Abstract
In the Drosophila optic lobes, the medulla processes visual information coming from inner photoreceptors R7 and R8 and from lamina neurons. It contains ~40,000 neurons belonging to over 70 different types. We describe how precise temporal patterning of neural progenitors generates these different neural types. Five transcription factors--Homothorax, Eyeless, Sloppy-paired, Dichaete and Tailless--are sequentially expressed in a temporal cascade in each of the medulla neuroblasts as they age. Loss of either Eyeless, Sloppy-paired or Dichaete blocks further progression of the temporal sequence. We provide evidence that this temporal sequence in neuroblasts, together with Notch-dependent binary fate choice, controls the diversification of the neuronal progeny. Although a temporal sequence of transcription factors had been identified in Drosophila embryonic neuroblasts, our work illustrates the generality of this strategy, with different sequences of transcription factors being used in different contexts.
Generation of neuronal diversity requires both spatial and temporal patterning of neural progenitors. Vertebrate neural progenitors transit through different competence states as they age, and thus generate a conserved order of different neural types1–4. Similarly, Drosophila neuroblasts (NB) generate differently fated progeny in a defined order5–10. A molecular mechanism of temporal specification has been identified in the Drosophila embryonic nerve cord where NBs sequentially express several transcription factors (TF) as they age: Hunchback (Hb), Krüppel (Kr), Pdm1/Pdm2 (Pdm), Castor (Cas), and Grainyhead (Grh)7, 11–13. This temporal cascade is necessary and sufficient for the specification of neuronal identities in multiple lineages of the nerve cord7–9, 11, 14–17. Does the same temporal gene cascade pattern neural progenitors in other systems? In Drosophila antennal lobe NBs, Kr defines one out of 40 fates of projection neurons18. In vertebrates, Ikaros, a mouse ortholog of Hb, is both necessary and sufficient for the early competence state of retinal progenitors19. However, a cascade of TFs analogous to that of Drosophila nerve cord NBs has not been reported elsewhere. Thus, it is still not clear whether this powerful mechanism is widely utilized in other systems. Here we address this question in the Drosophila medulla.
The medulla, containing ~40,000 neurons belonging to over 70 cell types, is the largest neuropil in the visual-processing center (optic lobe)20, 21. It is derived from a larval crescent-shaped neuroepithelium (NE) termed the Outer Proliferation Center (OPC). The single-layered NE cells of the OPC proliferate by dividing symmetrically. They are sequentially converted into medulla NBs in a wave of neurogenesis that initiates at the medial edge of the NE crescent and progresses laterally22–27 (Fig.1a,c). Each NB then divides asymmetrically multiple times to self-renew and to generate Ganglion Mother Cells (GMCs), which in turn divide once to produce medulla neurons22, 28, 29. The neuronal progeny of each NB form a chain, with newly generated neurons occupying the most superficial layer close to NBs and GMCs, and the first-born neurons occupying the deepest layer close to the medulla neuropil30, 31 (Fig.1c,d). Pioneering studies have identified several TFs specifying different subsets of medulla neuron types21, 30, 31. However, it was not clear how their expression in neurons is controlled to generate neuronal diversity.
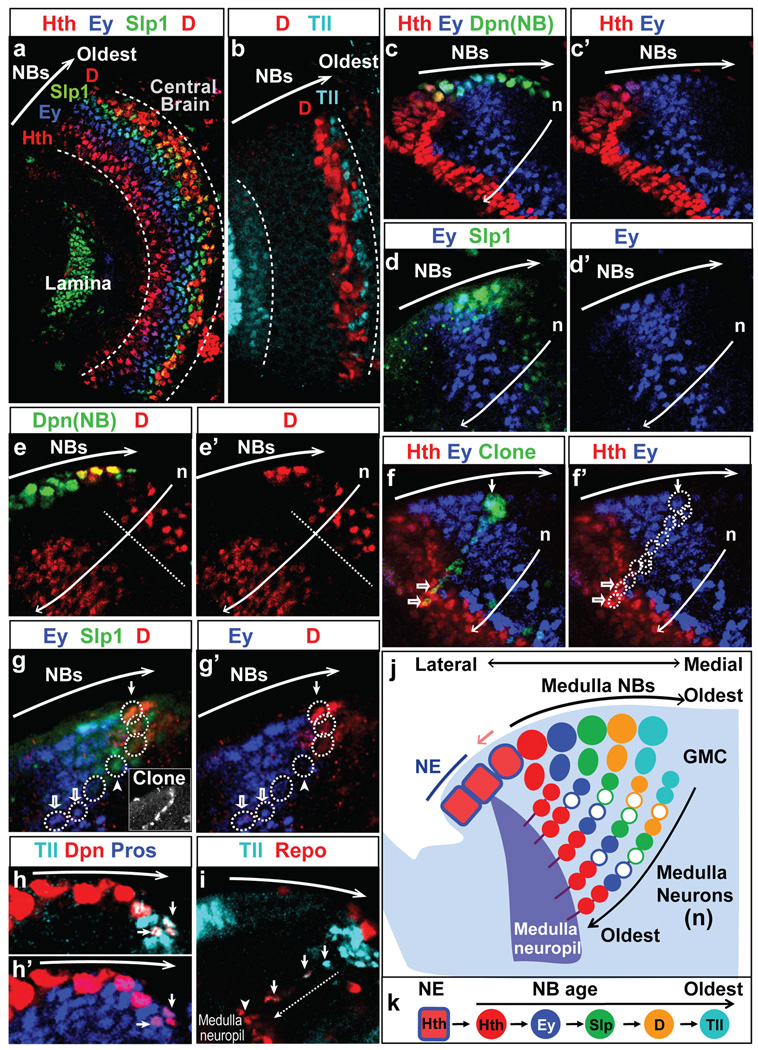
Figure 1. The developing medulla.
a. Model of a larval brain showing that the NE (blue) gives rise to the lamina on the lateral (L) side and to the medulla on the medial (M) side. A wave of neurogenesis (light red) converts NE cells (blue) into NBs (red). VNC: ventral nerve cord.
b. Surface view showing NE (Phalloidin, blue), medulla NBs (Dpn, red), and lamina neurons (Elav, purple).
c. Cross-section model showing NBs (red), GMCs (green), and neurons (purple). A single NB clone is shown by grey thick outlines.
d. Cross-section view showing the NE (DE-Cadherin, blue), medulla NBs (Dpn, red), medulla GMCs (Pros, green), medulla and lamina neurons (elav-Gal4>UASCD8-GFP, purple).
a-d. Small Red arrow: the wave of neurogenesis.
We found that five TFs, Homothorax (Hth), Eyeless (Ey), Sloppy-paired1 and 2 (Slp), Dichaete (D) and Tailless (Tll), are sequentially expressed in medulla NBs as they age. Ey, Slp and D are each required for turning on the next TF in the dividing NBs. Slp and D are also required for turning off the preceding TF. These TFs control the expression of downstream TFs that mark the identities of the neuronal progeny. Notch-dependent asymmetric division of GMCs further diversifies neuronal identities. Our identification of a novel temporal cascade of TFs distinct from the Hb->Kr->Pdm->Cas->Grh sequence suggests that TF-dependent temporal switching of neural progenitors is a common theme in neuronal specification, with different TF sequences being recruited in different contexts.
A temporal gene cascade in medulla NBs
In the developing medulla, the wave of conversion of NE into NBs makes it possible to visualize NBs at different temporal stages in one snapshot, with newly generated NBs on the lateral edge and the oldest NBs on the medial edge of the expanding crescent shaped NB region (Fig.1a,b). We conducted an antibody screen for TFs expressed in the developing medulla and identified five TFs, Hth, Ey, Slp1, D and Tll that are expressed in five consecutive stripes in NBs of increasing ages, with Hth expressed in newly differentiated NBs, and Tll in the oldest NBs (Fig.2a,b). This suggests that these TFs are sequentially expressed in medulla NBs as they age. Neighboring TF stripes show partial overlap in NBs with the exception of the D and Tll stripes, which abut each other. We and others had previously reported that Hth31 and Ey30 were expressed in medulla NBs, but they had not been implicated in controlling NB temporal identities. Hth and Tll also show expression in the NE.
Figure 2. A temporal sequence of TFs in medulla NBs.
a,b. Surface views showing that NBs sequentially express:
a. Hth (red), Ey (blue), Slp1 (green) and D (red).
b. D (red) and Tll (cyan).
c-i. Cross-section views showing the expression of the five TFs in NBs and their progeny.
c,c’. Hth (red), Ey (blue), Dpn(green).
d,d’. Ey (blue), Slp1 (green).
e,e’. D (red). The dashed line separates the two populations of D+ neurons (see text).
f,f’. In a NB clone (βGal, green in f, dashed circles in f’), the NB is Ey+ (blue, small arrow), while its progeny are Ey+ or Hth+ (red, open arrows).
g,g’. In a NB clone (βGal, white in inset), the NB is D+ (red, small arrow). It has generated Slp+ (green) GMCs (arrowhead), and Ey+ (blue) neurons (open arrows).
h,h’. The oldest NBs (small arrows) express Tll (cyan in h), Dpn (red) and nuclear Pros (blue in h’).
i. Tll+ NB progeny (small arrows) lose Tll (cyan), and turn on Repo (red) while migrating (along the dashed arrow) to become medulla neuropil glia (arrowhead).
j. Schematic model. For simplicity, the overlap between TFs is not shown; only one NB/GMC is shown for each stage. D expression in the deeper neuron population is not shown. Empty cells indicate a subset of neurons born during the Ey, Slp or D windows do not maintain the NB TF.
k. Model showing each NB sequentially expresses five TFs.
To ask whether each NB sequentially expresses the five TFs, we examined their expression in the NB progeny (Fig.1c,d). Hth, Ey and Slp1 are expressed in three different layers of neurons that correlate with birth-order, i.e. Hth in the first-born neurons of each lineage in the deepest layers; Ey or Slp1 in correspondingly more superficial layers, closer to the NBs. This suggests that they are born sequentially in each lineage (Fig.2c,c’,d,d’,j). D is expressed in two distinct populations of neurons. The more superficial population inherit D from D+ NBs (above dashed line, Fig.2e,e’). D+ neurons in deeper layers (corresponding to the Hth and Ey layers) turn on D expression independently and will be discussed later (below dashed line, Fig.2e.e’). We generated single NB clones and examined the expression of the TFs in the NB and its progeny. Single NB clones where the NB is at the Ey+ stage include Ey+ GMCs/neurons as well as Hth+ neurons (Fig.2f,f’). This indicates that Ey+ NBs have transited through the Hth+ stage and generated Hth+ neurons. Clones where the NB is at the D+ stage contain Slp1+ GMCs and Ey+ neurons (Fig.2g,g’), suggesting that D+ NBs have already transited through the Slp+ and Ey+ stages. This supports the model that each medulla NB sequentially expresses Hth, Ey, Slp1 and D as it ages, and sequentially produces neurons that inherit and maintain expression of the TF.
slp1 and slp2 are two homologous genes arranged in tandem and function redundantly in embryonic and eye development32, 33. Slp2 is expressed in the same set of medulla NBs as Slp1 (Supp. Fig.1a-a’’). We will refer to Slp1 and Slp2 collectively as Slp.
Tll is expressed in the oldest medulla NBs. The oldest Tll+ NBs show nuclear localization of Prospero (Pros) (Fig.2h,h’), suggesting that they undergo Pros-dependent cell-cycle exit at the end of their life, as in larval nerve cord and central brain NBs34. Tll+ NBs and their progeny express glial-cells-missing (gcm) (Supp.Fig.1b-b’), and the progeny gradually turn off Tll and turn on Repo, a glia-specific marker. These cells migrate towards deeper neuronal layers and take their final position as glial cells around the medulla neuropil (Fig.2i). Thus Tll+ NBs correspond to previously identified glioblasts between the optic lobe and central brain that express gcm and generate medulla neuropil glia35, 36. Clones where the NB is at the Tll+ stage contain Hth+ neurons and Ey+ neurons, among others (Supp. Fig.1c,c’), confirming that Tll+ NBs represent the final temporal stage of medulla NBs rather than a separate population of glioblasts. Therefore, these data clearly show that medulla NBs sequentially express five TFs as they age. The four earlier temporal stages generate neurons that inherit and maintain the temporal TF present at their birth, although a subset of neurons born during the Ey, Slp or D NB stages lose expression of the NB TF (Fig.2j). At the final temporal stage, NBs switch to glioblasts and then exit the cell cycle (Fig.2j,k).
Cross-regulations between temporal TFs
We examined whether cross-regulation among TFs of the NB temporal sequence contributes to the transition from one TF to the next. Loss of hth or its cofactor, extradenticle, does not affect the expression of Ey and subsequent progression of the NB temporal sequence (data not shown).
We generated ey null mutant clones using a BAC rescue construct recombined on an FRT chromosome in an eyJ5.71 null background. We also examined eyJ5.71 homozygous mutant larvae. In both cases, Slp expression is lost in NBs, along with neuronal progeny produced by Slp+ NBs, marked by the TF Twin of Eyeless (Toy, see below) (Fig.3a,a’, Supp.Fig.2a). However, NB division is not affected (Supp.Fig2b,b’), and Hth remains expressed in only the youngest NBs and first-born neurons (Fig.3b and data not shown). Targeted Ey RNAi using a VsxGal4 driver that is expressed in the central region of the NE and NBs37 gives the same phenotype (data not shown). This suggests that Ey is required to turn on the next TF, Slp, but is not required to repress Hth (Fig.3c).
Figure 3. Cross-regulations between TFs in the gene cascade.
a,a’. Surface view: in an eyJ5.71 mutant clone marked by lack of GFP (red) and of Ey (blue), Slp1 (green) is lost in NBs.
b. Cross-section view: in eyJ5.71 mutants, Hth (red) is only in the youngest NBs (Dpn marking all NBs, blue).
c. Summary model.
d-d”. Surface view: in slp mutant MARCM clones (GFP, white in d, dashed line in d’,d’’), NBs continue to express Ey (blue) and do not turn on D (red).
e. Summary model.
f-f’’. Surface view: in D mutant clones marked by lack of GFP (white in f, dashed line in f-f’’), NBs continue to express Slp1 (green) and do not turn on Tll (cyan).
g. Summary model.
h. Model summarizing cross-regulations between the five TFs. (*): sufficient but not required.
In clones of a deficiency mutation, slpS37A, that deletes both slp1 and slp2 33, NBs normally transit from Hth+ to Ey+, but older NBs maintain the expression of Ey and do not progress to express D or Tll (Fig.3d-e, Supp.Fig.2f,f’), suggesting that Slp is required to repress ey and activate D.
Similarly, in D mutant clones, NBs are also blocked at the Slp+ stage, and do not turn on Tll (Fig.3f-g), indicating that D is required to repress slp and activate tll.
Finally, in tll mutant clones, D expression is not expanded into oldest NBs, suggesting that tll is not required for NBs to turn off D (Supp.Fig.2j-j’’).
Thus, in the medulla NB temporal sequence, ey, slp and D, are each required for turning on the next TF. slp and D are also required for turning off the preceding TF (Fig.3h).
We also examined gain-of-function phenotypes of each gene. However, mis-expression of Hth, Ey, Slp1 or 2, or D in all NBs or in large NB clones is not sufficient to activate the next TF or repress the previous TF in NBs (Supp.Fig.2e-e’’,g-i’, and data not shown). Only mis-expressing tll in all NBs is sufficient to repress D expression (Supp.Fig.2k,k’).
In summary, cross-regulation among TFs is required for at least some of the transitions. We did not observe cross-regulation between hth and ey. Since ey is already expressed at low levels in the NE and in Hth+ NBs, an as yet unidentified factor might gradually up-regulate ey and repress hth to achieve the first transition. As tll is sufficient but not required to repress D expression, additional factors must act redundantly with Tll to repress D.
Notch-dependent binary fate choice
The temporal sequence of NBs described above could specify at least four neuron types plus glia (in fact more than 10 neuron types plus glia considering that NBs divide several times at each stage with overlaps between neighboring temporal TFs; see Discussion). As this is not sufficient to generate the 70 medulla neuron types, we asked whether another process increases diversity in the progeny neurons born from a NB at a specific temporal stage. Apterous (Ap) is known to mark about half of the 70 medulla neuron types21. In the larval medulla, Ap is expressed in a salt and pepper manner in subsets of neurons born from all temporal stages30 (Fig.4a,b). In the progeny from Hth+ NBs, all neurons appear to maintain Hth, with a subset also expressing Ap (Fig.4a). However, only half of the neurons born from NBs at other TF stages maintain expression of the NB TF. For instance, in the progeny of Ey+ NBs, Ey+ neurons are intermingled with about an equal number of Ey− neurons that instead express Ap (Fig.4a). NB clones contain intermingled Ey+ and Ap+ neurons (Fig.4d-d’’). This is also true for the progeny of Slp+ NBs: Slp1+ neurons are intermingled with Slp1−Ap+ neurons (Supp.Fig.3a). In the progeny of D+ NBs, D and Ap are co-expressed in the same neurons, and they are intermingled with neurons that express neither D nor Ap (above dashed line, Fig.4b). As mentioned above, neurons in deeper neuronal layers (corresponding to the Ey+ and Hth+ neuron layers, below dashed line, Fig.4b) also express D independently, and these neurons are Ap−. The expression of Ap is stable from larval to adult stages30 (Supp.Fig.3c,d).
Figure 4. Notch-dependent asymmetric division of medulla GMCs.
All panels are cross-section views with Ap (green) and Ey (blue).
a. A subset of Hth+ neurons (red) are Ap+ while Ey+ neurons are intermingled with Ap+ neurons.
b. D+ neurons above the dashed line co-express Ap; D expression below the dashed line is in Ap− neurons.
c,c’. Two daughters of a GMC are labeled by GFP (red). One is Ap+ and the other is Ap−.
d-d’’. Wild type tubGal4 MARCM clones marked by CD8-GFP (red) contain both Ap+ and Ey+ neurons.
e-f’’. Su(H) mutant MARCM clones (CD8-GFP, red in e, blue in f). e-e’’. Ap is lost and Ey expanded. f-f’’’. D (red) in NBs is not affected (open arrow) but D and Ap are lost in D+ NB progeny (above the dashed line, white arrow); the deeper layer of D expression in Ap− neurons (below the dashed line, star) is expanded.
g, A simplified schematic model.
The intermingling of Ap+ and Ap− neurons raised the possibility that asymmetric division of GMCs gives rise to one Ap+ and one Ap− neuron. We generated two-cell clones to visualize the two daughters of a GMC. In every case (n=11), one neuron is Ap+ and the other is Ap− (Fig.4c,c’; Supp.Fig.3b,b’), suggesting that asymmetric division of GMCs diversifies medulla neuron fates by controlling Ap expression.
Asymmetric division of GMCs in Drosophila involves Notch (N) dependent binary fate choice38–40. In the developing medulla, the N pathway is involved in the transition from NE to NB, and loss of Su(H), the transcriptional effector of N signaling, leads to faster progression of neurogenesis and NB formation24. However, Su(H) mutant NBs still follow the same TF sequence and generate GMCs and neuronal progeny (Supp.Fig.3e,f, Fig.4f-f’’, open arrow), allowing us to analyze the effect of loss of N function on GMC progeny diversification. Strikingly, neurons completely lose Ap expression in Su(H) mutant clones. All mutant neurons born during the Hth+ stage still express Hth, but not Ap, suggesting that the NON daughters of Hth+ GMCs are the neurons expressing both Ap and Hth (Supp. Fig.3h). In contrast to wild-type clones (Fig.4d-d’’), all Su(H) mutant neurons born during the Ey+ NB stage express Ey and none express Ap (Fig.4e-e’’). Similarly, all mutant neurons born during the Slp+ NB stage express Slp1 but lose Ap (Supp. Fig.3g,g’ and data not shown). These data suggest that, for Ey+ or Slp+ GMCs, the NOFF daughter maintains the NB TF expression, while the NON daughter loses this expression but expresses Ap. In the wild type progeny born during the D+ NB stage, Ap+ neurons co-express D. Both D and Ap are lost in Su(H) mutant clones in the D+ NB progeny (arrow in Fig.4f-f’’’), confirming that D is transmitted to the Ap+NON daughter of D+ GMCs. In contrast, the D+Ap− neurons in the deeper layers (corresponding to the NOFF progeny born during the Ey+ and Hth+ NB stages, see above) are expanded in Su(H) mutant clones at the expense of Ap+ neurons (star in Fig.4f-f’’’). Therefore, the deeper layer of D expression is turned on independently in the NOFF daughters of Hth+ and Ey+ GMCs.
Finally, in wild type, we observe a significant amount of apoptotic cells dispersed among neurons, suggesting that one daughter of certain GMCs undergoes apoptosis in some of the lineages (Supp. Fig.3i).
Together these data suggest that Notch-dependent asymmetric division of GMCs further diversifies neuronal identities generated by the temporal sequence of TFs (Fig.4g).
Temporal TFs control neural fates
How does the NB TF temporal sequence, together with the Notch-dependent binary fate choice, control neuronal identities in the medulla? We used TF markers specifically expressed in subsets of medulla neurons, but not in NBs, including Brain-specific homeobox (Bsh) and Drifter (Dfr)31, as well as other TFs identified in our antibody screen, e.g. Lim3 and Toy. Bsh is required and sufficient for the Mi1 cell fate41, and Dfr is required for the morphogenesis of nine types of medulla neurons, including Mi10, Tm3, TmY3, Tm27, Tm27Y31. We first investigated at which NB temporal stage these neurons were born by examining co-expression with the inherited NB TFs. We then examined whether the NB TFs regulate expression of these markers and neuron fates. The results for each NB stage are described below.
-Hth+ NB stage
Bsh is expressed in a subset of Hth+ neurons31 that also express Ap (Fig.5a,a’), suggesting that Bsh is in the NON daughter of Hth+ GMCs. Indeed, Bsh expression is lost in both Su(H) and hth mutant clones (Fig.5b,b’,c). Thus both Notch activity and Hth are required for specifying the Mi1 fate, consistent with the previous report that Hth is required for the Mi1 fate31. Ectopic expression of Hth in older NBs is also sufficient to generate ectopic Bsh+ neurons, although the phenotype becomes less pronounced in later parts of the lineage (Fig.5d). These data suggest that Hth is necessary and sufficient to specify early-born neurons, but the competence to do so in response to sustained expression of Hth decreases over time. This is similar to embryonic CNS NBs where ectopic Hb is only able to specify early-born neurons during a specific time window8, 42.
Figure 5. Hth and Ey are required for neuronal diversity.
All images are cross-section views of larval medulla.
a,a’. In wild type, Bsh (blue) is in neurons expressing both ap-LacZ (green), an enhancer trap that perfectly mimics Ap expression, and Hth (red).
b,b’. Bsh (blue), but not Hth (red) is lost in Su(H) mutant clones (GFP, green).
c. Bsh (blue) is lost in hthP2 mutant clones (GFP, green).
d. Bsh (blue) is ectopically expressed when UAS-GFP::Hth is driven by tubGal4 in a MARCM clone (GFP, red).
e, In wild type, Dfr (red) is expressed in two-three rows of Ap+ (green) neurons. There are also Dfr+Dac+(blue) Ap− neurons in a more superficial layer.
f. The Ap+Dfr+ neurons (below the dashed line) are intermingled with Ey+ (blue) neurons.
g,g’. Dfr expression (red) is lost in eyJ5.71 mutant clones marked by lack of GFP (green in g, dashed line in g’).
h,h’. Dfr+ (red) neurons are expanded in slp mutant clones (GFP, green). In this region there are very few Dfr+ Dac+ (blue) neurons. The expanded Dfr+ neurons do not express Dac.
- Ey+ NB stage
Lim3 is expressed in all Ap− progeny of both Hth+ and Ey+ NBs (Supp. Fig.4a-a’’, Fig.6i). Toy and Dfr are expressed in subsets of neurons born from Ey+ NBs, as indicated by their expression in the Ey+ neuron progeny layer. The most superficial row of Ey+Ap− neurons express Toy (and Lim3), suggesting that they are the NOFF progeny of the last-born Ey+ GMCs (Supp. Fig.4c,c’). Dfr is co-expressed with Ap in two or three rows of neurons that are intermingled with Ey+ neurons (Fig.5e,f), suggesting that they are the NON progeny from Ey+ GMCs (Fig.6i). In addition to these Ap+Dfr+ neurons, Dfr is also expressed in some later-born neurons that are Ap− but express another TF: Dachshund (Dac), in specific sub-regions of the medulla crescent31, 37 (Fig.5e).
Figure 6. Slp is required for neuronal diversity.
a-b’. Cross-section views of larval medulla, with Toy in red and Ap (or ap-lacZ) in blue.
a. In wild type, Toy+ neurons in the deeper layer are Ap−. The superficial Toy+ neurons are Ap+ and are intermingled with Slp1+ neurons (green).
b,b’. In slp mutant clones (GFP, green in b, dashed outline in b’), Toy+Ap+ neurons disappear.
c,d. Adult medulla with OrtC1-Gal4>UAS-CD8GFP (green), ap-lacZ (blue) and Toy (red). c. Horizontal view. d. View through the medulla cortex.
e,f. Flip-out clones in adults (OrtC1-Gal4, hsFLP, UAS-FRT-STOP-FRT-CD8GFP). Arrows point to neuron cell bodies.
e. Tm20. Photoreceptor axons in blue.
f. Tm5 and TmY10.
g,h. OrtC1-Gal4 MARCM clones in adults.
g. wt.
h. slp mutant.
i. Simplified model showing neuronal TF markers expressed in progeny of NBs of different stages. The lineage is approximate and does not take into account regional differences. The brackets for “D” indicate that D is not maintained in all NON progeny of D+ NBs.
We tested whether Ey in NBs regulates Dfr expression in neurons. As expected, Dfr expressing neurons are lost in ey null mutant clones (Fig.5g,g’), suggesting that they require Ey activity in NBs, even though Ey is not maintained in Ap+Dfr+ neurons. Furthermore, in slp mutant clones in which NBs remain blocked in the Ey+ state, the Ap+Dfr+ neuron population is expanded into later-born neurons (Fig.5h,h’) suggesting that the transition from Ey+ to Slp+ in NBs is required for shutting off the production of Ap+Dfr+ neurons. In addition, Ap+Dfr+ neurons are lost in Su(H) mutant clones (Supp.Fig4b). Thus Ey expression in NBs and the Notch pathway together control the generation of Ap+Dfr+ neurons.
- Slp+ and D+ NB stages
In addition to its expression with Ey in the NOFF progeny of the last-born Ey+ GMCs, Toy is also expressed in Ap+ (NON) neurons in more superficial layers generated by Slp+ and D+ NBs (Supp.Fig.4c,c’,d, Fig.6a,i). Consistently, in Su(H) mutant clones, we see an expansion of Toy+Ey+ neurons in the Ey progeny layer, followed by loss of Toy in the Slp and D progeny layer (Supp.Fig.4e).
We tested whether Slp is required for the NBs to switch from generating Toy+Ap− neurons, progeny of Ey+ NBs, to generating Toy+Ap+ neurons. Indeed, in slp mutant clones, the Toy+Ap+ neurons largely disappear, while Toy+Ap− neurons expand (Fig.6b,b’).
We examined Ap and Toy expression in specific adult neurons. OrtC1-Gal4 labels primarily Tm20 and Tm5 (Chi-Hon Lee, personal communication) plus a few TmY10 neurons, and these neurons express both Ap and Toy (Fig.6c-f). To examine whether Slp is required for the specification of these neuron types, we generated wild type or slp mutant MARCM clones by heat shocking for 1hr at early larval stage and analyzed the number of OrtC1-Gal4 marked neurons in the adult medulla. In wild type clones, OrtC1-Gal4 marks ~100 neurons (95.1±19.3, n=8) per medulla (Fig.6g, Supp. Fig.4f,h). In contrast, very few neurons (9.7±11.2, n=17) are marked by OrtC1-Gal4 in slp mutant clones (Fig.6h, Supp. Fig.4g,i). Slp is unlikely to directly regulate the ort promoter since Slp expression is not maintained in Ap+Toy+ neurons. Furthermore, the expression level of OrtC1-Gal4 in lamina L3 neurons (Chi-Hon Lee, personal communication) is not affected by slp mutation (Fig.6h). These data suggest that loss of Slp expression in NBs strongly affects the generation of Tm20 and Tm5 neurons.
In summary, our data show that the sequential expression of TFs in medulla NBs controls the birth-order dependent expression of different neuronal TF markers, and thus the sequential generation of different neuron types (Fig.6i).
Discussion
Although a temporal TF sequence that patterns Drosophila nerve cord NBs was reported more than a decade ago7, 12, it was not clear whether the same or a similar TF sequence patterns neural progenitors in other contexts3. Our identification of a novel temporal TF sequence patterning the Drosophila medulla suggests that temporal patterning of neural progenitors is a common theme for generating neuronal diversity, and that different TF sequences might be recruited in different contexts.
There are both similarities and differences between the two NB temporal sequences. In the Hb->Kr->Pdm->Castor->Grh sequence, ectopically expressing one gene is sufficient to activate the next gene, and repress the previous gene, but these cross-regulations are not necessary for the transitions, with the exception of Castor7, 11, 12, 15. In the Hth->Ey->Slp->D->Tll sequence, removal of Ey, Slp, or D does disrupt cross-regulations necessary for temporal transitions (except the Hth->Ey transition). However, in most cases these cross-regulations are not sufficient to ensure temporal transitions, suggesting that additional timing mechanisms or factors are required.
For simplicity, we represented the medulla NBs as transiting through five TF stages, while in fact the number of stages is clearly larger than five (Fig.6i). First, NBs divide more than once while expressing a given temporal TF, and each GMC can have different sub-temporal identities. Furthermore, there is significant overlap between subsequent temporal NB TFs: NBs expressing two TFs are likely to generate different neuron types from NBs expressing either one alone.
Although we are still investigating the complete lineage of medulla NBs, we show here how a novel temporal sequence of TFs is required to sequentially generate the diverse neurons that compose the medulla. The requirement for TF sequences in the medulla and in embryonic NBs suggests that this is a general mechanism for the generation of neuronal diversity. Interestingly, the mammalian orthologue of Slp1, Foxg1, acts in cortical progenitors to suppress early-born cortical cell fates43. Thus TF-dependent temporal patterning of neural progenitors might be a common theme in both vertebrate and invertebrate systems.
Methods Summary
We screened ~200 antibodies against TFs from various sources including: the polyclonal antibody collection against Drosophila segmentation proteins; various generous gifts from the Drosophila community; Developmental Studies Hybridoma Bank; as well as a collection of antibodies generated by the modENCODE project that were generously provided by Nicolas Nègre and Kevin White. Wild-type or mutant MARCM clones were generated by 37°C heat shocks at early larvae stages. Wandering 3rd instar larvae or adults were analyzed. For the generation of ey mutant clones, we used a BAC containing the ey genomic region inserted on chromosome 3L, recombined with FRT80B and Ubi-GFP, and crossed into an eyJ5.71 mutant background. This line was crossed with hs-Flp;; FRT80B, eyJ5.71/ In(4)ciD and the progeny was heat shocked for 1hr at 37°C 3 days before dissection of wandering 3rd instar larvae. Single NB clones were generated using AC225-Gal4, which is expressed in the NE to NB transition zone, driving UAS-FLP combined with act-FRT-STOP-FRT-nulacZ and tubGal80ts to provide temporal control. Two-cell clones were generated using two methods: Twin-spot MARCM10, or with pros-Gal4 (expressed in GMCs) driving UAS-FLP with ubi-FRT-STOP-FRT-nuGFP and tubGal80ts. Full methods are available in the online version
Methods
Antibodies and Immunostaining
We screened ~200 antibodies against TFs from various sources including: the polyclonal antibody collection against Drosophila segmentation proteins44; generous gifts from the Fly community; Developmental Studies Hybridoma Bank; as well as a collection of antibodies generated by the modENCODE45 project that were generously provided by Nicolas Negre and Kevin White. The positive ones among them, and other antibodies used in this work, include rabbit anti Slp1(1:200) and guinea pig anti Tll (1:200) (segmentation antibodies); rabbit anti-D (1:100)(ModENCODE); rabbit anti-Hth (1:500) (from Richard Mann), rat anti-Slp1(1:200) and rat anti-Slp2 (1:200) (from Ken Cadigan), guinea pig anti-D (1:50) (from John R. Nambu), rabbit anti-Dpn (1:500) (from Yuh-Nung Jan), guinea-pig anti-Dpn (1:1000) and guinea pig anti-Lim3 (1:250) (from Jim Skeath), rat anti-Ap (1:200) (from John Thomas), guinea pig anti-Bsh and rat anti-Dfr (from Makoto Sato), guinea pig anti-Toy (1:500) (from Uwe Walldorf); mouse anti-Ey (1:10) (from Patrick Callaerts and DSHB), mouse anti-Pros (1:10), mouse anti-Repo (1:50), 24B10 (1:20), Rat anti DE-Cadherin (1:20), Rat anti DN-Cadherin (1:50) and mouse anti-Dac (1:20) (all from DSHB); sheep anti-GFP (1:500, AbD Serotec), chick anti-beta-gal (1:200, Gallus Immunotech). Secondary antibodies are from Jackson or Invitrogen.
Immunostaining was done as described46 with a few modifications: Larval brains or adult brains were dissected in 1XPBS, and fixed in 4% Formaldehyde for 30 minutes (larval) or 45min (adult) on ice. Brains were incubated in primary antibody solution overnight at 4°C, washed three times and incubated in secondary antibody solution overnight at 4°C, washed three times and mounted in Slowfade. Images are acquired using a Leica SP5 confocal. Figures are assembled using Photoshop and Illustrator.
Genetics and fly strains
Canton S is used as wild type controls. To generate hth mutant MARCM clones, flies of y,w,hsFLP,UASCD8GFP; ; tubGal4, FRT82B tubGal80 /TM6B were crossed with FRT82B hthP2 /TM6B or FRT82B hth100-1 /TM6B flies (gifts from Richard Mann). To generate exd mutant MARCM clones, flies of FRT19A, tubGal80, hsFLP; UASLacZ /CyO; tubGal4 /TM6B were crossed with FRT19A exd1 /FM7C flies (gift from Richard Mann). The null mutant of ey: yw; eyJ5.71/In(4)ciD was obtained from Bloomington. To generate slp mutant MARCM clones, flies of y,w, hsFLP, UASCD8GFP; FRT40A tubGal80; tubGal4 /TM6B were crossed with FRT40A slpS37A /SM6-TM6B flies (gift from Andrew Tomlinson). To generate D mutant clones, y,w,hsFLP; ; FRT2A ubi-GFP was crossed with FRT2A D87 mutant flies (gift from John Nambu). To generate tll mutant clones, w; FRT82B tlll49 / TM3 (from Mitsuhiko Kurusu) was crossed with y,w,hsFLP,UASCD8GFP; ; tubGal4, FRT82B tubGal80 /TM6B flies. To generate wild type or Su(H) mutant MARCM clones, flies of y,w, hsFLP, UASCD8GFP; FRT40A tubGal80; tubGal4/TM6B were crossed with FRT40A or FRT40A Su(H)Δ47/CyO flies (gift from François Schweisguth). For these mutant clones, the progeny were heat shocked at 37°C at early larvae stage, and dissected at wandering 3rd instar stage or white pupae stage.
For targeted Ey RNAi, Vsx-Gal4 was used to drive two UAS-ey-RNAi transgenes (UAS-eyRNAi-JF02501 from Bloomington, and UAS-eyRNAi-kk107100 from VDRC stock center) together with UAS-Dcr2.
We used 1407a-Gal4 (an insertion into the inscuteable locus)47, combined with tubGal80ts to drive UAS-GFP::Hth, UAS-Ey (from Bloomington), UAS-Slp1(from Andrew Tomlinson), UAS-D (from John Nambu) or UAS-Tll (from Mitsuhiko Kurusu) in all NBs, and the progeny were shifted from 18°C to 29°C 4 days before dissection of the wondering 3rd instar larvae. For gain of function of Slp2, UAS-Slp2 (from Maria Leptin) was crossed with ywhsFLP; UAS-LacZ; act>y+>Gal4, and the progeny were heat shocked for 8 min at 37°C 3 days before dissection of the wondering 3rd instar larvae. For gain of function of Slp1, flies of yw; UAS-Slp1; FRT82B (from Andrew Tomlinson) was crossed with y,w,hsFLP,UASCD8GFP; ; tubGal4, FRT82B tubGal80 /TM6B flies, and the progeny were heat shocked for 1hr at 37°C 3 days before dissection of the wondering 3rd instar larvae.
To generate OrtC1-Gal4 wild type or slp mutant MARCM clones, virgin females of y,w,hsFLP,UASCD8GFP; FRT40A tubGal80/CyO; OrtC1-Gal4, UASCD8GFP/Tm2 (unpublished gift from Chi-Hon Lee) were crossed with FRT40A /CyO or FRT40A slpS37A /CyO males. The progeny were heat shocked at 37°C at early larval stage for 1hr, and the adult male progeny with the correct genotype were dissected and stained. To generate OrtC1-Gal4 flip-out clones, yw; OrtC1-Gal4 /CyO; OrtC1-Gal4 /TM3 (un-published gift from Chi-Hon Lee) were crossed with UAS-FRT-STOP-FRT-CD8::GFP, and the progeny were heat shocked at late pupal stage, and dissected in the adult stage.
Other strains used include “apmd544-Gal4”, “aprK568-lacZ” 48, “yw; act-FRT-STOP-FRT-lacZ; UASFLP”, and “UAS-Red-Stinger, UASFLP, ubi-FRT-STOP-FRT-NuGFP”(G-TRACE)49.
Generation of ey mutant clones by Bac-rescue
A BAC that contains the ey genomic region (CH321-01A12, BacPac Resources) was inserted by PhiC31 transgenesis on chromosome 3L in attP site PBac{y+-attP-3B}VK00031. The resulting transgenic flies were tested for rescue of the ey null allele eyJ5.71. Subsequently this ey BAC insertion was recombined with FRT80B (P{neoFRT}80B) and Ubi-GFP (P{Ubi-GFP(S65T)nls}3L). This chromosome arm was used to generate the strain yw, hs-Flp1.22;; FRT80B, eyBAC, Ubi-GFP/TM6B,Tb; eyJ5.71 that served as a wildtype copy of ey on the third chromosome. To generate mitotic clones this strain was crossed to flies with genotype hs-Flp1.22;; FRT80B; eyJ5.71/ In(4)ciD, ciD panciD svspa-pol , and the progeny were heat shocked for 1hr at 37°C 3 days before dissection of the wondering 3rd instar larvae. Clones in larvae that lacked both GFP fluorescence and staining with an anti-Ey antibody were further analyzed.
Generation of Single-NB clones
Larvae of the genotype AC225-Gal4 (which is expressed in the NE to NB transition), tubGal80ts, UASFLP, act-FRT-STOP-FRT-nuLacZ were grown at 18°C, and shifted to 29°C for 15min to inactivate tubGal80ts only in scattered newly-generated NBs, and after another 3–6 days at 18°C, the wandering 3rd instar larvae were dissected and stained.
Generation of two-cell clones
Two methods were used. One is Twin-spot MARCM10 (see Suppl. Figure 2 legend). The flies of elavGal4; FRT40A,UAS-mCD8::GFP,UAS-rCD2-miRNA /CyO,y+ were crossed with hsFLP; FRT40A,UAS-rCD2::RFP,UAS-GFPmiRNA /CyO,y+ (gifts from Tzumin Lee), and the progeny larvae were heat shocked at 37°C for 8min, and dissected 2 days later as wandering 3rd instar larvae; The other method which was used for Fig4c was to treat the larvae with the genotype of pros>Gal4 (that is expressed in GMCs), tubGal80ts, UASFLP, and ubi-FRT-STOP-FRT-nuGFP at 29°C for 1hr to inactivate tubGal80ts only in scattered GMCs, and to perform the staining 2 days later on wandering 3rd instar larvae. Only scattered GMCs flip out the STOP cassette, and transmit the GFP to the two daughters.
Supplementary Material
Acknowledgements
We thank the fly community and the modENCODE team for generous gifts of antibodies and fly stocks. Kevin White, Nicolas Negre, Daniel Vasiliauskas and Robert Johnston contributed to screening the modENCODE antibodies. Special thanks to Chi-Hon Lee for generously sharing unpublished information and the OrtC1-Gal4 line. We thank Richard Mann for suggestions and reagents; Desplan lab members for discussion and support, especially Robert Johnston, Daniel Vasiliauskas and Nathalie Neriec for critically reading the manuscript. This work was supported by NIH grant R01 EY017916 to C.D.; The Robert Leet and Clara Guthrie Patterson Trust Postdoctoral Fellowship to X.L.; The Canadian Institutes of Health Research (CIHR) to T.E.; fellowships from EMBO (ALTF 680-2009) and HFSPO (LT000077/2010-L) to C.B.; NIH grant GM058575 and a Career Development fellowship from the Leukemia and Lymphoma Society to R.V..
Footnotes
Author contributions
C.D. planned the project and analyzed the data together with X.L. and T.E.; T.E. X.L. and C.B. performed the antibody screen; X.L. conducted experiments with Hth, Ey, Slp and Tll neuroblasts as well as Ap and Notch pathway; T.E. analyzed the D neuroblasts; Z.C. generated the OrtC1-Gal4 flip-out and MARCM clones; R.V. generated the ey Bac rescue construct and stocks; S.V. examined Slp2 expression; A.C. identified the AC225-Gal4 line and J.M. defined its expression in the transition from NE to NB. The manuscript was written by X.L. and C.D. and all authors commented on it.
Author information:
The authors declare no competing financial interests.
References
- 1.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 2.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 3.Jacob J, Maurange C, Gould AP. Temporal control of neuronal diversity: common regulatory principles in insects and vertebrates? Development. 2008;135:3481–3489. doi: 10.1242/dev.016931. [DOI] [PubMed] [Google Scholar]
- 4.Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol. 2009;19:112–119. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 6.Akiyama-Oda Y, Hosoya T, Hotta Y. Asymmetric cell division of thoracic neuroblast 6-4 to bifurcate glial and neuronal lineage in Drosophila. Development. 1999;126:1967–1974. doi: 10.1242/dev.126.9.1967. [DOI] [PubMed] [Google Scholar]
- 7.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 8.Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- 9.Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- 10.Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kambadur R, et al. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- 13.Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- 15.Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 2006;20:2618–2627. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleary MD, Doe CQ. Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 2006;20:429–434. doi: 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran KD, Doe CQ. Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development. 2008;135:3491–3499. doi: 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao CF, Yu HH, He Y, Kao JC, Lee T. Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron. 2012;73:677–684. doi: 10.1016/j.neuron.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Fischbach KFD, APM The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475. [Google Scholar]
- 21.Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Curr Biol. 2008;18:553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- 24.Yasugi T, Sugie A, Umetsu D, Tabata T. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 2010;137:3193–3203. doi: 10.1242/dev.048058. [DOI] [PubMed] [Google Scholar]
- 25.Reddy BV, Rauskolb C, Irvine KD. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137:2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngo KT, et al. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev Biol. 2010;346:284–295. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. J Comp Neurol. 2003;455:417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- 29.Ceron J, Gonzalez C, Tejedor FJ. Patterns of cell division and expression of asymmetric cell fate determinants in postembryonic neuroblast lineages of Drosophila. Dev Biol. 2001;230:125–138. doi: 10.1006/dbio.2000.0110. [DOI] [PubMed] [Google Scholar]
- 30.Morante J, Erclik T, Desplan C. Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development. 2011;138:687–693. doi: 10.1242/dev.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasegawa E, et al. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development. 2011;138:983–993. doi: 10.1242/dev.058370. [DOI] [PubMed] [Google Scholar]
- 32.Grossniklaus U, Pearson RK, Gehring WJ. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992;6:1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- 33.Sato A, Tomlinson A. Dorsal-ventral midline signaling in the developing Drosophila eye. Development. 2007;134:659–667. doi: 10.1242/dev.02786. [DOI] [PubMed] [Google Scholar]
- 34.Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 35.Soustelle L, Giangrande A. Novel gcm-dependent lineages in the postembryonic nervous system of Drosophila melanogaster. Dev Dyn. 2007;236:2101–2108. doi: 10.1002/dvdy.21232. [DOI] [PubMed] [Google Scholar]
- 36.Colonques J, Ceron J, Tejedor FJ. Segregation of postembryonic neuronal and glial lineages inferred from a mosaic analysis of the Drosophila larval brain. Mech Dev. 2007;124:327–340. doi: 10.1016/j.mod.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Erclik T, et al. Combinatorial inputs from temporal and spatial axes generate neuronal diversity in the Drosophila medulla. (Submitted) [Google Scholar]
- 38.Skeath JB, Doe CQ. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- 39.Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin S, et al. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2010;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasegawa E, Kaido M, Takayama R, Sato M. Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe. Dev Biol. 2013 doi: 10.1016/j.ydbio.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 44.Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- 45.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morante J, Desplan C. Dissection and Staining of Drosophila Optic Lobes at Different Stages of Development. Cold Spring Harbor Protocols. 2011;2011 doi: 10.1101/pdb.prot5629. pdb.prot5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 48.Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM. apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- 49.Evans CJ, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.