Abstract
The Phe43 cavity is a mysterious feature in crystallographic structures of HIV-1 gp120-CD4 complexes. In this Structure issue, Acharya and colleagues provide structural explanations for the potent neutralization by CD4 mimetic miniproteins with chemical extensions that fit into this cavity.
The first X-ray crystallographic structures of HIV-1 gp120, initially solved for the HXBc2 laboratory-adapted strain (Kwong et al., 1998) and subsequently refined and extended to the YU2 primary isolate (Kwong et al., 2000) (Huang et al., 2004), revealed not only tantalizing insights into receptor interactions, but also a very puzzling structural feature. In the ternary complex of gp120 “core” bound to a fragment of CD4 and an Fab against a conserved determinant involved in coreceptor binding, an interfacial pocket of ~150 Å3 was evident at the nexus of three gp120 domains -the inner domain, the outer domain, and the bridging sheet; the pocket was capped by Phe43 of CD4, a residue previously shown to be critical for the gp120-CD4 binding interaction. The structural and functional significance of this so-called Phe43 cavity (Figure 1) was unclear, but its conservation and unlikely existence in the absence of CD4 suggested that it reflects large receptor-induced conformational changes in gp120. Beyond its relationship to gp120 function, the Phe43 cavity was recognized as a potentially useful target for antiviral strategies. Thus substituting a bulky hydrophobic tryptophanresidue (gp120 S375W) into the cavity partially stabilizes gp120 toward the CD4-bound state (Xiang et al., 2002), a feature that was explored for potential vaccine immunogen applications (Dey et al., 2007). HIV entry inhibitors that act by filling the Phe43 cavity were designed either as small organic molecules (Madani et al., 2008) (Curreli et al., 2012) or as engineered CD4 mimetic “miniproteins”pioneered by Vita and coworkers based on small protein scaffolds (Vita et al., 1998); the latter were taken through a series of structure-guided optimizations involving inclusion of Phe at position 23 of the miniproteins to mimic CD4 Phe43 in plugging the cavity (Martin et al., 2003), and culminating in addition of flexible hydrophobic extensions to this miniprotein residue that are capable of fitting into the cavity (Van Herrewege et al., 2008). The result was a dramatic increase in neutralization potency of HIV-1 pseudovirus infection, as much as >1000-fold for miniprotein M48U1 against some isolates.
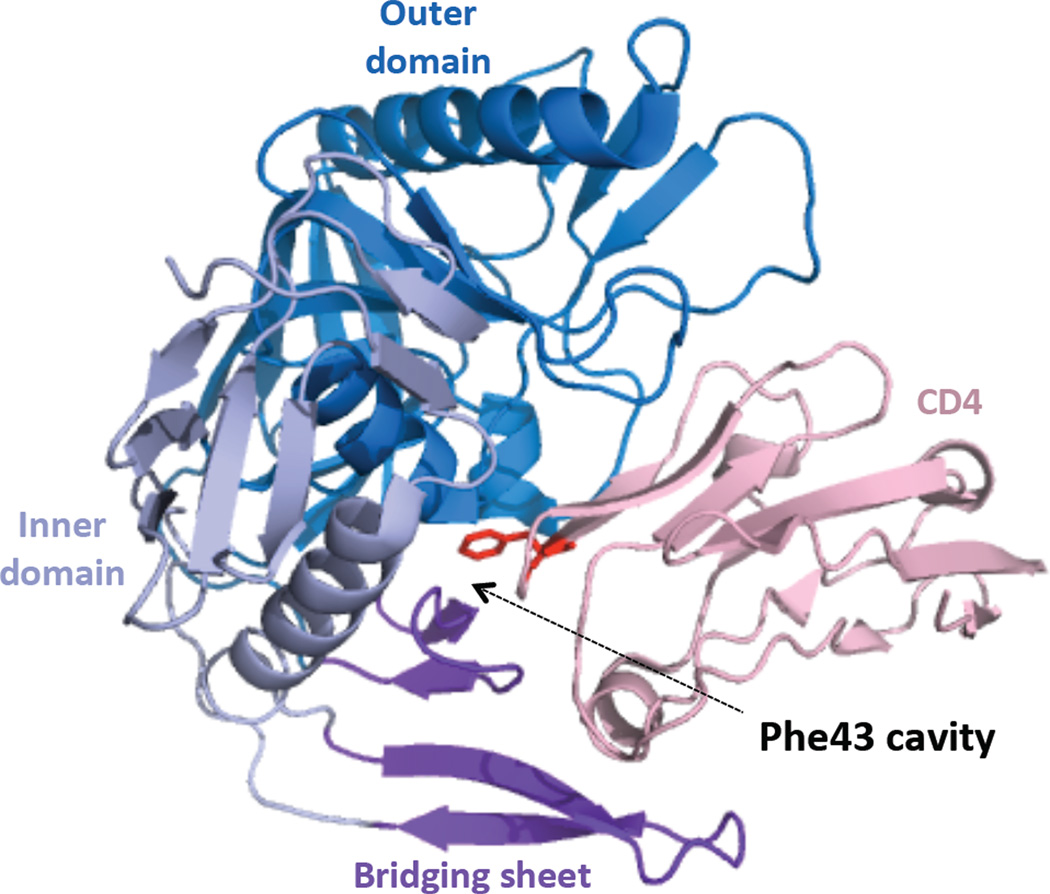
Figure 1. The Phe43 cavity.
CD4 (pink; only domain 1 is shown) binding induces formation of a ~150 Å3 interfacial pocket at the nexus of the gp120 inner domain (pale blue), outer domain (marine blue) and bridging sheet (purple); the phenyl ring of the Phe43 side chain (red) forms a lid to the cavity, and is the only contributing component from CD4. Generated using MacPyMOL from PDB 1RZK (Huang et al, 2004).
In this issue of Structure, Acharya et al. analyze the structural basis for the extreme neutralization potency of M48U1 as well as the related M48U7, which contains a different flexible hydrophobic extension at the same position. Surface plasmon resonance analyses indicated that M48U1, the more potent CD4 mimetic, bound to gp120 with extremely high affinity (KD = 0.15 nM), which reflected both a high association rate and extremely slow dissociation rate. Crystallographic analyses of these mimetics bound to a gp120 core protein (YU2 isolate) yielded interesting insights into their modes of action. Analyses of flexibilities of the extended moieties in the bound structures coupled with determination of combined fit parameters (shape complementarity and extent of surface burial) highlighted the superiority of M48U1 over M48U7 and other related miniproteins. The effects of the miniproteins on local conformation of gp120 were analyzed by comparison of the miniprotein-bound with the unliganded structures. The analyses revealed that when bound to the extension-containing mimetics, the Phe43 cavity resembled more closely that region in the unliganded (ground state) compared to the CD4-bound state; earlier mimetics lacking the extensions showed the opposite resemblance. The authors concluded that recognition of the ground state by M48U1 and M48U7 is more favorable energetically, contributing to their higher affinities. More expansive neutralization studies (180 isolate panel) revealed potent activity against all except those from Clade A/E. The presence of His at position 375 of gp120 of these isolates (instead of the canonical Ser) is interpreted to explain this resistance, since the His partially fills the Phe43 pocket thereby hindering access of the extensions (but not CD4 or the mimetics lacking the extensions). This is consistent with previous findings that resistance to M48U1 involves substitution of Ser375 with more bulky residues. The results are discussed in terms of a new mechanism of action of CD4 binding site ligands beyond the previously described avidity (multivalent forms) and avoidance of conformational change (e.g. mAb VRC01), namely optimization of fitting of the hydrophobic extensions within the interfacial Phe43 cavity.
These new agents and the structural elucidation of the mechanisms underlying their enhanced anti-HIV potencies promise to guide further design of novel neutralizing agents based on optimal fitting of extensions into the Phe43 cavity. The recent ex vivo and nonhuman primate studies with M48U1 as a vaginal microbicide to prevent HIV-1 sexual transmission (Dereuddre-Bosquet et al., 2012) are highly promising for the potential antiviral use of these miniproteins. Structure-guided design and analysis thus continue to pave the way for development of new CD4 mimetics that not only cap the Phe43 cavity, but also fit energetically favorable extensions deep within its boundaries.
Acknowledgements
Work in the authors’ laboratory is supported in part by the Intramural Program of the NIH, NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Curreli F, Choudhury S, Pyatkin I, Zagorodnikov VP, Bulay AK, Altieri A, Do Kwon Y, Kwong PD, Debnath AK. J Med Chem. 2012;55:4764–4775. doi: 10.1021/jm3002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereuddre-Bosquet N, Morellato-Castillo L, Brouwers J, Augustijns P, Bouchemal K, Ponchel G, Ramos OHP, Herrera C, Stefanidou M, Shattock R, et al. Plos Pathogens. 2012;8 doi: 10.1371/journal.ppat.1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, Li YX, Sodroski J, Kwong PD, Mascola JR, et al. Journal of Virology. 2007;81:5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, et al. Proc Natl Acad Sci USA. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N, Schon A, Princiotto AM, LaLonde JM, Courter JR, Soeta T, Ng D, Wang LP, Brower ET, Xiang SH, et al. Structure. 2008;16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, et al. Nature Biotechnology. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Van Herrewege Y, Morellato L, Descours A, Aerts L, Michiels J, Heyndrickx L, Martin L, Vanham G. Journal of Antimicrobial Chemotherapy. 2008;61:818–826. doi: 10.1093/jac/dkn042. [DOI] [PubMed] [Google Scholar]
- Vita C, Vizzavona J, Drakopoulou E, Zinn-Justin S, Gilquin B, Menez A. Biopolymers. 1998;47:93–100. doi: 10.1002/(SICI)1097-0282(1998)47:1<93::AID-BIP10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang LP, Hendrickson WA, Doyle ML, Sodroski J. Journal of Virology. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]