatitis virus (VSV-G), and GP from filoviruses (GP). As a control, we analyzed a reference homotrimer set assembled from the 3D complex database with viral fusion proteins removed (Levy et al., 2006). All of this data is normalized to report the number of pairs contained within 100 amino acids, to account for differences in the total number of residues in the various structures. The PDB structures that were analyzed are indicated in the legend (Figure 3). This analysis confirmed that HisCat pairs are prevalent in the prefusion conformation of HA and VSV-G and decrease as the post-fusion conformation is assumed (Figure 3). Since HisCat interactions are tolerated at neutral pH but not at low pH, one explanation for their enrichment in prefusion conformations relative to post-fusion conformations is that they are required for destabilizing the prefusion state as the pH decreases. HisCat interactions are also found in the reference sets since these interactions are tolerated at neutral pH. Interestingly, HisCat pairs are rare in the prefusion state of filoviruses, which is consistent with recent evidence indicating formation of the extended intermediate is pH-independent (Carette et al., 2011). Therefore, low pH may not play a direct role in destabilizing the prefusion conformation of GP by direct side chain protonation.
Figure 3. Histogram of median interactions in viral proteins and in reference data sets.
Using Rosetta Scientific Benchmarking ChargeCharge feature reporter, we catalogued the number of HisCat and AniAni interactions in both the pre- and post- fusion states of HA, VSV-G, and GP. The virus data set includes the following PDBs, HA prefusion (clade) 1JSD (H9), 1JSM (H5), 1RU7 (H1), 1RUY (H1), 1RVT (H1), 1RUZ (H1), 2WRH (H1), 3EYJ (H1), 3HTO (H1), 3KU3 (H2), 3LZG (H1), 3M5G (H7), 3QQB (H2), 3S11 (H5), 3SDY (H3), 3VUN (H3), 4F23 (H16), 4FNK (H3), 2YP7 (H3), influenza post-fusion 1HTM, VSV-G prefusion 2J6J biological assembly, VSV-G post-fusion 2CMZ, GP prefusion 3CSY, GP post-fusion 2EBO, 4G2K. All ligands and other proteins were removed from these PDBs prior to analysis. We analyzed a reference homotrimer dataset for comparison. The distance cutoff for HisCat and AniAni interactions were 6.5Å and 4Å respectively, and the number of interactions was normalized per 100 amino acids.
Our analysis revealed that AniAni pairs are significantly enriched in the post-fusion state of the viral fusion proteins. These residue pairs are very rarely found in both the reference data set and the prefusion conformations of the viral fusion proteins. One potential explanation for this observation is that AniAni interactions are destabilizing at neutral pH and thus critical to preventing formation of the post-fusion state under neutral conditions. Furthermore, AniAni interactions may be involved in regulating the precise timing of the collapse of the extended intermediate (Figure 3).
HisCat and AniAni pairs have a similar, though opposite, contributions to the free energy of folding of a protein; at one pH the pairing is tolerated, and at another pH the pair is repulsive. It is difficult to calculate the energetic contribution that electrostatic interactions have to the ΔG of a protein fold, due to uncertainties about desolvation energies, local dielectric constants, and variations in side chains pKas. Studies have found that electrostatic interactions can contribute as much as -7kcal/mol to ΔG and, as a consequence of Coulomb's law, repulsive forces are equally destabilizing (Kumar and Nussinov, 2001). In general, protein folds are typically only stabilized by ΔG values between -5 to -20 kcal/mol (Dill, 1990). What is clear is that repulsive interactions can substantially destabilize a protein fold since there are often multiple repulsive interactions found in each protein subunit.
Energetic control mechanisms
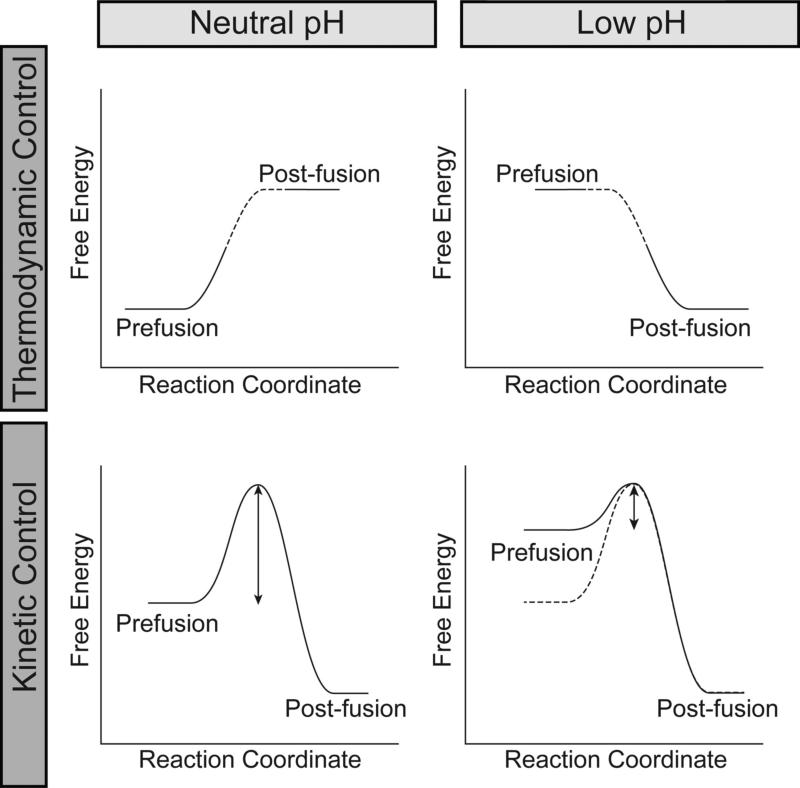
The reaction coordinates of viral fusion proteins are controlled by two distinct energetic mechanisms: 1) kinetic control, where there is a large activation barrier between the two conformations and this barrier decreases as a result of pH changes or 2) thermodynamic control, where a protein can exist in two distinct conformations and the energy minima are dictated by the pH (Figure 4) (Baker and Agard, 1994). Manifestations of these two mechanisms can be observed experimentally and these characteristics help to classify which energetic paradigm controls the conformational change. Fusion proteins regulated by thermodynamic control can undergo reversible conformational changes (Yao et al., 2003), while systems regulated by kinetic control undergo irreversible conformational changes (Ruigrok et al., 1986). The physical properties of HisCat and AniAni pairs contribute to these control mechanisms. Kinetic control relies on destabilization of the prefusion state, thereby decreasing the kinetic barrier that traps viral fusion proteins in the prefusion state (Figure 4). Thermodynamic control, on the other hand, relies upon destabilizing the post-fusion state at neutral pH and the prefusion state at low pH, allowing these proteins to function as a reversible switch. Here there is a prominent role for both HisCat and AniAni interactions to destabilizing the respective states. Exactly how these pairings contribute to kinetic and thermodynamic controls is still unclear though and further studies are warranted. Below we examine how HisCat and AniAni interaction function in three systems: HA from influenza A under kinetic control, protein G from VSV under thermodynamic control, and GP from filoviruses for which the mechanism is uncertain.
Figure 4.
Schematic of the thermodynamic and kinetic control models for pH-dependent conformational rearrangements
Influenza A virus HA1/HA2
Influenza A is a member of the Orthomyxoviridae family and causes respiratory tract infection in mammals and birds (Beigel et al., 2005). Influenza A contains two glycoprotein subunits, HA1 (surface) and HA2 (transmembrane), which facilitate viral entry. These proteins exist as a trimer of heterodimers (HA1/HA2) in the prefusion conformation. HA1 contains a glycan binding site that recognizes the influenza receptor, sialic acid, while HA2 is embedded in the membrane and contains the fusogenic subunit that promotes membrane fusion (Wilson et al., 1981). Unlike other viral proteins, receptor binding yields little conformational change and instead promotes viral endocytosis (Ha et al., 2001). Once inside the endosome HA2 undergoes a dramatic reorganization (Doms et al., 1985). Upon exposure to acidic pH, HA2 forms a highly stable extended α-helix, with a particularly impressive loop-to-helix transition in the hinge of the central stalk (Figure 6) (Bullough et al., 1994). In the prefusion state, HA2 is a compacted α-helix kinetically trapped by its association with HA1. HA2 conformational changes can be triggered in the absence of low pH by the addition of mild denaturants or heat, providing further support for the kinetic model (Ruigrok et al., 1986). Moreover, HA2 cannot adopt the prefusion conformation in the absence of HA1, again consistent with the kinetic model (Swalley et al., 2004). A consequence of the kinetic mechanism is that formation of the post-fusion state is irreversible therefore; it has been proposed that pH change predominantly destabilizes the HA1 interactions with itself and with HA2 (Huang et al., 2002). Indeed, as the virus enters the endosome and the vesicle matures, HA1 is predicted to acquire a greater cationic charge, destabilizing the HA1/HA1 interface and the interactions with HA2 (Kampmann et al., 2006; Mueller et al., 2008). Identifying precise residues that destabilize HA1 is challenging because of sequence drift in this highly mutable virus and limited structural data for each HA subtype. There are 17 known subtypes of influenza A HA and these subtypes can be divided into 4 clades based on evolutionary similarity. We generated consensus sequences for each HA subtype by aligning all available sequences in the Influenza Virus Resource (Bao et al., 2008) using the MUSCLE algorithm, then aligned each consensus sequence (Figure 5) (Edgar, 2004). This consensus sequence alignment coupled with solved structures of various HA subtypes, indicated in the legend, allowed us to identify various HisCat interactions which are conserved, as well as HisCat pairs which are subtype-unique (Figure 5).
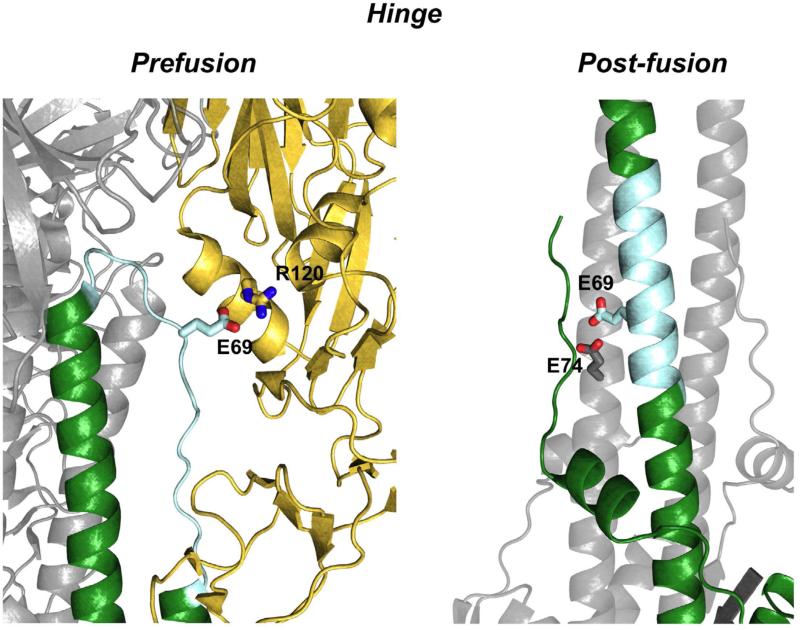
Figure 6. Hinge structural conformations in the prefusion and post-fusion conformation of Influenza HA.
Prefusion) The pH sensitive hinge (cyan) is depicted in the prefusion state interacting with HA1. A salt bridge between Glu69 of HA2 and Arg120 of HA1 is shown that stabilizes the heterodimer interface (1JSD) Hinge Post-Fusion) The hinge adopts an α-helical configuration where an AniAni interaction is formed between Glu69 and Glu74 on the adjacent chain (1HTM).
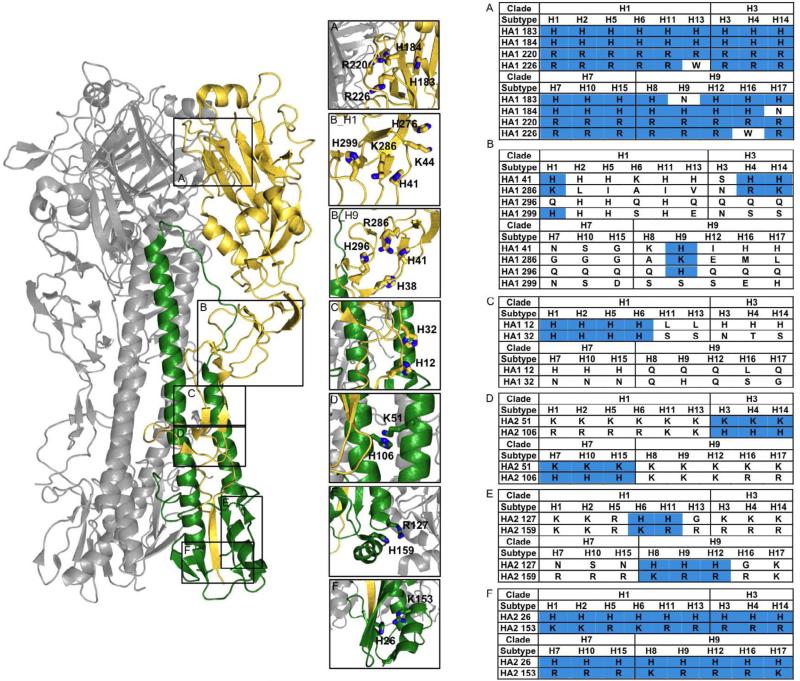
Figure 5. HisCat and AniAni interactions in influenza HA.
The prefusion state of HA, HA1 is shown in gold and HA2 is colored green. The prefusion state of HA contains numerous HisCat interactions, some of which are conserved, while others are subtype-specific. The residues occupying the position of the identified HisCat clusters, according to consensus sequence alignments generated for each HA subtype, are displayed in the tables on the right and residues that can participate in HisCat pairs are shaded blue. The numbering is consistent with the H1 clade and was determined from alignments to 1RU7 A) A conserved HisCat cluster at the head of HA1 found in all clades (1RU7). B_H1) The H1 HisCat cluster found in HA1 around the hinge (4F3Z). The interaction between Lys44 and H276 is only found in the H1 subtype, and for simplicity, is not included in the table. B_H9) The H9 HisCat cluster found in HA1 around the hinge. The interaction between His38 and His296 is only found in the H9 subtype, and for simplicity, is not included in the table. (1JSD). C) A HisCat interaction between His12 and His32, surrounding the fusions peptide pocket that is found in several H1 clade viruses. (1RUY). D) A HisCat cluster between Lys51 and His106 between the HA2 helices. This HisCat cluster is conserved in H3 and H7 clade viruses (4FNK). E) A HisCat cluster at the base of HA2 between His127 and Lys/Arg159 found in several H1 and H9 clade viruses (4FNK). F) A HisCat interaction at the base of HA2 between His26 and Lys/Arg153. This HisCat pair is conserved in all HA subtypes (1JSD).
The first HisCat cluster is located at the head of molecule adjacent to the HA1/HA1 interface (Figure 5A). Two cationic resides, His186 and Arg220, are absolutely conserved in all subtypes and these residues are between 4Å and 6Å of one another in various HA structures. Mutation of Arg220 to Ser increases the pH at which HA is triggered (Vanderlinden et al., 2010). Furthermore, mutations around this interface have been discovered that both increase and decrease the pH-dependence of the conformational change, demonstrating the interplay between stabilization and destabilization of the prefusion state (Daniels et al., 1985; Steinhauer et al., 1996). In most influenza strains, this HisCat cluster is surrounded by additional cations; a His is highly conserved at position 185 and other cations often cluster around this region (Figure 5A). Indeed, this HisCat cluster lies at the HA1/HA1 interface and molecular dynamics simulations have predicted that as the pH decreases the HA1/HA1 interface is destabilized, opening a channel for water to solvate HA2 (Huang et al., 2002, 2009; Korte et al., 2007).
The second HisCat interaction is found at the base of HA2. An α-helical segment that contains a conserved Lys or Arg at position 153 and a conserved His is found in a loop at position 26 (Figure 5F). During the transition to the post-fusion conformation this α-helical segment is dislodged and undergoes a large spatial rearrangement (Bullough et al., 1994). The electrostatic repulsive force from this HisCat cluster could provide some of the necessary force to translocate these α-helical segments during the structural rearrangement, although further analysis is necessary to confirm the role of this HisCat interaction in the fusion pathway.
The remaining HisCat clusters are found at various locations midway through the glycoprotein (Figure 5B,C). Interestingly, the location of these HisCat pairs is subtype-specific and we identified at least four different HisCat clusters in the various structures. Indeed, mutational studies in several HA subtypes have uncovered subtype-specific pH sensing mechanisms. For example, the H1 clade contains two histidines, His12 and His32, with side chains within 4Å (Figure 4C). Mutational studies that have varied His12 to Gln decrease the pH for which HA undergoes conformational rearrangement, while the addition of a His at position 23, normally a Tyr, increase the pH requirement (Reed et al., 2009, 2010). The H3 and H7 clades of the virus, on the other hand, contain a HisCat interaction between the side chains of Lys51 and His106 at the coiled-coil interface of HA2 (4Å). Variation of either of these residues results in a decrease in the pH of fusion (Thoennes et al., 2008). The H1 and H9 clades contain a cation at position 106, but this residue is oriented away from residue 51 (Thoennes et al., 2008).
Additionally, we identified several subtype specific HisCat clusters that have not been characterized experimentally. The H1 and H9 subtypes contain a large HisCat cluster in HA1 in the region surrounding the hinge. This cluster contains between 2 and 5 cationic residues in the different subtypes, although the exact location of these residues varies in subtypes (Figure 5B_H1, 5B_H9). H1 contains three cationic residues, His41, Lys286, and His299 within 5Å, while the side chains of His41 and Arg/Lys286 are found less then 3.5Å of each other in the H9 subtype (Figure 5B_H1, 5B_H9). Sequence alignments suggest that this interaction is also found in the H4 and H14 subtypes for which crystal structures are not available. Both the H1 and H9 subtypes contain an additional conserved HisCat interaction surrounding this region that is also subtype specific. Lys44 and His276, found in H1, are within 6Å and His38 and His296, from H9, are within 6Å. We identified an additional HisCat cluster found in several of the H1 and H9 clades at the base of HA2 between Arg/Lys at position 127 and His159 (Figure 5E). The different subtypes of HA are triggered at different threshold pH values, and the subtype variability of these HisCat clusters may contribute to these differences. Moreover, this deviation underscores the importance of additional structural and functional studies to pinpoint specific pH sensing regions of influenza HA proteins.
Loop to α-Helix Transition of the Hinge
The most dynamic structural transition in HA2 as it proceeds along the reaction coordinate, occurs in the hinge region (Figure 6). In the prefusion state, this 17 amino acid segment exists as an unstructured loop that makes extensive contacts with HA1. In the post-fusion conformation, the hinge forms an α-helix in the central triple stranded bundle. This interaction has been studied with a 33 amino acid peptide derived from this region (Carr and Kim, 1993). At neutral pH these peptides are unstructured monomers, but at pH 5 they form highly stable trimers. A single AniAni pair between two conserved anions, Glu69 and Glu74 (2.6Å), is hypothesized to account for this pH-dependence (Carr and Kim, 1993). Consequently, the behavior of the isolated hinge is more consistent with the thermodynamic model of conformational control, indicating that not all of the structural rearrangements can be well classified into one energetic model. Additional support for this notion comes from the Tm increase in HA2, ~15°C, as the pH decreases indicating that the post-fusion state is stabilized at low pH (Chen et al., 1995). However, the stability of the post-fusion conformation at neutral pH largely supports kinetic control.
Salt bridges between HA1 and HA2 have also been implicated in the pH-dependent conformational changes of HA (Rachakonda et al., 2007). Again, HA1 functions to kinetically trap HA2 in the prefusion arrangement and the hinge lies at the HA1/HA2 interface contacts. The hinge region has a strong anionic character. A salt bridge between Glu69, which forms an AniAni interaction in the post-fusion state, and Lys/Arg at position 120 on the cationic HA1 face stabilize this interface. As the pH decreases, the anionic character of the hinge decreases and this weakens the HA1/HA2 interface (Figure 6). Indeed, many mutations at the HA1/HA2 interface have been identified that alter the pH-dependence of this conformational change (Daniels et al., 1985; Godley et al., 1992; Korte et al., 2007; Rachakonda et al., 2007; Steinhauer et al., 1996). These observations demonstrate that the HA1/HA2 complex is thermodynamically tuned to function as a sensitive pH switch and variations to the Gibbs free energy of either the pre- or post-fusion state have functional consequences.
Vesicular Stomatitis Virus Protein G
VSV is a member of the Rhabdoviridae family of viruses that causes vesiculation and ulceration in the oral and nasal mucosa of cattle, horses, pigs and rarely other mammals (Letchworth et al., 1999). VSV-G is the fusion protein that allows VSV to enter cells and conformational changes in this protein are regulated by thermodynamic control. VSV-G undergoes reversible changes in response to pH variation, unlike the irreversible change displayed by HA. Indeed, VSV infectivity can be inactivated by low pH, and then subsequently recovered when exposed to neutral pH (Gaudin, 2000). VSV-G is composed of 4 domains and arranged as a trimer in the prefusion state on the viral surface, adopting the canonical class III glycoprotein fold (Roche et al., 2007). Domain 1 (DI) is composed of β-sheets, Domain II (DII) is α-helical, Domain III, (DIII) contains a pleckstrin homology fold, and Domain IV (DIV) contains the fusion peptide (Figure 7). Furthermore, DII can be divided into three sections that are non-continuous in the primary sequence, the N-heptad repeat (NHR), the Middle heptad repeat (MHR), and the C-heptad repeat (CHR). The MHR, CHR, and portions of the NHR are unstructured in the prefusion conformation and adopt the canonical 6 α-helix bundle in the post-fusion conformation (Fig. 7).
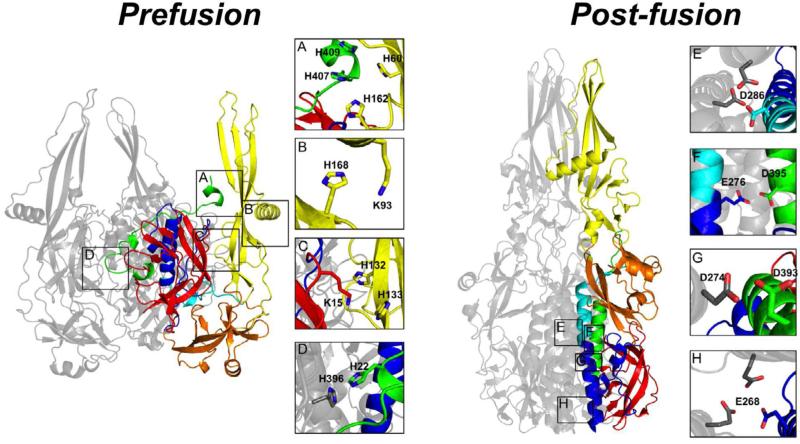
Figure 7. HisCat and AniAni interactions in VSV-G.
Prefusion) DI is red, DII is blue/green, DIII is orange, and DIV is yellow. DII can be divided into three sections that are non-continuous in the primary sequence, the NHR;blue, the MHR;Cyan, and the CHR;Green. In the compacted prefusion state a number of HisCat interactions are found, often between domains, and they are depicted in panels A-D (2J6J Biological assembly). Post-fusion) VSV-G elongates in the post-fusion state losing many of the HisCat interactions found in the prefusion state. In the post-fusion state many AniAni interactions form, occurring in DII and they are depicted in panels E-H (2CMZ).
In its prefusion state VSV-G is compact and there are extensive interactions between the four domains. Both DI and DII share large interfaces with DIV. It is well established that histidines are critical for the pH-dependent conformational changes in VSV-G and other class III viruses (Boo et al., 2012; Carneiro et al., 2003; Chang et al., 2012; Stauffer et al., 2007). The prefusion state of VSV also contains numerous clusters of HisCat pairs. Many of these clusters localize to interfaces between domains, in particular the interfaces between DIV and DI and DII. His407 and His409, from the unstructured region of the CHR of DII, are found 3.4Å and 5.5Å away from His162 and His60 from DIV (Figure 7A). Another HisCat pair is located between Lys93 and His168 in DIV (4Å) (Figure 7B). Likewise, Lys15 in DI is found nestled between His132 and His133 of DIV (5.2Å) (Figure 7C). The final HisCat interaction is inter-subunit located at the trimer interface; the side chains of His22 from the unstructured NHR and His396 of the unstructured CHR are found 3.5Å apart from one another (Figure 7D). Interactions that could destabilize the trimer interface of VSV-G are compelling in light of recent reports of a monomeric intermediate during the transition from the prefusion to the post-fusion trimer (Albertini et al., 2012). Furthermore, molecular dynamics studies have also identified His 132/Lys15 and His162/His407 as crucial repulsive interactions facilitating this pH-dependent conformational rearrangement of VSV-G (Rücker et al., 2012).
VSV-G undergoes a striking reorganization in response to the decreases in pH encountered in the endosome (Figure 7). DI, which occupies the interior of the protein at neutral pH, relocates to the exterior. DII, which is predominantly unstructured in the prefusion conformation, folds into a highly stable six α-helical bundle (Roche et al., 2006). The NHR and MHR form the extended three-helix core of the bundle, constituting the post-fusion trimer interface, and the CHR packs into the newly formed grooves. DIII, which occupies the base of the prefusion structure, packs against DII. Finally, DIV protrudes towards the host membrane, disengaging from DI and DII, eliminating many of the HisCat pairs. There is an increase in the number of AniAni interactions found in the post-fusion conformation (Figure 3). Four AniAni interactions are formed per subunit, all found in DII, two of which are found at the trimer interface. Both Asp286 and Glu268 of the MHR and NHR respectively contact the cognate residue in the opposing subunits (Figure 7E,H). Another two AniAni interactions occur between the core three-helix bundle and the outer three α-helices. Asp274 and Glu276 of the NHR respectively pair with Asp395 and Asp393 from the CHR (Figure 7F,G). Given the exquisite stability of viral six-helix bundles, mechanisms that disfavor this conformation at neutral pH are likely. Furthermore, AniAni interactions are found at the trimer interface, providing another compelling reason for the oligomeric change in protein G as it progresses through the reaction coordinate (Albertini et al., 2012).
In the post-fusion state, VSV-G also contains a number of pH-dependent salt bridges between His and Asp or Glu. One such interaction is of particular note because it occurs between His residues that were involved in a HisCat interaction in the prefusion conformation. His132 and His133 participate in a salt bridge with D145 of DIV in the low pH conformation. Mechanisms that take advantage of stabilizing interactions in both the prefusion and post-fusions states could provide VSV-G with its unique ability to undergo reversible conformational changes.
Filovirus GP
Filoviruses, comprised of Ebolaviruses and Marburgviruses, are extremely pathogenic, causing a highly fatal hemorrhagic fever in humans and non-human primates (Miller and Chandran, 2012). Embedded in the membrane of the filamentous viral particle is the fusion machinery, glycoprotein GP, comprised of the subunits GP1 and GP2. These proteins are arranged as the obligate trimer of dimers. Two natural genetic filovirus isolates have been characterized structurally, Zaire Ebolavirus (EBOV) and Lake Victoria Marburgvirus (MARV). Filoviruses have a distinct fusion mechanism from the aforementioned viruses; the prefusion state is not destabilized by acidic pH, even though the virus has been demonstrated to enter the endosome (Nanbo et al., 2010; Takada et al., 1997). This may provide an explanation for the rarity of HisCat interactions in the prefusion conformation of GP (Figure 3). Once inside the endosome, GP1 is cleaved by endosomal proteases (Chandran et al., 2005). In the prefusion state the GP resembles a chalice and upon triggering GP2 adopts a short six-helix bundle (Fig 8) (Lee et al., 2008). Acidic pH alone is not sufficient to trigger the cleaved intermediate; although recent work suggests that cleaved GP treated with acidic pH and denaturants or heat can induce membrane binding in purified, soluble ectodomain protein (Brecher et al., 2012). Thus, the prefusion state is not metastable, in the classical sense, and some other triggering mechanism is required. Recently, an endosomal receptor for EBOV has been identified, Nieman Pick Complex 1, suggesting that receptor binding triggers the first transition in the reaction coordinate; formation of the extended intermediate (Carette et al., 2011).
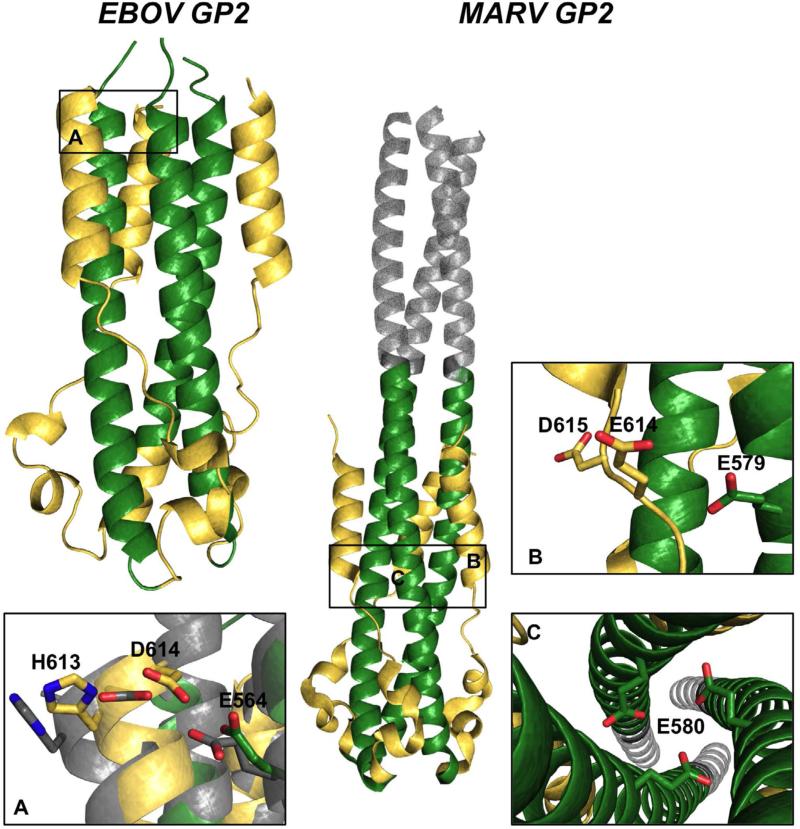
Figure 8. AniAni interactions in GP2 post-fusion conformations.
EBOV GP2) The 6-helix bundle of ebola GP2 is depicted, the CHR is colored gold and the NHR is colored green (2EBO). Panel A shows the AniAni interaction found between the CHR and the NHR. In grey is an overlay of the neutral pH structure (1EBO) and the low pH structure (2EBO). MARV GP2) MARV GP2 post-fusion state is colored akin to EBOV GP2, with the GCN4 trimerization tag colored grey (4G2K). AniAni pairs that make up the anionic stripe can be seen in panels B and C.
Despite evidence that the prefusion conformation is not destabilized by endosomal pH, acidic pH does indeed play a prominent role in filovirus fusion. Previously, it was thought that low pH was only necessary to promote proteolytic cleavage (i.e., low pH does not play a direct role on GP conformation); however, there is now evidence demonstrating that low pH has a significant role in stabilizing the post-fusion state. A designed six-helix bundle derived from EBOV and the full length GP2 protein from MARV are both stabilized in acidic pH (Tm increase of ~40°C as the pH decreases from 6.1-4.8) (Harrison et al., 2011, 2012). These pH effects are predominantly the result of AniAni interactions. In EBOV GP2, an AniAni pair is found between Asp 614 in the CHR and Glu 564 in the NHR (Figure 8A). Intriguingly, EBOV GP2 has been crystallized in the post-fusion state at both neutral and acidic pH. At neutral pH the GCN4 trimerization tag was appended to N-terminus of GP2 to promote trimerization and stability (Malashkevich et al., 1999; Weissenhorn et al., 1998). The majority of these two crystal structures superimpose very well, with an RMSD score of 1.08Å between the helical bundles. Of note, however, the last helical turn, where the AniAni interaction is found, does not overlay well and the RMSD for this segment alone is 1.85Å (Figure 8A). In the neutral pH structure, the oxygens on the side chains of these two acidic groups are 0.7Å further apart then at neutral pH (Figure 8). There is also salt bridge between His 613 and Asp 614 that is displaced by 0.7Å in neutral pH crystal structure, further implicating a role for pH-dependent salt bridges at low pH. When these anionic residues are removed from the designed 6-helix bundle, the pH-dependent stability was completely abrogated (Harrison et al., 2011). Full length MARV GP2 was shown to have identical pH-dependent behavior (40°C increase in thermal stability from pH 6.3 to 4.8). By varying both Glu 579 and Glu 580 to the neutral analog glutamine, the stability of GP2 at neutral pH increased by 22°C, although this variant still displayed greater thermostability at low pH (Figure 8B,C) (Harrison et al., 2012). Examination of the recently determined crystal structure of MARV GP2 reveals that these residues are found among an anionic stripe running across the midsection of the GP2 bundle, with Glu580 facing the trimer interface, interacting with the cognate amino acid. Additionally, Glu579 forms an AniAni interaction with Glu614 in the CHR (Figure 8B) (Koellhoffer et al., 2012). Surprisingly, varying either of these residues alone to their neutral counterparts did not increase the thermostability at neutral pH, suggesting a concerted mechanism is responsible for these pH effects (Harrison et al., 2012).
The fusion peptide from EBOV has been shown to undergo pH-dependent conformational rearrangements as well. Filoviruses contain an internal fusion peptide, which is found one helical turn upstream of Glu564 in the NHR. A peptide derived from this region displayed a marked increase in α-helicity at pH 5.5 and induced rapid fusion of liposomes at this pH (Gregory et al., 2011). The lowest energy NMR representations of the peptide at the two different pHs show that the α-helical region is flanked by two anionic residues, Glu540 and Glu545, which are much closer spatially, 12Å to 5Å, as the peptide is compacted at low pH (Gregory et al., 2011). Taken together, these results suggest that pH plays a critical role in the formation of the post-fusion conformation of GP2, but there is no evidence that pH is a factor in the transition from the prefusion state to the extended intermediate. Indeed, the structurally related ASLV is known to undergo a unique two-step fusion mechanism; receptor binding triggers the first step, likely formation of the prefusion intermediate, while the second step, collapse of the prefusion intermediate, is triggered by pH change (Cardone et al., 2012; Matsuyama et al., 2004).
The energetic mechanism that regulates the GP2 conformational change is still unclear. It is interesting that the post-fusion state is stabilized by acidic pH, which is consistent with the thermodynamic mechanism, but that the pH change is not sufficient to trigger the conformational change in the glycoprotein – receptor binding and proteolysis are also required. The GP reaction coordinate demonstrates that the two energetic controls may not be mutually exclusive and individual steps along the pathway may have different energetic controls.
Concluding Remarks
Here we outline the role of electrostatic interactions that govern transitions through the reaction coordinate of pH-dependent viral fusion proteins. Analysis of three dimensional structures coupled with mutational data present a unique paradigm; HisCat interactions found in the prefusion state destabilize this conformation as the pH decreases, while AniAni interactions prevent formation of the post-fusion state until the appropriate pH is encountered. Yet, there are still a number of unanswered questions concerning the details of these mechanisms. Elucidating the thermodynamic contributions of these pairs is required to clarify the role of electrostatics in facilitating these conformational changes. This may be challenging since fusion studies are often performed with viruses and mutating conserved residues in the glycoproteins often result in fusion incompetent viruses. Viral fusion proteins are segregated by two energetic mechanisms, thermodynamic and equilibrium, that govern these conformational changes. However, the exact contribution of these residue pairings to these two mechanisms is, at this point, speculative. Taken together, HisCat and AniAni interactions, in addition to pH-dependent salt bridges, form the basis for the pH sensitive switching mechanisms observed in viral fusion proteins. This paradigm will serve to guide future structural studies aimed at uncovering the thermodynamic requirements of pH sensitive switching in viral fusion proteins.
Highlights.
– Viral fusion proteins undergo pH-dependent conformational changes.
– Electrostatic repulsion between side chains mediates structural transitions.
– Histidine-cation (HisCat) and anion-anion (AniAni) interactions are common.
Acknowledgments
J. R. L. and B. A. K. acknowledge funding from the NIH (R01-AI090249 and R01-GM073960). J.S.H was supported in part by an NIH molecular Biophysics Training Grant (T32-GM008572) and an NIH postdoctoral training grant through the Lineberger Comprehensive Cancer Center at U.N.C. (T32-CA009156), J. F. K. is supported by Medical Scientist Training Program T32-GM007288.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertini A. a, Mérigoux C, Libersou S, Madiona K, Bressanelli S, Roche S, Lepault J, Melki R, Vachette P, Gaudin Y. Characterization of monomeric intermediates during VSV glycoprotein structural transition. PLoS Pathogens. 2012;8:e1002556. doi: 10.1371/journal.ppat.1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D, Agard D. a. Influenza hemagglutinin: kinetic control of protein function. Structure. 1994;2:907–910. doi: 10.1016/s0969-2126(94)00091-3. [DOI] [PubMed] [Google Scholar]
- Baldwin RL. Energetics of protein folding. J. Mol Biol. 2007;371:283–301. doi: 10.1016/j.jmb.2007.05.078. [DOI] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, De Jong MD, Lochindarat S, Nguyen TKT, Nguyen TH, Tran TH, et al. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- Boo I, TeWierik K, Douam F, Lavillette D, Poumbourios P, Drummer HE. Distinct roles in folding, CD81 receptor binding and viral entry for conserved histidine residues of hepatitis C virus glycoprotein E1 and E2. Biochem. J. 2012;443:85–94. doi: 10.1042/BJ20110868. [DOI] [PubMed] [Google Scholar]
- Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J. Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough P, Hughson F, Skehel J, Wiley D. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Cardone G, Brecher M, Fontana J, Winkler DC, Butan C, White JM, Steven AC. Visualization of the Two-Step Fusion Process of the Retrovirus Avian Sarcoma/Leukosis Virus by Cryo-Electron Tomography. J. Virol. 2012;86:12129–12137. doi: 10.1128/JVI.01880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro F. a, Stauffer F, Lima CS, Juliano M. a, Juliano L, Da Poian AT. Membrane fusion induced by vesicular stomatitis virus depends on histidine protonation. J. Biol. Chem. 2003;278:13789–13794. doi: 10.1074/jbc.M210615200. [DOI] [PubMed] [Google Scholar]
- Carneiro F. a, Vandenbussche G, Juliano M. a, Juliano L, Ruysschaert J-M, Da Poian AT. Charged residues are involved in membrane fusion mediated by a hydrophilic peptide located in vesicular stomatitis virus G protein. Mol. Membr. Biol. 2006;23:396–406. doi: 10.1080/09687860600780892. [DOI] [PubMed] [Google Scholar]
- Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanel-Vos C, Kielian M. A Conserved Histidine in the ij Loop of the Semliki Forest Virus E1 Protein Plays an Important Role in Membrane Fusion. J. Virol. 2004;78:13543–13543. doi: 10.1128/JVI.78.24.13543-13552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Hackett B, Winter CC, Buchholz UJ, Dutch RE. Potential electrostatic interactions in multiple regions affect HMPV F-mediated membrane fusion. J. Virol. 2012;86:9843–9853. doi: 10.1128/JVI.00639-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wharton S. a, Weissenhorn W, Calder LJ, Hughson FM, Skehel JJ, Wiley DC. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc. Natl. Acad. Sci. USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Zimmerberg J, Kozlov MM. Membranes of the world unite! J. Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland WW. Low-barrier hydrogen bonds and enzymatic catalysis. Arch. Biochem. Biophys. 2000;382:1–5. doi: 10.1006/abbi.2000.2011. [DOI] [PubMed] [Google Scholar]
- Daniels RS, Downie JC, Hay a J., Knossow M, Skehel JJ, Wang ML, Wiley DC. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- Dill K. Dominant Forces in Protein Folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Doms R, Helenius A, White J. Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J. Biol. Chem. 1985;260:2973–2981. [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y. Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Sub-cellular Biochem. 2000;34:379–408. doi: 10.1007/0-306-46824-7_10. [DOI] [PubMed] [Google Scholar]
- Godley L, Pfeifer J, Steinhauer D, Ely B, Shaw G, Kaufmann R, Suchanek E, Pabo C, Skehel JJ, Wiley DC. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- Gregory SM, Harada E, Liang B, Delos SE, White JM, Tamm LK. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc. Nat. Acad. Sci. USA. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Nat. Acad. Sci. USA. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Castañeda C. a, Schlessman JL, Sue GR, Isom DG, Cannon BR, García-Moreno E,B. The pK(a) values of acidic and basic residues buried at the same internal location in a protein are governed by different factors. J. Mol. Biol. 2009;389:34–47. doi: 10.1016/j.jmb.2009.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JS, Higgins CD, Chandran K, Lai JR. Designed protein mimics of the Ebola virus glycoprotein GP2 α-helical bundle: stability and pH effects. Protein Sci. 2011;20:1587–1596. doi: 10.1002/pro.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JS, Koellhoffer JF, Chandran K, Lai JR. Marburg virus glycoprotein GP2: pH-dependent stability of the ectodomain α-helical bundle. Biochemistry. 2012;51:2515–2525. doi: 10.1021/bi3000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Opitz R, Knapp E-W, Herrmann A. Protonation and stability of the globular domain of influenza virus hemagglutinin. Biophysical J. 2002;82:1050–1058. doi: 10.1016/S0006-3495(02)75464-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Korte T, Rachakonda PS, Knapp E-W, Herrmann A. Energetics of the loop-to-helix transition leading to the coiled-coil structure of influenza virus hemagglutinin HA2 subunits. Proteins. 2009;74:291–303. doi: 10.1002/prot.22157. [DOI] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Lin K, Strick N, Neurath A. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. The Role of histidine residues in low-pH-mediated viral membrane fusion. Structure. 2006;14:1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Kielian M, Rey F. a. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Donald JE, Grigoryan G, Leser GP, Fadeev AY, Lamb R. a, DeGrado WF. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Nat. Acad. Sci. USA. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koellhoffer JF, Malashkevich VN, Harrison JS, Toro R, Bhosle RC, Chandran K, Almo SC, Lai JR. Crystal Structure of the Marburg Virus GP2 Core Domain in Its Postfusion Conformation. Biochemistry. 2012;51:7665–7675. doi: 10.1021/bi300976m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte T, Ludwig K, Huang Q, Rachakonda PS, Herrmann A. Conformational change of influenza virus hemagglutinin is sensitive to ionic concentration. Eur. Biophys. J. 2007;36:327–335. doi: 10.1007/s00249-006-0116-0. [DOI] [PubMed] [Google Scholar]
- Kozlov MM, McMahon HT, Chernomordik LV. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 2010;35:699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nussinov R. Fluctuations in ion pairs and their stabilities in proteins. Proteins. 2001;43:433–454. doi: 10.1002/prot.1056. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A, O'Meara MJ, Tyka M, Jacak R, Song Y, Kellogg EH, Thompson J, Davis IW, Pache R. a, Lyskov S, et al. Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchworth GJ, Rodriguez LL, Del cbarrera J. Vesicular stomatitis. Veterinary J. 1999;157:239–260. doi: 10.1053/tvjl.1998.0303. [DOI] [PubMed] [Google Scholar]
- Levy ED, Pereira-Leal JB, Chothia C, Teichmann S. a. 3D complex: a structural classification of protein complexes. PLoS Compt. Biol. 2006;2:e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Kielian M. E1 mutants identify a critical region in the trimer interface of the Semliki forest virus fusion protein. J. Virol. 2009;83:11298–11306. doi: 10.1128/JVI.01147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozach P-Y, Huotari J, Helenius A. Late-penetrating viruses. Curr. Opin. Virol. 2011;1:35–43. doi: 10.1016/j.coviro.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Malashkevich VN, Schneider BJ, McNally ML, Milhollen M. a, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc. Nat. Acad. Sci. USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Delos SE, White JM. Sequential Roles of Receptor Binding and Low pH in Forming Prehairpin and Hairpin Conformations of a Retroviral Envelope Glycoprotein. J. Virol. 2004;78:8201–8209. doi: 10.1128/JVI.78.15.8201-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Ann. Rev. Bioc. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- Miller EH, Chandran K. Filovirus entry into cells - new insights. Curr. Opin. Virol. 2012;2:206–214. doi: 10.1016/j.coviro.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EH, Harrison JS, Radoshitzky SR, Higgins CD, Chi X, Dong L, Kuhn JH, Bavari S, Lai JR, Chandran K. Inhibition of Ebola virus entry by a C-peptide targeted to endosomes. J. Biol. Chem. 2011;286:15854–15861. doi: 10.1074/jbc.M110.207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DS, Kampmann T, Yennamalli R, Young PR, Kobe B, Mark AE. Histidine protonation and the activation of viral fusion proteins. Biochem. Soc. Trans. 2008;36:43–45. doi: 10.1042/BST0360043. [DOI] [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathogens. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop DB. Follow the protons: a low-barrier hydrogen bond unifies the mechanisms of the aspartic proteases. Acc. Chem. Res. 2001;34:790–797. doi: 10.1021/ar000184m. [DOI] [PubMed] [Google Scholar]
- Pessi A, Langella A, Capitò E, Ghezzi S, Vicenzi E, Poli G, Ketas T, Mathieu C, Cortese R, Horvat B, et al. A general strategy to endow natural fusion-protein-derived peptides with potent antiviral activity. PloS One. 2012;7:e36833. doi: 10.1371/journal.pone.0036833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z-L, Zheng Y, Kielian M. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J. Virol. 2009;83:4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachakonda PS, Veit M, Korte T, Ludwig K, Böttcher C, Huang Q, Schmidt MFG, Herrmann A. The relevance of salt bridges for the stability of the influenza virus hemagglutinin. FASEB J. 2007;21:995–1002. doi: 10.1096/fj.06-7052hyp. [DOI] [PubMed] [Google Scholar]
- Reed ML, Yen H-L, DuBois RM, Bridges O. a, Salomon R, Webster RG, Russell CJ. Amino acid residues in the fusion peptide pocket regulate the pH of activation of the H5N1 influenza virus hemagglutinin protein. J. Virol. 2009;83:3568–3580. doi: 10.1128/JVI.02238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ML, Bridges O. a, Seiler P, Kim J-K, Yen H-L, Salomon R, Govorkova E. a, Webster RG, Russell CJ. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J. Virol. 2010;84:1527–1535. doi: 10.1128/JVI.02069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, Bressanelli S, Rey F. a, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Roche S, Rey F. a, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Rücker P, Wieninger S. a, Ullmann GM, Sticht H. pH-dependent molecular dynamics of vesicular stomatitis virus glycoprotein G. Proteins. 2012;80:2601–2613. doi: 10.1002/prot.24145. [DOI] [PubMed] [Google Scholar]
- Ruigrok RW, Martin SR, Wharton S. a, Skehel JJ, Bayley PM, Wiley DC. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virol. 1986;155:484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. 2009;83:1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer F, De Miranda J, Schechter MC, Carneiro F. a, Salgado LT, Machado GF, Da Poian AT. Inactivation of vesicular stomatitis virus through inhibition of membrane fusion by chemical modification of the viral glycoprotein. Antiviral Res. 2007;73:31–39. doi: 10.1016/j.antiviral.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Steinhauer D. a, Martín J, Lin YP, Wharton S. a, Oldstone MB, Skehel JJ, Wiley DC. Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc. Nat. Acad. Sci. USA. 1996;93:12873–12878. doi: 10.1073/pnas.93.23.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalley S, Baker B, Calder L. Full-Length Influenza Hemagglutinin HA2 Refolds into the Trimeric Low-pH-Induced Conformation. Biochemistry. 2004;185:5902–5911. doi: 10.1021/bi049807k. [DOI] [PubMed] [Google Scholar]
- Takada a, Robison C, Goto H, Sanchez A, Murti KG, Whitt M. a, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc. Nat. Acad. Sci. USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoennes S, Li Z-N, Lee B-J, Langley W. a, Skehel JJ, Russell RJ, Steinhauer D. a. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virol. 2008;370:403–414. doi: 10.1016/j.virol.2007.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden E, Göktas F, Cesur Z, Froeyen M, Reed ML, Russell CJ, Cesur N, Naesens L. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J. Virol. 2010;84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Carfí A, Lee K, Skehel J, Wiley D. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Hinz A, Gaudin Y. Virus membrane fusion. FEBS Letters. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Delos S. Structures and Mechanisms of Viral Membrane Fusion Proteins: Multiple Variations on a Common Theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I, Skehel J, Wiley D. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Yao Y, Ghosh K, Epand RF, Epand RM, Ghosh HP. Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low pH but not by heat or denaturant. Virol. 2003;310:319–332. doi: 10.1016/s0042-6822(03)00146-6. [DOI] [PubMed] [Google Scholar]