Abstract
OBJECTIVE
The purpose of this study was to develop and characterize a human antibody in a single-chain antibody fragment format (scFv) that is directed specifically against claudin-3 (CLDN3).
STUDY DESIGN
The synthetic ETH-2 Gold human antibody phage display library was used to select scFv specific against CLDN3. scFv binding properties were analyzed by surface plasmon resonance; specificity was confirmed with enzyme-linked immunosorbent assay, immunofluorescence, and flow cytometry on a panel of ovarian and uterine serous carcinoma cell lines.
RESULTS
Surface plasmon resonance studies indicated scFv H6 to be the clone with the highest affinity against CLDN3 (KD of 23.60 nmol/L). scFv H6 efficiently stained CLDN3-expressing cells and recognized its epitope in enzyme-linked immunosorbent assay that was performed with uterine serous papillary carcinoma native protein extract, which suggested that a conformational epitope is recognized by this antibody. Cell surface immunofluorescence with laser scanning confocal microscopy confirmed the specific binding to the native membrane CLDN3.
CONCLUSION
scFv H6 may represent a novel antitumor agent against chemotherapy-resistant ovarian and serous papillary carcinomas and other human malignancies that overexpress CLDN3.
Keywords: claudin-3, ovarian carcinoma, phage display library, scFv, uterine serous tumor
Claudins represent a large family of integral membrane proteins that are located within the tight junctions of epithelia and endothelia. Acting as a diffusion barrier to movement of proteins and lipids within the plasma membrane, tight junctions play a critical role in the establishment and the maintenance of cell polarity.1 Alterations of these functions are involved deeply in cancer cell biology, and the loss of tight junction integrity may play an important role in the loss of cohesion, invasiveness, and lack of differentiation that are observed typically in tumor cells.2–4 Several studies have reported recently that specific claudin family members are overexpressed in a wide variety of cancer types5; in particular, claudin-3 and -4 have been reported to be expressed at high levels in epithelial ovarian cancer of all subtypes and in breast, prostate, and pancreatic tumors.6,7 Both claudin-3 and -4 recently have also been reported to be highly differentially expressed in uterine serous papillary carcinoma (USPC), which is an aggressive form of endometrial cancer.8
Claudin-3 is a tetraspan protein with N and C termini that are located in the cytoplasm and 4 hydrophobic regions and with 2 predicted extracellular loops9 that offer promising targets for antibody-based therapy. Claudins are involved critically in the organization of epithelial tissue architecture and interact in both homotypic and heterotypic ways7,10; thus, they may be barely accessible to antibodies in well-structured epithelia. In contrast, in tumors, claudins do not form classic tight junctions as found in normal epithelial but become exposed on tumor cells as “free” claudins that are amenable to extracellular antibody binding, which makes them potential candidates for cancer immunotherapy.
A variety of claudin-specific antibodies that are available commercially all recognize the intracellular C-terminal part of claudins, which makes them useful reagents for the detection of claudin expression in fixed or denatured tumor tissue but not on intact cells in situ or in vivo. Moreover, although not suitable for the development of human therapeutics, polyclonal chicken antibodies that specifically recognize extracellular claudin loops and selectively bind to the cell surface of intact cells have been developed recently.11 Phage antibody display is 1 of the best alternatives for the production of recombinant human antibodies in single-chain fragment variable (scFv) format for research, clinical, and therapeutic applications. This is specifically true when dealing with the development of antibodies or fraction of antibodies that is directed against targets that are endowed with high homologic findings among different species, such as claudins.11 scFv fragments, which consist of only 1 heavy-chain variable domain and 1 light-chain variable domain that is linked covalently by a short peptide linker, contain the complete antigen-binding site of an antibody, with the same potential monomeric binding affinity as the parental monoclonal antibody.12 In comparison with murine monoclonal antibodies that are produced by the classic hybridoma techniques, human antibodies that were obtained by selection with the antibody phage display library against target antigens did not induce harmful immune response in patients and maintained an intact binding site and exhibited rapid tumor penetration and rapid systemic clearance.13
This article describes the selection from an ETH-2 synthetic phage display library14 of a new scFv format against the second extracellular loop of claudin-3 (2CL3) that selectively binds to the cell surface of intact tumor cells that overexpress claudin-3. Several antibody fragments have been selected from the library; 1 anti-2CL3 scFv format was chosen for further characterization because of its consistent reactivity to claudin-3-expressing human cancer cell lines. The characteristics of this scFv format make it an ideal candidate to be tested for in vivo tumor targeting.
Materials and Methods
Bacterial strains
Bacterial Escherichia coli suppressor strain TG1 (supE, hsd Δ5, thi Δ [lac-proAB] F′[traD36 proAB+lacIq lacZΔM15]) and HB2151 nonsupressor strain (nalr thi-1 ara Δ [lac-proAB] F′ [proAB+lacIq lacZΔM15]) were used for phage antibody production and large-scale soluble scFv production.
2CL3 peptide
A predictive model of claudin protein, with 4 transmembrane segments with 2 extracellular loops and N and C termii that are located in the cytoplasm has been proposed.13 One peptide (2CL3) of 31 amino acids (PVSWSANTIIRDFYN-PVVPEAQKREMGAGLY) that corresponds to the second loop of claudin-311 was chosen for biopanning and generation of an scFv format. In vitro synthesis and N-biotinylation of 2CL3 peptide was performed at Alpha Diagnostics International (San Antonio, TX).
Cell lines
Three primary ovarian serous papillary carcinoma (OSPC) and 2 USPC cell lines were used in this work; these cell lines have been characterized previously and tested positive for claudin-3 expression by quantitative reverse transcriptase-polymerase chain reaction.8,15–17 Briefly, OSPC-1 was established from a metastatic tumor sample; OSPC-2 and OSPC-3 were established from primary OSPC specimens. Similarly, USPC-2 was obtained from a primary endometrial site, and USPC-4 was established from an intraabdominal metastatic site. Tumor cell lines were cultured in RPMI 164 medium (Invitrogen, Grand Island, NY) that was supplemented with 10% fetal bovine serum, 200 μg/mL penicillin, and 200 μg/mL streptomycin at 37°C, 5% CO2. Peripheral blood lymphocytes (PBL) that were obtained from healthy donors were evaluated as control cell lines.
scFv format selection by ETH-2 Gold phage display library
The ETH-2 Gold library, a modified version of the antibody library of Pini et al,18 is a synthetic human recombinant antibody library that consists of > 109 possible antibody combinations in an scFv format that are displayed on the surface of filamentous phage.18 Selection of scFv format from the ETH-2 library was directed against biotinylated 2CL3 peptide. An aliquot of ETH-2 library that contained approximately 1012 transducing units was incubated with 3 mL of 50 nmol/L biotinylated 2CL3 in phosphate-buffered saline solution (PBS) 6% milk for 1 hour at room temperature with gentle shaking. The bound phage were captured with 100 μL of streptavidin-coated M-280 Dynabeads (Dynal Bio-tech ASA, Oslo, Norway) that were previously blocked 1 hour in PBS 4% milk, separated with a magnetic device (MPC-1; Dynal Biotech ASA), and washed 10 times with PBS-T (0.05% Tween 20 in PBS) and 10 times with PBS. Bound phages were eluted with 1 mL of 100 mmol/L triethylamine; the solution was neutralized immediately by the addition of 0.5 mL of 1 mol/L tris-HCl pH 7.4. Eluted phages were used to infect exponentially growing E coli TG1 cells and were amplified for the next round of panning. Five rounds of panning were carried out in solution against biotinylated peptide 2CL3. Phages from the fifth selection were used to infect exponentially growing TG1 and HB2151 cells, which should favor the production of soluble antibody fragments. scFv expression was induced from single colonies by isopropyl-beta-D-thiogalactopyranoside (1 mmol/L final concentration), and the supernatants that contained soluble scFv fragments were recovered and tested for specific 2CL3 recognition.
Screening of supernatants by enzyme-linked immunosorbent assay (ELISA)
Supernatants of individual colonies that had been induced for scFv expression were screened for binding to biotinylated 2CL3 by ELISA, essentially as described by Viti et al.19 StrepMax streptavidin-coated plates (ABgene, Surrey, UK) were coated with 1 μg/well of biotinylated 2CL3 in PBS, and bound scFv fragments were detected by means of anti-myc tag antibody 9E10 (Roche Diagnostics, Indianapolis, IN) followed by an antimouse horseradish peroxidase (HRP)-conjugated antibody (Sigma Chemical Company, St. Louis, MO). The immunoreaction was developed with 3,31–5,51-tetramethylbenzidine (soluble BM blue POD substrate; Roche Diagnostics) and stopped by adding 1 mol/L sulfuric acid. Results were expressed after the absorbance at 450 nm was considered. The most reactive clones were chosen for further characterization.
Sequencing of scFv antibody genes
Nucleic acid sequencing of scFv antibody genes was carried out on a genetic analyzer (ABI Prism 3130; Applied Bio-systems, Foster City, CA) with the Big Dye Terminator Cycle Sequencing Kit (version 1.1; Applied Biosystems). Templates for termination reactions were DNA miniprep. The primer that was used for sequencing was fdseq long (5′-GACGTTAGTAAATGAATTTTCTG-TATGAGG-3′) annealing downstream of the scFv gene.
Soluble scFv purification
scFv from selected bacterial clones were purified by affinity chromatography with Protein A Sepharose Fast Flow resin (Amersham Biosciences AB, Uppsala, Sweden), according to the manufacturer’s instructions. The resin was packed into a chromatography column (Bio-Rad Laboratories, Hercules, CA); antibody supernatant was loaded onto the column, and the resin was washed with PBS, 100 mmol/L NaCl, 0.1% Tween 20, 0.5 mmol/L ethylenediaminetetraacetic acid (EDTA), followed by washing with PBS, 500 mmol/L NaCl, 0.5 mol/L EDTA. When buffer OD280 was washed after the column was < 0.05, antibody was eluted in 0.7 mL fractions with 100 mmol/L triethylamine. Immediately after elution, pH was neutralized by adding 0.3 mL 1 mol/L Tris-HCl pH 7.4 to each fraction. Purity of every fraction was evaluated by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fractions with the highest protein concentration were pooled, dialyzed against PBS overnight at 4°C, and stored at −80°C in small aliquots. Specificity of purified scFv fragments for 2CL3 was determined by ELISA (as described earlier) with biotinylated 2CL3 for well coating and scFv as primary antibodies.
Surface plasmon resonance
Purified scFv binding properties were analyzed by surface plasmon resonance with a BIAcore X instrument (BIAcore AB, Uppsala, Sweden). Antigen-coated chips were prepared according to the manufacturer’s instructions by injecting 270 nmol/L biotinylated 2CL3 onto a streptavidin-coated sensor chip (Sensor Chip SA; BIAcore AB). Sensorgrams for kinetic measurements were generated by the injection of soluble scFv (at 4 concentrations that ranged from 25–300 nmol/L) in HBS-EP buffer (BIAcore AB 10 mM Hepes, 150 mM NaCl, 3 mM EDTA, 0.005% Tween-20;). Kinetic data were collected, modeled, and fitted for each antibody concentration by means of BIAevaluation software (version 3.1; BIAcore AB).
ELISA on cell lysate
Whole cell lysate was prepared by ice-cold lysis buffer-extraction of 1 × 106 USPC-4 cells and PBL. For membrane cell lysate, 1 × 106 USPC-4 cells were washed and scraped in ice-cold PBS, pelleted, and incubated in hypotonic buffer (100 mmol/L Tris-HCl pH 7.4, 1 mmol/L EDTA, 1 mmol/L ethylene glycol tetraacetic acid) for 10 minutes on ice. After centrifugation at 2000 rpm for 10 minutes, the small pellet that contained the membrane fraction was resuspended in 70 μL of STEN buffer (2 mmol/L EDTA, 150 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.6, 0.05% NP-40), incubated for 1 hour on ice, and centrifuged to recover supernatant, which was enriched with membrane proteins. The protein concentrations in whole extract and in membrane extract were measured with the Bradford method with the use of the Bio-Rad protein assay (Bio-Rad Laboratories). ELISA was performed on 96-well plates that were incubated at 4°C with 7 μg/well of cell extracts in PBS. Soluble scFv in blocking buffer at a final concentration of 400 ng/well was added to each well and incubated for 1 hour at room temperature. After being washed, a 1:1500 dilution of anti-myc tag antibody 9E10 (Roche Diagnostics, Indianapolis, IN) in blocking buffer was added for 1 hour at room temperature, followed by a 1:1000 dilution of an antimouse HRP-conjugated antibody (Sigma Chemical Company). The immunoreaction was developed with 3,31–5,51-tetramethylbenzidine (soluble BM blue POD substrate; Roche Diagnostics), stopped by the addition of 1 mol/L sulfuric acid, and read with an ELISA reader.
Fluorescence-activated cell sorting (FACS) analysis
FACS analysis was performed with scFv against a panel of human cell lines that highly overexpress claudin-3 and against normal control cells that were negative for claudin-3 expression. Briefly, approximately 2.5 × 105 cells were resus-pended with 50 μL PBS 1% bovine serum albumin (BSA) that contained 4 μg of primary scFv antibody and were incubated for 1 hour at room temperature. After being washed, cells were resus-pended with 100 μL PBS 1% BSA solution that contained 0.7 μg of an anti-myc tag antibody 9E10 (Roche Diagnostics) for 1 hour at 4°C, followed by washing and incubation with 2 μg of fluorescin isothiocyanate (FITC)-conjugated goat antimouse immunoglobulin G (Jackson Immuno Research, West Grove, PA) for 30 minutes at 4°C. Finally, cells were washed with PBS, resuspended in 500 μL PBS, and immediately analyzed by FAC-Scan (BD Biosciences, San Diego, CA).
Cell surface immunofluorescence
USPC-4 cells and control PBL that were obtained from healthy volunteers by density gradient centrifugation (Lympholyte Cell Separation Media; Cedarlane Laboratories Limited, Burlington, NC) were washed and aliquoted. The 2 × 105 cells were incubated directly with 0.5 μg/mL of purified antibody that had been diluted in 100 μL of PBS 1% BSA for 1 hour. Cells were then resuspended with 100 μL PBS 1% BSA solution that contained 0.7 μg of an anti-myc tag antibody 9E10 (Roche Diagnostics) for 1 hour, followed by incubation with Alexa Fluor 568-labeled goat antimouse immunoglobulin G (H + L) antibody (Invitrogen, Grand island, NY) that was diluted 1:200 in 100 μL PBS 1% BSA for 30 minutes. After being stained, cells were fixed with 2% paraformaldehyde in PBS for 30 minutes on ice, washed, and absorbed to polylysine-coated cover slips. Cells were embedded in mounting medium (DAKO Corp, Carpinteria, CA) and analyzed with the confocal microscopy (FV 200 Olympus Fluoview confocal laser-scanning microscope; Olympus Corporation, Tokyo, Japan). For immunofluorescence with adherent cells, USPC-4 cells were grown for 48 hours on sterile chamber slides (Lab-Tek II Chamber Slide System; Nalge Nunc International, Rochester, NY). After the medium was discarded, cells were incubated directly with 0.5 μg/mL of purified antibody that had been diluted in 100 μL of PBS 1% BSA and processed as described earlier for cells in suspension. After being stained, cells were fixed with 2% paraformaldehyde in PBS for 30 minutes on ice, washed, embedded in mounting medium, and analyzed with the confocal microscopy. For control images, cells were counterstained with phalloidin FITC-labeled (Sigma Chemical Company) before being fixed with paraformaldehyde. After being stained with Alexa Fluor 568-labeled goat anti-mouse, cells were incubated with 0.2 U/well of phalloidin FITC-labeled for 10 minutes to allow the dye to bind filamentous actin. Then cells were washed with PBS and fixed as described earlier.
RESULTS
Selection of 2CL3-specific scFv antibodies was accomplished with the use of the ETH-2 Gold phage display library.
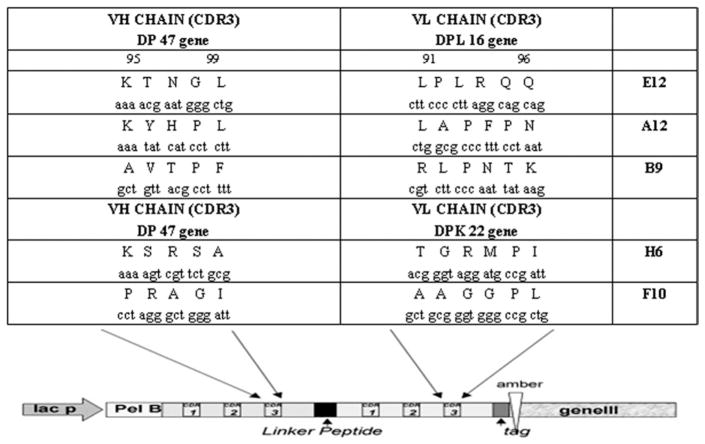
Bacterial supernatants that were obtained from 180 individual colonies of TG1 E coli strain and from 90 individual colonies of HB2151 E coli strain that were infected with phages from the fifth round of panning were used to detect binding to biotinylated 2CL3 in a primary ELISA screening, as described.19 Sixty-three of 180 scFv (35%) that expressed TG1 E coli supernatants and 17 of 90 scFv (20%) that expressed HB2151 E coli supernatants that were examined were found to react specifically with the antigen. All ELISA positive clones were probed for binding to streptavidin in a further ELISA, and no cross-reactivity was observed (data not shown). Thirty-six clones that gave a strong ELISA signal (OD450 > 0.4, color development after 10 minutes) were analyzed by sequencing. According to the complementarity-determining region 3 (CDR3) sequences of the variable light (VL) and variable heavy (VH) chains, the clones were subgrouped into 5 different groups with identical sequence, which was represented by scFv HB2151 (H6), scFv TG1 (F10), scFv TG1 (A12), scFv HB2151 (E12), and scFv HB2151 (B9). Both the human germline genes that were used to construct the light chain variable region of the library were represented: the scFv HB2151 (H6) and scFv TG1 (F10) VL chains were derived from the DPK-22 germline gene, whereas the scFv TG1 (A12), scFv HB2151 (E12), and scFv HB2151 (B9) VL chains were derived from the DPL-16 germline gene (Figure 1).
FIGURE 1. CDR3 sequences of selected scFv.
A schematic representation of the single-chain antibody fragment format (scFv) antibody gene as M13 pIII fusion protein is also illustrated.
CDR3, complementarity-determining region 3.
Romani. Development of a human scFv against claudin-3. Am J Obstet Gynecol 2009.
Biochemical characterization of the selected scFv antibodies
For scFv purification, the 5 representative antibody fragments were cultured for large-scale production and purified by affinity chromatography from bacterial supernatant. scFv H6, E12, and B9 were expressed with the nonsuppressor host HB2151 E coli; scFv F10 and A12 were expressed with the TG1 E coli strain. All scFv fragments were expressed in soluble form in the supernatant, without significant differences between the bacterial strains, and purified with protein A. The fragments all showed the expected molecular masses, and their purity was > 90%, as confirmed by SDS-PAGE analysis (data not shown). Maximum yield of the purified products ranged from 75–80 μg soluble antibody/liter of culture.
All 5 purified scFv specifically recognize 2CL3 peptide, as detected in ELISA. To evaluate their binding activity, ELISA was carried out with the 5 scFv fragments at different quantities (25, 50, 100, 200, 400, 800, and 1600 ng of pure scFv/well) against biotinylated 2CL3. All the antibodies showed dose-dependent reactivity (Figure 2); scFv H6 was the most reactive, even at low concentrations compared with other scFv fragments. As discerned from Figure 2, F10 clone seems to be unreactive at all the concentrations that were tested and was not considered. Therefore, the remaining 4 clones were selected for further analysis.
FIGURE 2. Reactivity of scFv antibodies against 2CL3 peptide.
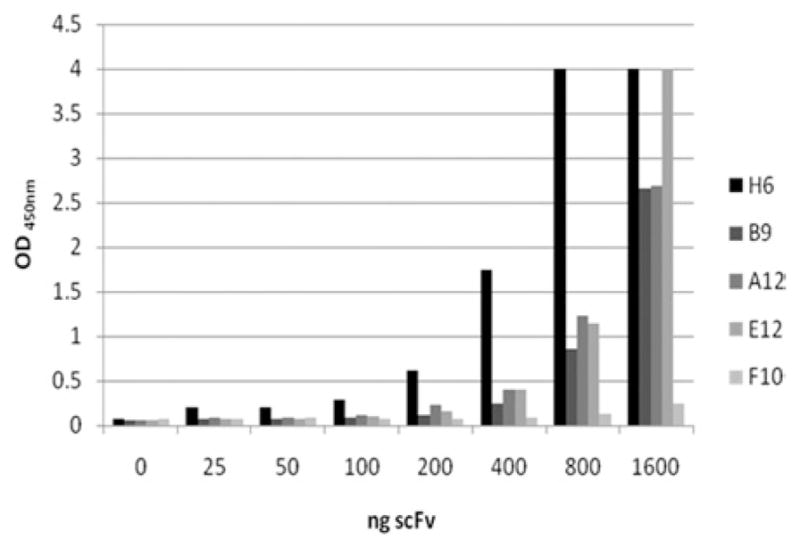
Histogram show the results of the enzyme-linked immunosorbent assay that was carried out with the biotinylated second extracellular loop of claudin-3 (2CL3) as antigen and the different single-chain antibody fragment format (scFv) as primary antibodies at the indicated quantities (25, 50, 100, 200, 400, 800, 1600 ng/well).
OD, optical density.
Romani. Development of a human scFv against claudin-3.
Am J Obstet Gynecol 2009.
2CL3 peptide binding affinity of purified scFv antibodies
Surface plasmon resonance studies provided quantitative measures of scFv-antigen binding and dissociation kinetics. Purified scFv were used for the determination of the affinity constants on biotinylated 2CL3-coated chip with a BIAcore X instrument (BIAcore AB). The results for Kon, Koff, and KD determinations are shown in the Table. All clones exhibited different binding profiles; the affinities ranged from 1.27 mmol/L to 23.60 nmol/L, which indicated a poor to moderately high affinity. The 2 clones with the slowest dissociation constant were scFv H6 with KD of 23.60 nmol/L and scFv B9 with KD of 76.00 nmol/L. Unexpectedly, scFv A12 and E12 clones, which showed high reactivity in ELISA, bound to 2CL3 with affinity in the micromolar-millimolar range because of a fast dissociation and a slow association constant, respectively. Given the low affinity of the A12 and E12 clones, they were discarded for further analysis. scFv H6 and B9 were identified as the best clones and were then evaluated in more detail.
TABLE.
Kinetic values of selected single-chain antibody fragments
| Clone | Kon (M-1s-1) | Koff (s-1) | KD (mol/L) |
|---|---|---|---|
| H6 | 2.63E + 04 | 6.23E − 04 | 23.60 nmol/L |
| B9 | 2.62E + 05 | 2.00E − 02 | 76.00 nmol/L |
| A12 | 1.04E + 04 | 1.65E − 02 | 1.59 μmol/L |
| E12 | 3.86E + 01 | 4.92E − 03 | 1.27 mmol/L |
Rate constants were deduced from the BIAcore curves; the dissociation constant KD was calculated as Koff divided by Kon. E, exponential function; M-1s-1, unit of measure of the association constant Kon; s-1, unit of measure of the dissociation constant Koff.
Romani. Development of a human scFv against claudin-3. Am J Obstet Gynecol 2009.
Binding specificity of the anti-2CL3 scFv fragments
The 2CL3 specificity of selected antibody clones was verified in an ELISA assay, in parallel with irrelevant proteins (Figure 3). Both scFv H6 and B9 exhibited highly selective binding properties to the desired antigen, because no meaningful cross-reaction was observed with the heterologous antigens (Figure 3). Indeed, the scFv reactivity was 17–40 times lower than reactivity against the specific antigen 2CL3. scFv H6 was 2.5 times more reactive than scFv B9 at all the concentrations that were tested. ELISA on native cell extracts (Figure 4) indicates that scFv H6 recognizes the 2CL3 epitope in USPC-4 native cellular extract, which suggests that not only the linear, but also the conformational epitope, is recognized by the antibody. Because transmembrane proteins such as claudin-3 can be difficult to visualize because of their relative low abundance in total cell lysates and their hydrophobic nature, we enriched the membrane-bound protein fraction that was extracted from cell culture in some experiments. Consistent with this view and as shown in Figure 4, the intensity of the binding of scFv H6 on a membrane extract was significantly higher than the value that was obtained on total extract. No unspecific binding was observed on PBL, which are reported to express low levels of claudin-3 (Figure 4).7,8 Negative control without primary antibody shows no reaction.
FIGURE 3. Specificity of scFv H6 and B9 for 2CL3 peptide.
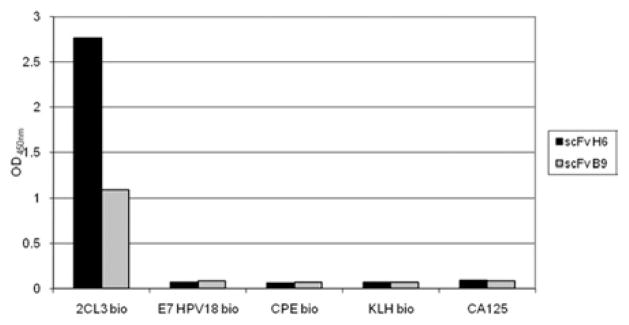
Enzyme-linked immunosorbent assay signal of scFv H6 and B9 on heterologous antigen shows that 2CL3 is the only antigen that was recognized.
CA125, recombinant CA125 protein; CPE bio, biotinylated recombinant Clostridium perfringens enterotoxin A full-length native protein; E7 HPV18 bio, biotinylated recombinant E7 oncoprotein of human papilloma virus type 18; KLH bio, biotinylated recombinant keyhole limpet hemocyanin protein; OD, optical density.
Romani. Development of a human scFv against claudin-3. Am J Obstet Gynecol 2009.
FIGURE 4. Cell lysate enzyme-linked immunosorbent assay.
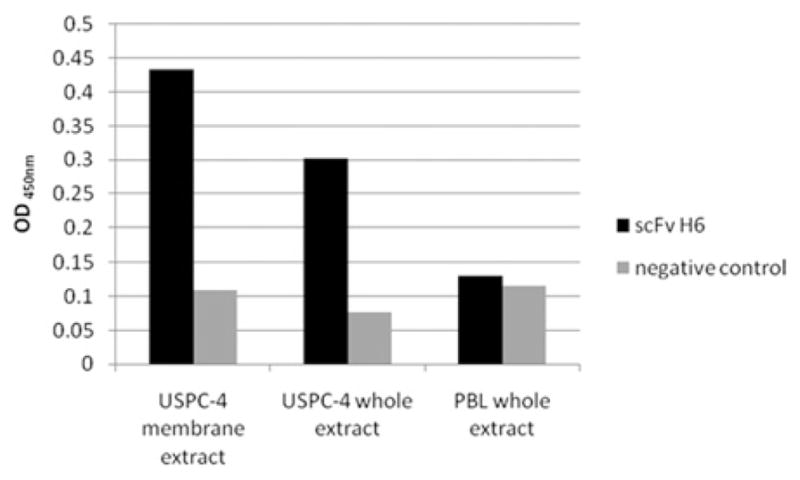
Binding of single-chain antibody fragment format (scFv) H6 to whole cell extracts and membrane cell extract of uterine serous papillary carcinoma (USPC)-4 primary cell line and peripheral blood lymphocytes (PBL). Negative control with secondary antibodies anti-myc tag 9E10 and antimouse horseradish peroxidase conjugated is also shown.
OD, optical density.
Romani. Development of a human scFv against claudin-3.
Am J Obstet Gynecol 2009.
Reactivity of scFv to CLDN3-expressing cells
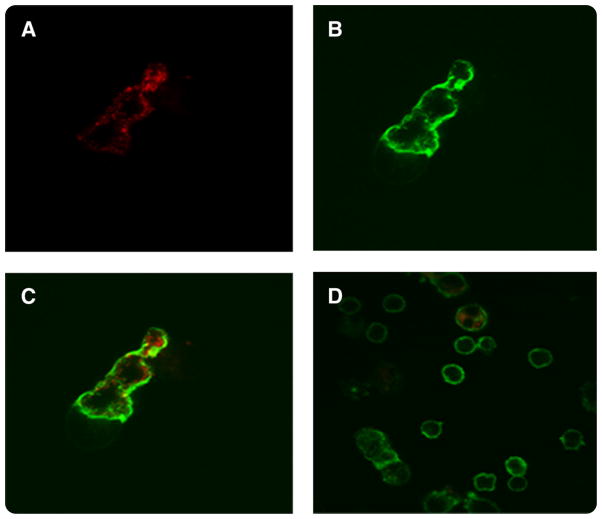
The binding of scFvs to native 2CL3 epitope of CLDN3-expressing human cell lines was assessed by flow cytometry with soluble purified scFv. scFv H6 and B9 were tested for their ability to recognize the 2CL3 epitope on a panel of un-fixed human cell that included 3 ovary cancer cell lines (OSPC-1, OSPC-2, OSPC-3), 2 endometrial cancer cell lines (USPC-2, USPC-4), and PBL that had been obtained from healthy donors. Although scFv H6 was found to be reactive with all cell lines, albeit at varying intensities, likely because of a different accessibility to the epitope by the scFv (Figure 5), no significant binding was found for scFv B9. Importantly, no scFv H6 binding was evident on PBL that expressed claudin-3 at a low level (Figure 6). The surface binding of 2CL3-specific scFv was validated further by immunofluorescence. Unfixed USPC-4 cells in suspension were incubated with scFv H6; binding to cell surface-expressed claudin-3 was analyzed with a laser scanning confocal microscope. As shown in Figure 7, binding of scFv H6 to the cellular surface of tumor cells was consistent. The analysis of the scFv H6-stained cells revealed a cell surface-specific staining pattern, with claudin-3 found distributed as focal and localized spots on the cell surface. Positive control of membrane staining with phalloidin shows the same binding pattern. Figure 7, D shows control images with PBL in suspension that were double stained with scFv H6 and phalloidin. No unspecific binding of scFv H6 to the cells was detected.
FIGURE 5. Binding profiles of scFv H6 and B9 to claudin-3 expressing human cell lines.
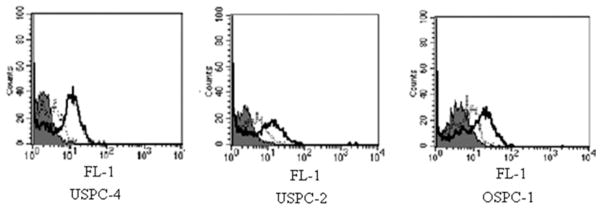
Shown is the binding of single-chain antibody fragment format (scFv) H6 (bold line) and scFv B9 (dashed line) to uterine serous papillary carcinoma (USPC)-2 and -4 and ovarian serous papillary carcinoma (OSPC)-1 cell lines. Histograms for negative control with secondary antibodies anti-myc 9E10 and antimouse-fluorescin isothiocyanate are shown in solid gray.
FL-1, green channel in which the fluorescin isothiocyanate fluorochrome is detected.
Romani. Development of a human scFv against claudin-3. Am J Obstet Gynecol 2009.
FIGURE 6. Reactivity of scFv H6 with human cell lines.
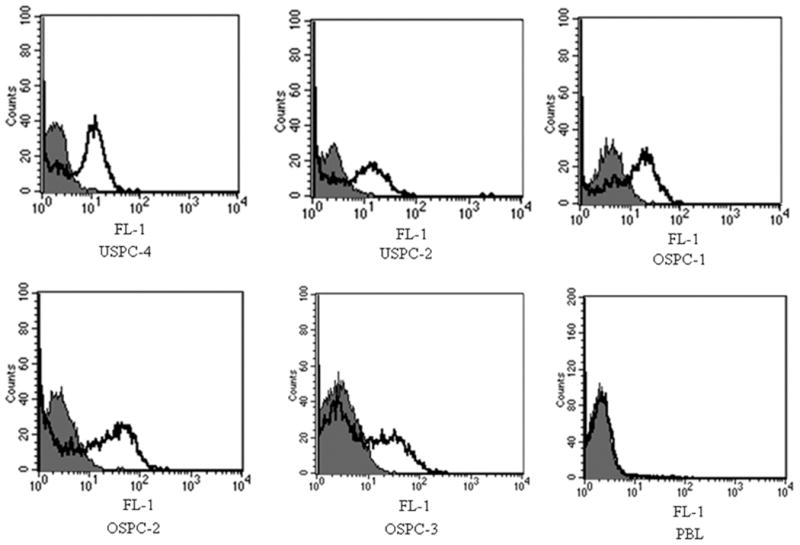
Fluorescence-activated cell sorting analysis of single-chain antibody fragment format (scFv) H6 binding to unfixed human endometrial cancer lines (uterine serous papillary carcinoma [USPC-2, -4]) and ovary cancer lines (OSPC-1, OSPC-2, OSPC-3) that express claudin-3. Human peripheral blood lymphocytes (PBL) were used as negative controls. Bold lines indicate the level of reactivity of single-chain antibody fragment format H6; solid gray histograms represent background binding level of the secondary antibodies anti-myc 9E10 and antimouse-fluorescin isothiocyanate without the primary antibody.
FL-1, green channel in which the fluoroscin isothiocyanate fluorochrome is detected.
Romani. Development of a human scFv against claudin-3. Am J Obstet Gynecol 2009.
FIGURE 7. Immunofluorescence of scFv H6 with USPC-4 cells and PBL analyzed by laser scanning confocal microscopy.
Representative micrographs of single-chain antibody fragment format (scFv) membrane staining (red) and phalloidin fluorescin isothiocyanate membrane staining (green) of unfixed cells in suspension. A, Labeling of uterine serous papillary carcinoma-4 (USPC-4) cells with scFv H6 and Alexa Fluor 568-labeled goat antimouse. B, Labeling of USPC-4 cells with phalloidin fluorescin isothiocyanate. C, Double immunofluorescence staining of USPC-4 with scFv H6 and phalloidin fluorescin isothiocyanate labeled. D, Double immunofluorescence staining of peripheral blood lymphocytes (PBL) with scFv H6 and phalloidin fluorescin isothiocyanate labeled.
Romani. Development of a human scFv against claudin-3. Am J Obstet Gynecol 2009.
Comment
The identification and characterization of tumor-specific and/or differentially expressed markers remains a major priority for the development of novel target cancer therapies against ovarian and biologically aggressive endometrial carcinomas. Of particular interest among the spectrum of overexpressed molecules are those that are located at the cell surface, because they are readily accessible and can be used to target cancer cells with highly specific ligands, such as monoclonal antibodies. One of the major limitations in the use of monoclonal antibodies in human tumor therapy is the accessibility of antibodies to tumor cells at advanced stages of cancer, because they are generally too large to penetrate sizable tumor masses.20 Moreover, the use of murine antibodies in humans is limited because of their high immunogenicity and consequent induction of human antimouse antibody responses, which can severely limit the efficacy of antibody therapy. In contrast, antibody phage display technology is a strategy that can be used to isolate tumor-specific antibodies in an scFv format, which is able to bind their cognate antigens in the cellular context, for diagnostic or therapeutic uses in vivo.21 The scFv fragment, in which the variable domain of the heavy and light chains is joined by a peptide link, is the smallest fragment that still contains the intact binding site. Because of their human origin, antibodies that are isolated from phage display human antibody libraries can be used directly without concern about their immunogenicity in vivo. In addition, their small size may increase dramatically their capacity to penetrate solid tumor masses, even in areas far from blood vessels.22
We report for the first time the generation of a recombinant completely human antibody fragment that is specific to the 2CL3, a membrane protein that is crucial for tight junction formation and function and found highly differentially expressed in several human cancer lines, including ovarian and uterine serous tumors.5 Although their functional role in cancer progression remains unclear, the differential expression of claudin proteins between tumor and normal cells, in addition to their membrane localization, makes them potential candidates for target cancer therapy. In this study, antibodies were isolated from the ETH-2 Gold synthetic phage display library,14 which is a human recombinant antibody library that consists of > 109 possible antibody combinations in an scFv format that are displayed on the surface of filamentous phage. The sequencing of selected scFvs demonstrated that several antibodies had identical sequences and could be grouped into 5 classes. Members from each group were chosen and evaluated in more detail. The biophysical properties and the overall stability of scFv fragments depend on the primary structure and on the combination of VH and VL in the scFv format.23 Among the selected antibodies that specifically recognized claudin-3, scFv H6 exhibited the combination of DPK22 VL with DP47 VH, which is considered to be 1 of the most stable conformations.24–26
The ability of the antibody to bind 2CL3 was determined with standard binding assays, such as ELISA, immunofluorescence, and flow cytometric analysis. The scFv H6 clone efficiently stained claudin-3-expressing cells and recognized its epitope in ELISA that was performed with USPC-4 native protein extract. FACS analysis and cell surface immunofluorescence of USPC-4 cells with laser scanning confocal microscopy confirmed the specific binding to the native membrane claudin-3. BIAcore analysis (BIAcore AB) was used for the determination of the kinetic constant of the scFv binding to the antigen. The affinities (KD) of the selected scFvs oscillated between 1.27 mmol/L and 23.60 nmol/L.
Because most purified scFv fragments are reported to be a mixture of monomers and dimers, with different sizes and pharmacokinetics,27 these data should be confirmed on monomeric fraction to better understand the real binding properties of this scFv in the targeting of tumors. However, the analysis of purified scFv H6 that was performed by native PAGE indicated the it does not spontaneously form dimers, because most of it appeared in monomeric form (data not shown), which makes us quite confident that the KD value is close to the real value.
Compared with the other clones, scFv H6 shows the slowest off rate, which is typically the major kinetic mechanism that results in higher affinity.28 The binding affinities of this scFv are in a range for efficient in vivo antigen binding; recent reports have shown that affinities beyond 10 nmol/L do not significantly increase tumor retention of scFvs.29,30 Moreover, the moderate affinity of the anti-2CL3 scFv fragment against its target antigen might turn out to be a crucial asset for its safety and potential activity in vivo, because of the known expression of the claudin-3 receptor not only in malignant tissues but also in some normal tissues, which includes the gut, lungs, and kidney.31
In conclusion, we have developed and characterized the first completely human single-chain antibody using a synthetic phage display library, which specifically targets claudin-3. This new immunoreagent meets all criteria for a potential anticancer compound; it is human, hence, poorly immunogenic, and it binds specifically and with good affinity to the extracellular domain of claudin-3, which is a receptor that is expressed highly and differentially on the surface of multiple biologically aggressive human cancers, including ovarian and USPC. Moreover, its small molecular size should provide for efficient tissue penetration, yet give rapid plasma clearance. Taken together, these results warrant further evaluation of scFv H6 as a potential novel antitumor agent against ovarian and uterine serous tumors refractory to standard treatment modalities.
Acknowledgments
This study was supported in part by Grants from the Angelo Nocivelli, Berlucchi, and Camillo Golgi Foundations, Brescia, Italy; NIH R01 CA122728-01A2 to A.D.S.; and Grants 501/A3/3 and 0027557 from the Italian Institute of Health (A.D.S.); and by NIH Research Grant CA-16359 from the National Cancer Institute. 0002-9378/$36.00
We thank Dr Gaetana Lanzi for the confocal microscopy analysis of the data.
References
- 1.Van Itallie CM, Anderson JM. The molecular physiology of tight junction pores. Physiology. 2004;19:331–8. doi: 10.1152/physiol.00027.2004. [DOI] [PubMed] [Google Scholar]
- 2.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–6. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 3.Morita K, Furuse M, Fujimoto K, et al. Claudin multi gene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–6. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukita S, Furuse M, Itho M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–93. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 5.Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57:919–28. doi: 10.1016/j.addr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Rangel LBA, Argarwal R, D’Souza T, et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenoma. Clin Cancer Res. 2003;9:2567–75. [PubMed] [Google Scholar]
- 7.Hewitt KJ, Argarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santin AD, Zhan F, Cané S, et al. Gene expression fingerprint of uterine serous papillary carcinoma: identification of novel molecular markers for uterine serous cancer diagnosis and therapy. Br J Cancer. 2005;92:1561–73. doi: 10.1038/sj.bjc.6602480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukita S, Furuse M. Claudin-based barrier in simple and stratified cellular sheets. Curr Opin Cell Biol. 2002;14:531–6. doi: 10.1016/s0955-0674(02)00362-9. [DOI] [PubMed] [Google Scholar]
- 10.Tsukita S, Furuse M. Pores in the wall: claudins constitute tight junction strands containing aqueous pores. J Cell Biol. 2000;149:13–6. doi: 10.1083/jcb.149.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Offner S, Hekele A, Teichmann U, et al. Epithelial tight junction proteins as potential antibody targets for pancarcinoma therapy. Cancer Immunol Immunother. 2005;54:431–45. doi: 10.1007/s00262-004-0613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird RE, Hardmann KD, Jacobson J, et al. Single-chain antigen-binding proteins. Science. 1988;242:423–6. doi: 10.1126/science.3140379. (erratum in: Science 1989; 244:409) [DOI] [PubMed] [Google Scholar]
- 13.Weiner LM. Building better magic bullets improving unconjugated monoclonal antibody therapy for cancer. Nature Reviews Cancer. 2007;7:701–6. doi: 10.1038/nrc2209. [DOI] [PubMed] [Google Scholar]
- 14.Silacci M, Brack S, Schirru G, et al. Design, construction and characterization of a large synthetic human antibody phage display library. Proteomics. 2005;5:2340–50. doi: 10.1002/pmic.200401273. [DOI] [PubMed] [Google Scholar]
- 15.Santin AD, Zhan F, Bellone S, et al. Discrimination between uterine serous papillary carcinomas and ovarian serous papillary tumours by gene expression profiling. Br J Cancer. 2004;90:1814–24. doi: 10.1038/sj.bjc.6601791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santin AD, Bellone S, Ravaggi A, et al. Induction of tumor-specific CD83 cytotoxic T lymphocytes by tumor lysatepulsed autologous dendritic cells in patients with uterine serous papillary cancer. Br J Cancer. 2002;86:151–7. doi: 10.1038/sj.bjc.6600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bignotti E, Tassi RA, Calza S, et al. Differential gene expression profiles between tumor biopsies and short-term primary cultures of ovarian serous carcinomas: identification of novel molecular biomarkers for early diagnosis and therapy. Gynecol Oncol. 2006;103:405–16. doi: 10.1016/j.ygyno.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 18.Pini A, Viti F, Santucci A, et al. Design and use of a phage display library: human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem. 1998;273:21769–76. doi: 10.1074/jbc.273.34.21769. [DOI] [PubMed] [Google Scholar]
- 19.Viti F, Nilsson F, Demartis S, et al. Design and use of phage display library for the selection of antibodies and enzymes. Methods Enzymol. 2000;326:480–505. doi: 10.1016/s0076-6879(00)26071-0. [DOI] [PubMed] [Google Scholar]
- 20.Turatti F, Mezzanzanica D, Nardini E, et al. Production and validation of the pharmacokinetics of a single-chain fragment of the MGR6 antibody for targeting of tumors expressing HER-2. Cancer Immunol Immunother. 2001;49:679–86. doi: 10.1007/s002620000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoogenboom H. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–16. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 22.Colcher D, Goel A, Pavlinkova G, Beresford G, Booth B, Batra SK. Effect of genetic engineering on the pharmacokinetics of antibodies. Q J Nucl Med. 1999;43:132–9. [PubMed] [Google Scholar]
- 23.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 24.Chames P, Hoogenboom HR, Henderikx P. Selection of antibodies against biotinylated antigens. Methods Mol Biol. 2002;178:147–57. doi: 10.1385/1-59259-240-6:147. [DOI] [PubMed] [Google Scholar]
- 25.Ewert S, Huber T, Honegger A, et al. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325:531–53. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 26.Accardi L, Donà MG, Di Bonito P, Giorgi C. Intracellular anti-E7 human antibodies in single-chain format inhibit proliferation of HPV16-positive cervical carcinoma cells. Int J Cancer. 2005;116:564–70. doi: 10.1002/ijc.21052. [DOI] [PubMed] [Google Scholar]
- 27.Viti F, Tarli L, Giovannoni L, et al. Increased binding affinity and valence of recombinant antibody fragments lead to improved targeting of tumoral angiogenesis. Cancer Res. 1999;59:347–52. [PubMed] [Google Scholar]
- 28.Martinez-Torrecuadrada J, Cifuentes G, Lopez-Serra P, et al. Targeting the extracellular domain of fibroblast growth factor receptor 3 with human single-chain Fv antibodies inhibits bladder carcinoma cell line proliferation. Clin Cancer Res. 2005;11:6280–90. doi: 10.1158/1078-0432.CCR-05-0282. [DOI] [PubMed] [Google Scholar]
- 29.Adams GP, Schier R, McCall AM, et al. High affinity restricts the localization and tumor penetration of single-chain fv antibody molecules. Cancer Res. 2001;61:4750–5. [PubMed] [Google Scholar]
- 30.Winthrop MD, DeNardo SJ, Albrecht H, et al. Selection and characterization of anti-MUC-1 scFvs intended for targeted therapy. Clin Cancer Res. 2003;9(suppl):3845s–53s. [PubMed] [Google Scholar]
- 31.Michl P, Buchholz M, Rolke M, et al. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology. 2001;121:678–84. doi: 10.1053/gast.2001.27124. [DOI] [PubMed] [Google Scholar]