Abstract
Background and Purpose
The proteasome is a multi-subunit complex that proteolytically cleaves proteins. The replacement of the constitutive proteasome subunits β1, β2, and/or β5 with the IFNγ-inducible subunits LMP2, MECL1, and/or LMP7 results in the ‘immunoproteasome’. The inducible subunits change the cleavage specificities of the proteasome, but it is unclear whether they have functions in addition to this. The purpose of the present study was to determine the role of the proteasome in general, as well as LMP7 and MECL1 specifically, with regard to cytokine production by activated primary splenocytes.
Methods
A LMP7/MECL1-null mouse was engineered to determine the roles of these subunits in cytokine production. Isolated splenocytes from wild-type and LMP7/MECL1–/– mice were treated with lactacystin and activated with PMA and ionomycin and subsequently cytokine mRNA levels were quantified.
Results
The present study demonstrates that LMP7/MECL1 regulates the expression of IFNγ, IL4, IL10, IL2Rβ, GATA3, and t-bet. In contrast, the regulation of IL2, IL13, TNFα, and IL2Rα by the proteasome appears to occur independently of LMP7/MECL1.
Conclusions
Collectively, the present study demonstrates that LMP7 and MECL1 regulate cytokine expression, suggesting this system represents a novel mechanism for the regulation of cytokines and cytokine signaling.
Key Words: Cytokines, GATA3, LMP7, MECL1, Proteasomes, Splenocytes
Introduction
Proteasomes are large multi-subunit protein complexes responsible for the majority of non-lysosomal proteolysis occurring in cells [1,2]. Structurally, proteasomes are comprised of 14 distinct subunits which are subdivided into α subunits (α1–α7) and β subunits (β1–β7). All α subunits are assembled to form a 7-membered ring; similarly, the β subunits also assemble to form a ring. The proteolytic activity of the proteasome can be traced to 3 specific β subunits, β1 (Y), β2 (Z), and β5 (X), each of which has distinct proteolytic cleavage specificities. The activity of β1 is described as post-acidic, because it cleaves peptide bonds after acidic residues; similarly, β2 and β5 subunits mediate trypsin-like and chymotrypsin-like activities, respectively. The proteasome, as a whole, can exist as either a 20S or 26S structure. The 20S proteasome has the capacity to cleave proteins to a relatively limited extent in an ATP-independent fashion but cannot efficiently cleave folded proteins. The 26S proteasome, in contrast, is comprised of the 20S proteasome capped at both ends by 19S regulatory structures that confer the ability to cleave folded proteins, but proteolytic activity is dependent on ATPase activity [1,2].
In addition to the constitutively expressed proteasome, distinct inducible proteasomal subunits can replace components of the constitutively expressed proteasome to form what has been termed the ‘immunoproteasome’ [1,2,3,4]. There are 3 inducible β subunits, again constituting the proteolytically active subunits of the proteasome. These include the low-molecular-mass protein (LMP) 2 (β1i), multi-catalytic endopeptidase complex-like (MECL) 1 (β2i), and LMP7 (β5i), which replace the constitutively expressed subunits, β1, β2, and β5, respectively. The incorporation of the inducible proteasomal subunits is thought to selectively modify the cleavage specificity of the proteasome to enhance effectiveness of antigen processing and presentation [5]. This is thought to be largely due to the replacement of β1 with LMP2 [2,6]. The purpose of induction of LMP7 and MECL1, however, remains unclear. Although the presumed contribution of the induced LMP7 and MECL1 proteolytic subunits lies in the enhancement of antigen processing, whether this is the sole function of these subunits or whether LMP7 and MECL1 have additional functions on leukocyte activity and response remains to be determined.
Results of recent studies have provided strong evidence that bone marrow-derived dendritic cells lacking one or more of the inducible proteasomal subunits had diminished ability to present specific influenza epitopes to CD8+ T cells. The impaired antigen presentation resulted in decreased numbers of interferon (IFN)γ-producing CD8+ T cells in response to the influenza epitopes [6]. Another group demonstrated diminished IFNγ induction in response to Toxoplasma gondii by CD8+ T cells in LMP7-null mice, despite comparable numbers of activated CD8+ T cells [7].
Results of our own previous studies have revealed that proteasome proteolytic activity in primary macrophages, as well as in the RAW264.7 macrophage cell line, can be modified in vitro in response to stimulation with lipopolysaccharide (LPS) [8,9,10]. Moreover, we have demonstrated that LPS-induced production of inflammatory mediators and Toll-like receptors (TLRs), including tumor necrosis factor (TNF)α, interleukin (IL)6, IL12 p40, IL12 p35, cyclooxygenase 2, inducible nitric oxide synthase, and TLR2, are all dependent upon a fully functional proteasome [11]. With regard to T cells, we have also found that proteasome inhibitors have differential effects on cytokine production between Jurkat T cells and primary murine splenocytes [12]. Collectively, the results from these studies demonstrate that the proteasome can function to regulate cytokine production by immune cells.
There is strong evidence to support the conclusion that the inducible proteasomal subunits can influence cytokine production by macrophages, T cells, and possibly other immune cell types. In macrophages, it would appear that modulation of the proteasome itself can directly influence cytokine production. In other immune cell types, such as lymphocytes, modulating effects have not been defined, and the extent to which modulation of cytokine production is due to defective antigen processing/presentation versus direct effects of the immunoproteasome itself on cytokine induction remains to be determined. The purpose of the present study was, therefore, to determine the role of the proteasome in modulating cytokine production in splenocytes activated with phorbol-12-myristate-13-acetate (PMA)/ionomycin, thus bypassing antigen processing and presentation.
Materials and Methods
Materials
Lactacystin was purchased from A.G. Scientific (San Diego, Calif., USA). All other reagents were purchased from Sigma Chemical Co. (St. Louis, Mo., USA).
Animals and Cell Culture
LMP7/MECL1 double-null mice were generated as described [13]. Age-matched control mice (C57BL/6) were purchased from Jackson Laboratories (Bar Harbor, Me., USA). All studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals adopted by the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee at the University of Missouri Kansas City. Spleens were isolated aseptically and processed into single-cell suspensions (5 × 106 cells/ml) using standard procedures. Cells were cultured in RPMI 1640 supplemented with 100 units penicillin/ml, 100 units streptomycin/ml, 50 μmol/l 2-mercaptoethanol, and 2% bovine calf serum.
Real-Time PCR
Freshly isolated splenocytes (5 × 106 cells/ml) were treated with lactacystin as described in the figure legends. To assess gene expression, total RNA was isolated from treated splenocytes, using RNeasy Mini Kits (Qiagen, Valencia, Calif., USA), following the manufacturer's protocol. The relative expression levels of the target genes were determined by SYBR green real-time PCR using predesigned Quantitect Primer Assays following the manufacturer's protocol (Qiagen). The CT values of each target gene were first normalized to the CT value of the housekeeping gene, ribosomal protein, rpL13a. Fold change in each target gene was then calculated relative to the control, the wild-type background (BKG) group, using the ΔΔCT method of relative quantification as described in ‘Critical Factors for Successful Real-Time PCR’ (available online at: www.qiagen.com/literature/brochures).
Statistical Analysis
The mean ± SE was determined from triplicate samples for each treatment group in the individual experiments. Homogeneous data were evaluated by two-way parametric analysis of variance (ANOVA), and the Holm-Sidak post hoc analysis was used to compare response of cells in treatment groups to the cells maintained as vehicle (VH) controls when significant differences were observed [14]. In addition, the Holm-Sidak test was used to compare differences in response between wild-type and LMP7/MECL1-null genotypes. Statistical analyses were performed using SigmaStat 3.0 (Systat Software, Inc., San Jose, Calif., USA).
Results
Differential Regulation of TH1 Cytokines by LMP7 and MECL1 in Freshly Isolated Murine Splenocytes
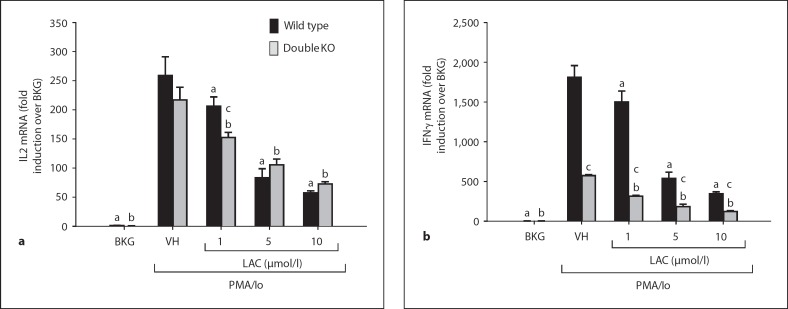
Results of previously published studies have provided evidence for decreased TH1 cytokine production in LMP7/MECL1 double-null mice in response to influenza antigenic epitopes and T. gondii[7]. To distinguish between effects of LMP7/MECL1 on antigen processing and direct effects on cytokine induction, splenocytes were isolated from LMP7/MECL1 double-null mice as well as wild-type mice and activated with PMA and ionomycin ex vivo. Viability was not affected by either lactacystin treatment or LMP7/MECL1 deficiency (data not shown). Production of the TH1 cytokine IL2 was not found to be significantly different between wild-type and LMP7/MECL1-null splenocytes activated with PMA/ionomycin (fig. 1), suggesting that these proteasome subunits do not contribute significantly to the induction of IL2. However, the proteasome inhibitor lactacystin inhibited IL2 production similarly in wild-type and LMP7/MECL1-null splenocytes. The inhibition of IL2 by lactacystin is consistent with previous reports of proteasome inhibitors in activated T cells [15]. In contrast to IL2, steady-state mRNA for the prototypical TH1 cytokine IFNγ was significantly diminished in activated splenocytes from LMP7/MECL1-null mice compared to wild-type splenocytes, although lactacystin suppressed IFNγ transcription by both wild-type and LMP7/MECL1-null splenocytes. Collectively, the data suggest that proteasome proteolytic activity promotes the transcription of both IL2 and IFNγ in wild-type mice, but only IFNγ, not IL2, is dependent upon the expression of LMP7 and MECL1.
Fig. 1.
Effect of lactacystin on PMA/ionomycin (Io)-stimulated TH1 cytokine production in splenocytes derived from wild-type and LMP7/MECL1 double-knockout mice. Freshly isolated splenocytes (5 × 106 cells/ml) from wild-type or LMP7/MECL1 double-knockout mice were treated with lactacystin (1-10 μmol/l) or VH (0.1% ethanol) for 30 min followed by activation of the cells with 40 nmol/1 PMA/0.5 μmol/l Io. Cells were harvested 6 h later, the RNA isolated and analyzed for either IL2 (a) or IFNγ (b). Cellular viability was ≥80% for all treatment groups (assessed by trypan blue exclusion). The results are presented as fold induction over wild-type BKG and are the mean ± SE of triplicate cultures. a p < 0.05 vs. the wild-type VH group, b p < 0.05 vs. the LMP7/ MECL1-null VH group,c p < 0.05 between wild-type and LMP7/MECL1-null splenocytes with respect to the BKG, VH, or LAC treatment group (two-way ANOVA with two-tailed Holm-Sidak post hoc analysis).
The Inducible Proteasomal Subunits LMP7 and MECL1 Contribute to the Transcription of the TH2 Cytokines IL4 and IL10, but Not IL13
Given that LMP7 and MECL1 contribute to the induction of the TH1 cytokine IFNγ in activated splenocytes, the contribution of these two proteasomal subunits to influence TH2 cytokine production in response to PMA/ionomycin was also assessed. Similar to IFNγ, induction of IL4 transcription was almost completely suppressed in activated LMP7/MECL1 double-null splenocytes relative to the wild type, suggesting a critical role for these subunits in the regulation of IL4 (fig. 2a). In addition, lactacystin suppressed IL4 mRNA expression in a concentration-dependent manner in wild-type splenocytes. In contrast to IL4, there was little difference in IL13 mRNA expression between activated splenocytes from wild-type versus LMP7/MECL1-null mice (fig. 2b). Lactacystin suppressed IL13 induction similarly in activated splenocytes from both genotypes. Activated LMP7/MECL1-null splenocytes also produced significantly less IL10 compared to activated wild-type splenocytes (fig. 2c). Interestingly, lactacystin treatment of LMP7/MECL1-null splenocytes caused an increase in IL10 mRNA levels, which was unexpected. Overall, the results indicate that LMP7 and/or MECL1 have differential effects on cytokine production that do not appear to be attributable to skewed TH1/TH2 differentiation.
Fig. 2.
Effect of lactacystin on PMA/ionomycin (Io)-stimulated TH2 cytokine production in splenocytes derived from wild-type and LMP7/MECL1 double-knockout mice. Freshly isolated splenocytes (5 × 106 cells/ml) from wild-type or LMP7/MECL1 double-knockout mice were treated with lactacystin (1-10 μmol/l) or VH (0.1% ethanol) for 30 min followed by activation of the cells with 40 nmol/1 PMA/0.5 μmol/l Io. Cells were harvested 6 h later, the RNA isolated and analyzed for either IL4 (a), IL13 (b), or ILIO (c). Cellular viability was > 80% for all treatment groups (assessed by trypan blue exclusion). The results are presented as fold induction over wild-type BKG and are the mean ± SE of triplicate cultures. a p < 0.05 vs. the wild-type VH group, b p < 0.05 vs. the LMP7/MECL1-null VH group, and c p < 0.05 between the wild-type and LMP7/MECL1-null splenocytes with respect to the BKG, VH, or LAC treatment group (two-way ANOVA with two-tailed Holm-Sidak post hoc analysis).
Induction of TNFα and IL6 Is Independent of LMP7 and MECL1 in Activated Splenocytes
Because transcription of IL10, an anti-inflammatory cytokine, is markedly reduced in LMP7/MECL1 double-null mice, we predicted that the induction of pro-inflammatory cytokines, such as TNFα and IL6, previously shown to be counter-regulated by IL10 [16,17], would be greatly increased in the LMP7/MECL1 null cells. Surprisingly, there was relatively little difference between the two genotypes in the induction of TNFα and IL6, suggesting the lack of a significant contribution of LMP7 and/or MECL1 to these responses (fig. 3). Whereas lactacystin pretreatment significantly suppressed TNFα mRNA expression in both wild-type and LMP7/MECL1-null splenocytes, it had little effect on IL6 production in splenocytes from either genotype. Taken together, these data demonstrate that TNFα induction by PMA/ionomycin in primary splenocytes is proteasome dependent, but unlike IFNγ and IL4, is not specifically dependent on the LMP7/MECL1 subunits.
Fig. 3.
Effect of lactacystin on PMA/ionomycin (Io)-stimulated pro-inflammatory cytokine production in splenocytes derived from wild-type and LMP7/MECL1 double-knockout mice. Freshly isolated splenocytes (5 × 106 cells/ml) from wild-type or LMP7/ MECL1 double-knockout mice were treated with lactacystin (1-10 μmol/l) or VH (0.1% ethanol) for 30 min followed by activation of the cells with 40 nmol/1 PMA/0.5 μmol/l Io. Cells were harvested 6 h later, the RNA isolated and analyzed for either TNFα (a) or IL6 (b). Cellular viability was >80% for all treatment groups (assessed by trypan blue exclusion). The results are presented as fold induction over wild-type BKG and are the mean ± SE of triplicate cultures. a p < 0.05 vs. the wild-type VH group, b p < 0.05 vs. the LMP7/MECL1-null VH group, and c p < 0.05 between the wild-type and LMP7/MECL1-null splenocytes with respect to the BKG, VH, or LAC treatment groups (two-way ANOVA with two-tailed Holm-Sidak post hoc analysis).
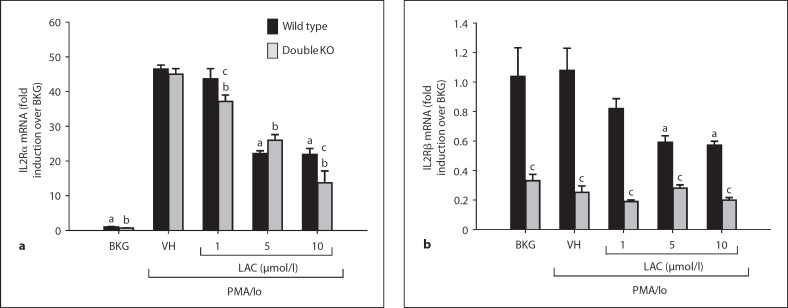
Lactacystin Suppresses mRNA Expression of IL2 Receptors α and β in Activated Splenocytes
Results of our previous studies provided evidence that lactacystin pretreatment suppresses transcription of IL2 by PMA/ionomycin-activated wild-type splenocytes. Given that IL2 also regulates its receptors, we predicted that the mRNA expression of the IL2 receptors (IL2Rs) would be inhibited by lactacystin. Although lactacystin suppressed the mRNA expression of both IL2Rα and IL2Rβ in wild-type splenocytes, only IL2Rα induction was inhibited by lactacystin in LMP7/MECL1-null splenocytes (fig. 4). In addition, IL2Rα induction was comparable between wild-type and LMP7/MECL1-null splenocytes, whereas IL2Rβ expression was markedly reduced in LMP7/MECL1-null splenocytes compared to wild-type splenocytes. Collectively, the data suggest that mRNA expression of IL2Rα and IL2Rβ is proteasome dependent, similar to IL2 itself. The data also demonstrate that LMP7 and/or MECL1 contribute to IL2Rβ but not IL2Rα expression.
Fig. 4.
Effect of lactacystin on IL2 receptor expression in activated splenocytes derived from wild-type and LMP7/MECL1 double-knockout mice. Freshly isolated splenocytes (5 × 106 cells/ml) from wild-type or LMP7/MECL1 double-knockout mice were treated with lactacystin (1-10 μmol/l) or VH (0.1% ethanol) for 30 min followed by activation of the cells with 40 nmol/1 PMA/0.5 μmol/l ionomycin (Io). Cells were harvested 6 h later, the RNA isolated and analyzed for either IL2Rβ (a) or IL2Rß (b). Cellular viability was >80% for all treatment groups (assessed by trypan blue exclusion). The results are normalized to wild-type BKG and are the mean ± SE of triplicate cultures. a p < 0.05 vs. the wild-type VH group, b p < 0.05 vs. the LMP7/MECL1-null VH group, andc p < 0.05 between the wild-type and LMP7/MECL1-null splenocytes with respect to the BKG, VH, or LAC treatment groups (two-way ANOVA with two-tailed Holm-Sidak post hoc analysis).
Expression of t-bet and GATA3 Is Decreased in LMP7/MECL1-Null Splenocytes
Thus far, our findings support the conclusion of decreased IFNγ and IL4 induction in activated LMP7/MECL1-null splenocytes compared to wild-type splenocytes. Accordingly, the expression of the key transcription factors that function to regulate IFNγ and IL4, specifically, t-box expressed in T cells (t-bet) and GATA-binding protein (GATA)3, respectively, was evaluated in LMP7/MECL1-null and wild-type splenocytes. Both constitutive and induced mRNA levels of t-bet and GATA3 were decreased in LMP7/MECL1-null splenocytes compared to the wild type (fig. 5). The differences in GATA3 induction were more marked than those in the induction of t-bet, with a 67% decrease in GATA3 induction and a 20% decrease in t-bet induction in LMP7/MECL1 double-null splenocytes compared to wild-type splenocytes. Lactacystin caused significant inhibition of t-bet transcription, but only a trend toward decreased GATA3 transcription in wild-type splenocytes was noted. Lactacystin also caused significant inhibition of t-bet mRNA expression in LMP7/MECL1-null splenocytes that was concentration dependent. In contrast, lactacystin had little effect on GATA3 transcription in LMP7/MECL1-null splenocytes.
Fig. 5.
Effect of lactacystin on inducible transcription factor expression in activated splenocytes derived from wild-type and LMP7/MECL1 double-knockout mice. Freshly isolated splenocytes (5 × 106 cells/ml) from wild-type or LMP7/MECL1 doubleknockout mice were treated with lactacystin (1–10 μmol/l) or VH (0.1% ethanol) for 30 min followed by activation of the cells with 40 nmol/l PMA/0.5 μmol/l ionomycin (Io). Cells were harvested 6 h later, the RNA isolated and analyzed for either t-bet (a) or GATA3 (b). Cellular viability was ≥80% for all treatment groups (assessed by trypan blue exclusion). The results are presented as fold induction over wild-type BKG and are the mean ± SE of triplicate cultures. a p < 0.05 vs. the wild-type VH group, b p < 0.05 vs. the LMP7/MECL1-null VH group, and c p < 0.05 between the wild-type and LMP7/MECL1-null splenocytes with respect to the BKG, VH, or LAC treatment groups (two-way ANOVA with twotailed Holm-Sidak post hoc analysis).
LMP7 and MECL1 Do Not Regulate the Transcription Factors YY1 and STAT1
Because ying-yang 1 (YY1) has been reported to regulate IFNγ and IL4 and is itself regulated by the proteasome [18,19,20], the expression of YY1 was investigated in wild-type and LMP7/MECL1-null splenocytes. In addition, we also investigated the expression of signal transducer and activator of transcription (STAT)1, another transcription factor that regulates t-bet and IFNγ [21] and is regulated by the proteasome [22]. In contrast to t-bet and GATA3, neither lactacystin nor LMP7/MECL1 had substantial effects on YY1 or STAT1 mRNA levels (fig. 6).
Fig. 6.
Effect of lactacystin on YY1 and STATI expression in activated splenocytes derived from wild-type and LMP7/MECL1 double-knockout mice. Freshly isolated splenocytes (5 × 106 cells/ml) from wild-type or LMP7/MECL1 double-knockout mice were treated with lactacystin (1-10 μmol/l) or VH (0.1% ethanol) for 30 min followed by activation of the cells with 40 nmol/1 PMA/0.5 μmol/l ionomycin (Io). Cells were harvested 6 h later, the RNA isolated and analyzed for either YY1 (a) or STATI (b). Cellular viability was >80% for all treatment groups (assessed by trypan blue exclusion). The results are normalized to wild-type BKG and are the mean ± SE of triplicate cultures. a p < 0.05 vs. the wild-type VH group,b p < 0.05 vs. the LMP7/MECL1-null VH group, and c p < 0.05 between the wild-type and LMP7/MECL1-null splenocytes with respect to the BKG, VH, or LAC treatment groups (two-way ANOVA with two-tailed Holm-Sidak post hoc analysis).
Discussion
The present studies are the first to demonstrate that the inducible proteasomal subunits LMP7 and MECL1 play a role in the mRNA expression of particular cytokines, such as IFNγ, IL4, and IL10, but not others, such as IL2 and IL13. In addition, our studies demonstrate that expression of the cytokine-regulating transcription factor GATA3 is also regulated by LMP7 and/or MECL1. The current studies are fully consistent with reports that mice deficient in LMP7 and/or MECL1 have reduced cytokine production in response to influenza epitopes and T. gondii [6,7]. Collectively, the aforementioned studies demonstrate that LMP7 and MECL1 regulate the production of IFNγ, IL4, IL10, IL2Rβ, t-bet, and GATA3 and, importantly, that these effects are independent of antigen processing and presentation.
Interestingly, IL6 is not detectably influenced by lactacystin treatment, which is in contrast to what we have observed previously in LPS-activated macrophages [8]. We have, however, previously reported the differential inhibitory effect of lactacystin on cytokine production in LPS-activated macrophages in which IL1β and IL6 were suppressed by lactacystin whereas TNFα was relatively refractory [11]. Collectively, our studies suggest that the sensitivity of cytokine transcription to suppression by lactacystin appears to be specific to the cell type and/or mode of activation.
Although induction of IFNγ is already markedly decreased in LMP7/MECL1-null splenocytes, it is further suppressed by lactacystin (fig. 1). The aforementioned findings suggest that, although IFNγ is regulated to a large extent by LMP7 and/or MECL1, other proteasomal proteases are likely involved in IFNγ regulation. Although levels of t-bet expression are lower in LMP7/MECL1-null splenocytes (fig. 5), the decreased expression is relatively modest in comparison to the extent of reduction in IFNγ production (fig. 1). Furthermore, t-bet expression levels were further decreased to a substantial extent by lactacystin in LMP7/MECL1-null splenocytes. Collectively, these findings suggest that although t-bet is regulated to a limited extent by LMP7 and/or MECL1, it is largely regulated by one or more of the other proteasomal proteases.
Interestingly, IL10 induction is markedly diminished in LMP7/MECL1-null splenocytes but actually increases upon lactacystin treatment, suggesting a non-LMP7/non-MECL1 protease is negatively regulating IL10 (fig. 2). The lack of effect of lactacystin on wild-type splenocytes may be the result of inhibition of multiple proteasomal proteases by lactacystin, some of which are involved in the upregulation (LMP7/MECL1) and others in the downregulation of IL10 expression. Alternatively, the effect of lactacystin on IL10 mRNA levels may be due to indirect or off-target effects of lactacystin.
Previous studies suggest that in addition to the constitutive proteasome and the immunoproteasome, there are also ‘intermediate’ proteasomes that are assembled from a combination of constitutively expressed and inducible subunits. Specifically, intermediate proteasomes with subunits β1, β2, and LMP7 as well as β2, LMP2, and LMP7 have been identified in various human tissue types, including the liver, kidney, and gastrointestinal tract [23]. It is not clear whether intermediate proteasomes are expressed in the MECL1/LMP7-null mice. It is also unclear what effect intermediate proteasomes would have on cytokine production in our model. Further studies will be needed to determine the role of intermediate proteasomes in cytokine induction by activated immune cells.
Recent studies suggest multiple roles for LMP7 in immune cells. In addition to its role in antigen processing, LMP7 has also been shown to be involved in cell stress and immune function. With respect to oxidative stress, LMP7 has been shown to be important for the clearance of poly-ubiquitinated oxidized proteins [24]. These studies demonstrate that increased levels of poly-ubiquitinated oxidized proteins in LMP7-null mice result in an increase in protein aggregates and a subsequent decrease in cell viability, suggesting a role for LMP7 in cell survival. With respect to cytokine production, another recent study shows inhibition of LMP7 with a selective inhibitor decreased IFNγ production in anti-CD3/anti-CD28-activated PBMCs, which is consistent with the current studies [25]. In addition, inhibition of LMP7 had no effect on the expression of IL2Rα (CD25), which is also consistent with our studies. In contrast to the effect of the LMP7 inhibitor on IFNγ production, there was little difference in IFNγ production between splenocytes from wild-type and LMP7–/– mice, which the authors attributed to compensatory mechanisms [25]. The decrease in IFNγ production by splenocytes from LMP7/MECL1–/– mice in comparison to the wild type suggests that such compensatory mechanisms are no longer functional in the double-knockout mice or, alternatively, that MECL1 also plays a role in the regulation of IFNγ production.
There is growing evidence to suggest a role for the immunoproteasome in the activation of nuclear factor (NF) κB. LMP2, in particular, has been implicated in the regulation of NFκB activation. A recent study showed decreased IκB degradation in LPS-stimulated B cells from LMP2–/– mice [26]. In addition, suppression of NFκB activation in TNFα-stimulated splenocytes from NOD mice is associated with impaired LMP2 expression [27]. Similarly, this group has also found decreased NFκB binding and IκB degradation in T2 cells, a human lymphocyte cell line that lacks the genes for LMP7 and LMP2 [28]. Collectively, these studies suggest a role for LMP2 in NFκB activation. In contrast, the current study demonstrates no difference in the induction of TNFα, a gene regulated by NFκB, in splenocytes from wild-type and LMP7/MECL1–/– mice, suggesting NFκB activation is not dependent on the LMP7 and/or MECL1 subunits specifically.
The present study provides evidence to support the conclusion that the expression of most cytokines induced in PMA/ionomycin-activated splenocytes is regulated by LMP7, MECL1, and/or one of the other proteasomal proteases. This is likely because the proteasome is critical for the activation of transcription factors, such as NFκB, that function to regulate multiple cytokines [29]. Furthermore, it has been reported that a number of negative regulators of cytokine induction, such as hypoxia-inducible factor 1α and nuclear erythroid factor 2-related factor 2, are degraded by the proteasome [30,31,32]. Thus, the proteasome is responsible both for the activation of positive regulators, as well as the degradation and elimination of negative regulators of gene expression in general and cytokine induction in particular.
The current data demonstrate that LMP7/MECL1 contribute to the induction of IFNγ, t-bet, IL4, IL10, and GATA3 in activated splenocytes. IFNγ is the signature cytokine produced by TH1 cells, and t-bet is a key transcription factor involved in TH1 differentiation. In contrast, IL4 and IL10 are produced by TH2 cells, and GATA3 is a key TH2-polarizing transcription factor. Overall, this suggests that signaling through the immunoproteasome may be responsible for providing a cellular state that is permissive for the differentiation of T cells. In contrast, the early events of T-cell activation do not appear to be impacted by the LMP7/MECL1 subunits insofar as induction of IL2 and IL2Rα (CD25) are comparable between wild-type and LMP7/MECL1-null splenocytes.
Overall, the current data suggest that specific protease subunits within the proteasome may function to regulate one or more signaling factors that are distinct in the regulation of specific cytokines. Whereas it is established that the activity of different kinases and phosphatases dictate which cytokines are induced or suppressed in activated leukocytes, the present studies demonstrate that the activity of different proteases in the proteasome can similarly regulate cytokine/cytokine receptor expression in activated splenocytes. Collectively, these findings support the conclusion that the proteasomal proteases play a critical role in the transcription of cytokines, their receptors, as well as regulators of cellular differentiation which may ultimately determine the initiation, direction, and cessation of immune responses.
Acknowledgements
This work was supported in part by National Institutes of Health grants, GM050870 (N.Q.), AI054962 (N.Q.), RR016475 (C.E.R.), and ES018885 (C.E.R.). The authors thank Dr. Xiaoyu Tan, Julia Reis, and Jing Shen for technical assistance.
References
- 1.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 2.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Waters JB, Fruh K, Peterson PA. Proteasomes are regulated by interferon gamma: implications for antigen processing. Proc Natl Acad Sci USA. 1992;89:4928–4932. doi: 10.1073/pnas.89.11.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel PM. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 5.Groettrup M, Khan S, Schwarz K, Schmidtke G. Interferon-gamma inducible exchanges of 20S proteasome active site subunits: why? Biochimie. 2001;83:367–372. doi: 10.1016/s0300-9084(01)01251-2. [DOI] [PubMed] [Google Scholar]
- 6.Pang KC, Sanders MT, Monaco JJ, Doherty PC, Turner SJ, Chen W. Immunoproteasome subunit deficiencies impact differentially on two immunodominant influenza virus-specific CD8+ T cell responses. J Immunol. 2006;177:7680–7688. doi: 10.4049/jimmunol.177.11.7680. [DOI] [PubMed] [Google Scholar]
- 7.Tu L, Moriya C, Imai T, Ishida H, Tetsutani K, Duan X, Murata S, Tanaka K, Shimokawa C, Hisaeda H, Himeno K. Critical role for the immunoproteasome subunit LMP7 in the resistance of mice to Toxoplasma gondii infection. Eur J Immunol. 2009;39:3385–3394. doi: 10.1002/eji.200839117. [DOI] [PubMed] [Google Scholar]
- 8.Qureshi N, Perera PY, Shen J, Zhang G, Lenschat A, Splitter G, Morrison DC, Vogel SN. The proteasome as a lipopolysaccharide-binding protein in macrophages: differential effects of proteasome inhibition on lipopolysaccharide-induced signaling events. J Immunol. 2003;171:1515–1525. doi: 10.4049/jimmunol.171.3.1515. [DOI] [PubMed] [Google Scholar]
- 9.Qureshi N, Vogel SN, Van Way C 3rd, Papasian CJ, Qureshi AA, Morrison DC. The proteasome: a central regulator of inflammation and macrophage function. Immunol Res. 2005;31:243–260. doi: 10.1385/IR:31:3:243. [DOI] [PubMed] [Google Scholar]
- 10.Reis J, Tan X, Yang R, Rockwell CE, Papasian CJ, Vogel SN, Morrison DC, Qureshi AA, Qureshi N. A combination of proteasome inhibitors and antibiotics prevents lethality in a septic shock model. Innate Immun. 2008;14:319–329. doi: 10.1177/1753425908096855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen J, Reis J, Morrison DC, Papasian C, Raghavakaimal S, Kolbert C, Qureshi AA, Vogel SN, Qureshi N. Key inflammatory signaling pathways are regulated by the proteasome. Shock. 2006;25:472–484. doi: 10.1097/01.shk.0000209554.46704.64. [DOI] [PubMed] [Google Scholar]
- 12.Rockwell CE, Qureshi N. Differential effects of lactacystin on cytokine production in activated Jurkat cells and murine splenocytes. Cytokine. 2010;51:12–17. doi: 10.1016/j.cyto.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caudill CM, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. T cells lacking immunoproteasome subunits MECL-1 and LMP7 hyperproliferate in response to polyclonal mitogens. J Immunol. 2006;176:4075–4082. doi: 10.4049/jimmunol.176.7.4075. [DOI] [PubMed] [Google Scholar]
- 14.Sidak Z. Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc. 1967;62:626–633. [Google Scholar]
- 15.Berges C, Haberstock H, Fuchs D, Miltz M, Sadeghi M, Opelz G, Daniel V, Naujokat C. Proteasome inhibition suppresses essential immune functions of human CD4+ T cells. Immunology. 2008;124:234–246. doi: 10.1111/j.1365-2567.2007.02761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajasingh J, Bord E, Luedemann C, Asai J, Hamada H, Thorne T, Qin G, Goukassian D, Zhu Y, Losordo DW, Kishore R. IL-10-induced TNF-alpha mRNA destabilization is mediated via IL-10 suppression of p38 MAP kinase activation and inhibition of HuR expression. FASEB J. 2006;20:2112–2114. doi: 10.1096/fj.06-6084fje. [DOI] [PubMed] [Google Scholar]
- 17.Kanters E, Pasparakis M, Gijbels MJ, Vergouwe MN, Partouns-Hendriks I, Fijneman RJ, Clausen BE, Forster I, Kockx MM, Rajewsky K, Kraal G, Hofker MH, de Winther MP. Inhibition of NF-κB activation in macrophages increases atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2003;112:1176–1185. doi: 10.1172/JCI18580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye J, Cippitelli M, Dorman L, Ortaldo JR, Young HA. The nuclear factor YY1 suppresses the human gamma interferon promoter through two mechanisms: inhibition of AP1 binding and activation of a silencer element. Mol Cell Biol. 1996;16:4744–4753. doi: 10.1128/mcb.16.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walowitz JL, Bradley ME, Chen S, Lee T. Proteolytic regulation of the zinc finger transcription factor YY1, a repressor of muscle-restricted gene expression. J Biol Chem. 1998;273:6656–6661. doi: 10.1074/jbc.273.12.6656. [DOI] [PubMed] [Google Scholar]
- 20.Guo J, Casolaro V, Seto E, Yang WM, Chang C, Seminario MC, Keen J, Georas SN. Yin-Yang 1 activates interleukin-4 gene expression in T cells. J Biol Chem. 2001;276:48871–48878. doi: 10.1074/jbc.M101592200. [DOI] [PubMed] [Google Scholar]
- 21.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 22.Kim TK, Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 23.Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, Bousquet-Dubouch MP, Theate I, Parmentier N, Van den Eynde BJ. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci USA. 2010;107:18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, Kruger E. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 25.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, Shwonek P, Parlati F, Demo SD, Bennett MK, Kirk CJ, Groettrup M. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 26.Hensley SE, Zanker D, Dolan BP, David A, Hickman HD, Embry AC, Skon CN, Grebe KM, Griffin TA, Chen W, Bennink JR, Yewdell JW. Unexpected role for the immunoproteasome subunit LMP2 in antiviral humoral and innate immune responses. J Immunol. 2010;184:4115–4122. doi: 10.4049/jimmunol.0903003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T, Faustman D. NOD mice are defective in proteasome production and activation of NF-κB. Mol Cell Biol. 1999;19:8646–8659. doi: 10.1128/mcb.19.12.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi T, Faustman D. Essential role of human leukocyte antigen-encoded proteasome subunits in NF-κB activation and prevention of tumor necrosis factor-α-induced apoptosis. J Biol Chem. 2000;275:5238–5247. doi: 10.1074/jbc.275.7.5238. [DOI] [PubMed] [Google Scholar]
- 29.Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 30.Lukashev D, Klebanov B, Kojima H, Grinberg A, Ohta A, Berenfeld L, Wenger RH, Ohta A, Sitkovsky M. Cutting edge: hypoxia-inducible factor 1α and its activation-inducible short isoform I.1 negatively regulate functions of CD4+ and CD8+ T lymphocytes. J Immunol. 2006;177:4962–4965. doi: 10.4049/jimmunol.177.8.4962. [DOI] [PubMed] [Google Scholar]
- 31.Sitkovsky M, Lukashev D. Regulation of immune cells by local-tissue oxygen tension: HIF1α and adenosine receptors. Nat Rev Immunol. 2005;5:712–721. doi: 10.1038/nri1685. [DOI] [PubMed] [Google Scholar]
- 32.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]