Summary
Transcription by RNA polymerase II (Pol II) is a tightly controlled process critical to normal cellular metabolism. Understanding how transcriptional regulation is orchestrated has mainly involved identifying and characterizing proteins that function as transcription factors. During the past decade, however, an increasing number of long non-coding RNAs (lncRNAs) have been identified as transcriptional regulators. This revelation has spurred new discoveries, novel techniques, and paradigm shifts, which together are redefining our understanding of transcriptional control and broadening our view of RNA function. Here we summarize recent discoveries concerning the role of lncRNAs as regulators of mammalian mRNA transcription, with a focus on key concepts that are guiding current research in the field.
Keywords: transcription, RNA polymerase II, long non-coding RNA, chromatin, modular scaffold, nuclear compartmentalization
What was once a category comprised mostly of rRNAs and tRNAs, ncRNAs that function in gene expression are increasing both in number and in the diversity of biological systems they control. This deluge of newly discovered functional ncRNAs has been partially driven by deep-sequencing technologies that enabled the discovery that much of mammalian genomes are transcribed into a variety of different ncRNAs. The challenge has become categorizing these ncRNAs, understanding when and where they are transcribed, and determining if individual ncRNAs have a biological function. Meeting these goals has spawned new frontiers of discovery in varied fields of biology.
Here we focus on nuclear long ncRNA (lncRNA) regulators of mammalian mRNA transcription (long typically refers to >200 nt). This represents a relatively new, but rapidly growing class of ncRNAs. Although their mechanisms of action are still being characterized, regulatory themes are beginning to emerge. Below we discuss different mechanisms by which mammalian lncRNAs control transcription, summarizing representative recent discoveries. These categories do not encompass all discovered mechanisms by which lncRNAs control mRNA transcription; however, embody some of the better understood regulatory themes and current concepts in the field.
lncRNAs controlling chromatin structure
Perhaps the most frequently observed mechanism by which lncRNAs control transcription is by mediating changes in chromatin modifications at specific regions of the genome, which in turn controls the transcriptional state of genes in those regions. The emerging model is that lncRNAs interact directly with chromatin modifying complexes that either place or remove modifications on the histone proteins that constitute chromatin (Figure 1A). Most examples to date are lncRNAs that interact with histone methyltransferase complexes, which add methyl groups to specific locations on histone tails. Interaction with the lncRNA is critical for the placement of the methyl marks at specific regions of the genome. Knockdown of the lncRNA results in loss of the methyl marks. The prevailing model is that the lncRNAs target the chromatin modifying complexes to specific regions of the genome, although the mechanism of targeting is not yet understood [1, 2].
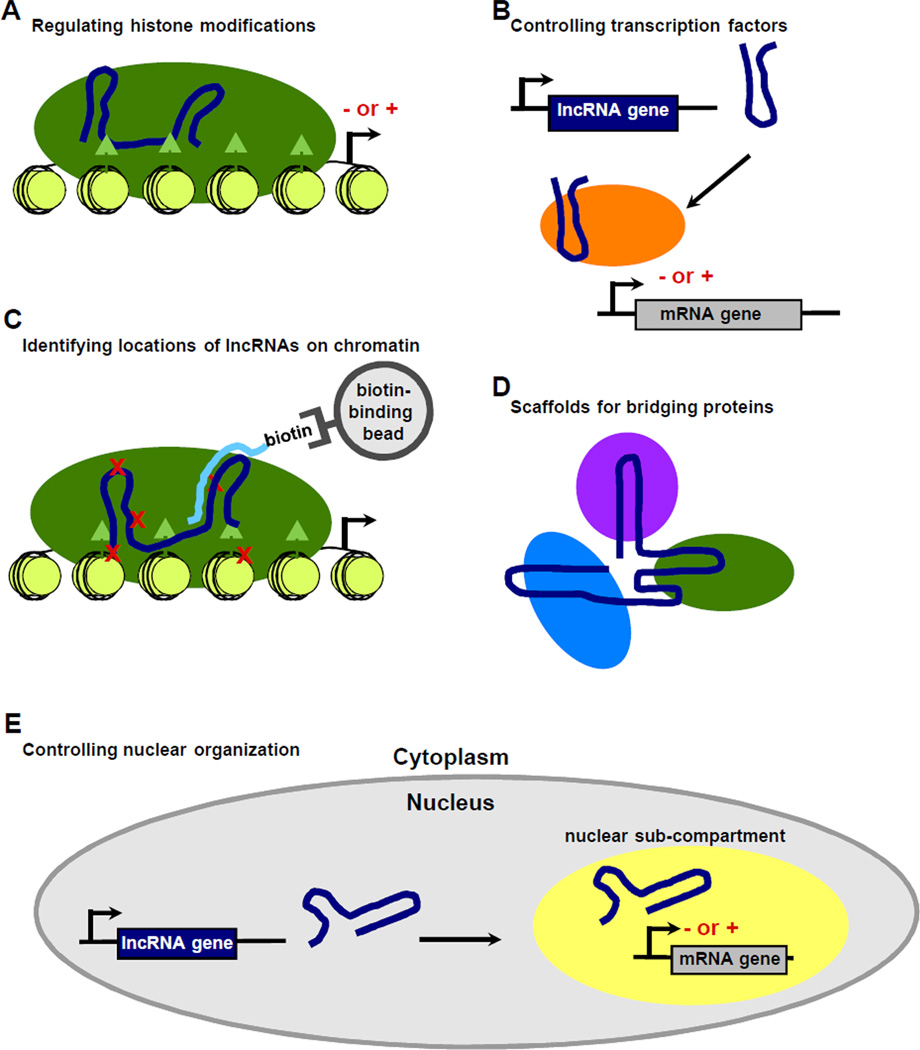
Figure 1.
lncRNAs that control transcription do so by a variety of mechanisms. (A) lncRNAs (blue) can bind to histone modifying complexes (green oval) and control the placement of histone modifications at specific regions of the genome. The histone modifications (triangles) can have a stimulatory (+) or inhibitory (−) effect on transcription. (B) lncRNAs can bind to and alter the activity of protein regulators of transcription, represented as an orange oval. They can be activating (+) or repressive (−). (C) Techniques have been developed to determine where on the genome an lncRNA localizes. Biotinylated oligos (light blue) that are antisense to lncRNAs (dark blue) can be used to enrich for lncRNA-associated chromatin that is purified from crosslinked (red X's) cells. A histone modifying complex is represented by the green oval, with histone modifications shown as triangles. (D) lncRNAs can serve as scaffolds to which multiple chromatin modifying or transcriptional regulatory proteins bind, indicated by the different colored ovals. (E) The nucleus in mammalian cells is compartmentalized, which is thought to facilitate transcriptional regulation. lncRNAs can be key structural components of sub-nuclear structures, including nuclear speckles, perinucleolar compartments, and paraspeckles.
Although similar in function, lncRNAs that regulate chromatin do so in a diversity of biological and disease systems. For example, recent work identified DBE-T, an lncRNA transcribed specifically in patients with facioscapulohumeral muscular dystrophy (FSHD) [3]. DBE-T recruits the histone methyltransferase ASH1L to the genomic locus previously understood to control FSHD. There, ASH1L places activating methyl marks on histones (e.g. tri-methylation of lysine 4 on histone H3, H3K4me3), leading to expression of genes that are otherwise silent in healthy individuals.
Other disease states, including cancer, have also been linked to lncRNAs that control chromatin [4–8]. One example is ANRIL (antisense noncoding RNA in the INK4 locus), which is transcribed in the antisense orientation from a locus that encodes three tumor suppressor proteins. ANRIL interacts with two histone methyltransferase complexes, PRC1 and PRC2 (polycomb repressive complexes 1 and 2), to confer repressive marks on chromatin and silence expression of the tumor suppressor proteins, which favors a state of proliferation [9, 10].
lncRNAs that control the state of chromatin are not limited to disease systems, but have also been shown to contribute to normal developmental processes. Indeed, the first discovered were those involved in controlling allele-specific gene expression. These include Xist, which silences of one of the X chromosomes in females (reviewed in [11]), as well as Kcnq1ot1 and Air, which control the allele-specific silencing of sets of genes at specific loci (reviewed in [12]). Each of these lncRNAs is thought to function by recruiting histone modifying complexes to a specific allele, or in the case of Xist, an entire chromosome. More recently, Kncq1ot1 was also found to regulate DNA methylation during allele-specific silencing by interacting with the DNA methyl transferase DNMT1 [13].
Other examples of lncRNAs that control the transcriptional state of chromatin include HOTTIP (HOXA transcript at the distal tip) and HOTAIR (Hox anti-sense intergenic RNA), which function in the development and establishment of body segmentation by regulating genes transcribed from Hox loci. HOTTIP binds to the histone methyl transferase WDR5/MLL to recruit it to the HOXA locus and activate transcription by conferring the H3K4me3 mark [14]. By contrast, HOTAIR represses transcription by interacting with the PRC2 complex and recruiting it the HOXD locus where it places the H3K27me3 mark [15, 16]. HOTAIR has also been shown to represses transcription in metastatic cancers by reprogramming PRC2 occupancy genome-wide [5, 7]. When HOTAIR was knocked out of mice, no significant phenotype nor changes in the chromatin state of HOXD locus were observed, indicating that either mouse and human HOTAIR have different functionalities or that other mechanisms exist to control this locus in vivo [17].
Chromatin-regulating lncRNAs are also thought to function in pluripotency and differentiation. Indeed, dozens of lncRNAs in mouse embryonic stem (ES) cells impact lineage-specific gene expression and exit from the pluripotent state upon knockdown [18]. Moreover, many associate with chromatin modifying complexes, and knocking down either the lncRNA or the chromatin modifying complex with which it interacts resulted in significantly overlapping changes in gene expression [18]. A recent study identified lncRNAs in human ES cells that interact with subunits of PRC2, in addition to transcription factors that control either pluripotency (SOX2) or neurogenesis (REST) [19]. Indeed, knockdown of these lncRNAs impacted either pluripotency or neuronal differentiation. In other studies, transcription of the Oct4 gene, whose protein product is important for the self-renewal of ES cells, was controlled by an lncRNA transcribed from an Oct4 pseudogene locus [20]. The proposed mechanism of regulating Oct4 transcription involves interaction of the lncRNA with PRC2 and the G9a chromatin modifying complex, as well as other factors. Ultimately lncRNAs could emerge as widespread epigenetic control factors as more lncRNAs that interact with histone modifying complexes in a diversity of systems continue to be discovered.
lncRNAs controlling transcription factors
Several lncRNAs function as effector molecules that bind to and control the activity of proteins that themselves are transcription factors (Figure 1B). These lncRNAs are also referred to as trans-regulators (or regulators that function in trans). Moreover, they have the capacity to regulate many different genes, and in some cases, entire transcriptional programs.
Gas5 (growth-arrest-specific transcript 5) is an lncRNA that controls the action of the transcriptional regulatory protein glucocorticoid receptor (GR) [21]. The lncRNA folds into a structure that mimics the DNA element to which GR typically binds, thereby inhibiting GR transcription. As another example, PANDA (P21 associated ncRNA DNA damage activated) is a recently discovered lncRNA whose transcription is induced by p53 in response to DNA damage [22]. PANDA associates with a transcription factor protein named NF-YA and inhibits the induction of apoptotic genes upon DNA damage. Interestingly, in response to DNA damage, p53 activates a second lncRNA named lincRNA-p21, which binds to hnRNP-K [6]. This interaction is thought to mediate transcriptional inhibition of many genes in response to DNA damage.
Not all trans-regulatory lncRNAs are inhibitory. SRA (steroid receptor RNA activator) was first discovered as an lncRNA coactivator because it enhanced transcription by nuclear hormone receptors [23, 24]. Later it was found to coactivate transcription by MyoD, which functions in muscle cell differentiation [25]. Through the years other studies have revealed SRA to be a multifaceted lncRNA that also functions as part of corepressor complexes, as well as complexes involved in gene insulation [26–28].
Some lncRNAs target general transcription factors that function at most, if not all, genes. The transcription factor P-TEFb (positive transcription elongation factor b), which regulates the transition to productive transcript elongation, is negatively controlled by sequestration in a multi-subunit complex held together by 7SK RNA. The interaction between P-TEFb and 7SK RNA is reversible and controlled by other proteins in the complex, as well as cellular signals [29, 30]. lncRNAs have also been found to bind to and regulate Pol II itself. Mouse B2 RNA and human Alu RNA bind directly to Pol II, assemble into complexes at promoters, and inhibit transcription by blocking contacts between the polymerase and promoter DNA [31, 32]. The interaction between B2 RNA and Pol II also causes a conformational change in complexes at the promoter that prevents phosphorylation of the C-terminal domain on the largest subunit of Pol II, which is a critical signal for events in early transcription [33].
Determining the locations of lncRNA occupancy on the genome
Seminal to answering the question of how lncRNAs regulate transcription, or any process that occurs on the genome, is determining where on the genome these molecules localize. Early methods to localize ncRNAs on specific regions of chromatin included RNA TRAP (tagging and recovery of associated proteins) and ChOP (chromatin oligo-precipitation). RNA TRAP tags chromatin-associated lncRNAs in fixed cells by first annealing modified oligos antisense to the lncRNA [34, 35]. A horseradish peroxidase-conjugated antibody then recognizes the modified oligo and catalyzes a localized covalent deposition of a biotin tag on the surrounding chromatin proteins, which can be visualized by immunofluorescence or precipitated to allow the DNA to be probed by PCR. The ChOP assay uses biotinylated oligos antisense to lncRNAs to precipitate the lncRNAs and associated chromatin from formaldehyde crosslinked cells [31, 36]. PCR is then used to probe for specific locations on the genome with which the lncRNAs associated.
Two related techniques were recently developed to localize lncRNAs genome-wide. These techniques, termed ChIRP (chromatin isolation by RNA purification) and CHART (capture hybridization analysis of RNA targets), also take advantage of biotinylated oligos antisense to the lncRNA of interest [37, 38]. In general, the antisense oligos are added to a solution of fragmented chromatin prepared from crosslinked cells. The network of crosslinks allows the lncRNA, its bound proteins, and the associated chromatin to be isolated when the oligos are pulled out of solution using biotin-binding beads (Figure 1C). Deep sequencing of the oligo-enriched chromatin reveals the regions of the genome with which the target lncRNA was associated. Although similar in principle, the ChIRP and CHART techniques differ in their details, such as the crosslinking reagent and the number of antisense oligos used. CHART-seq data has shown that chromatin eluted with RNase H (specific for the hybrid between the lncRNA and the antisense DNA oligo used in purification) results in sequencing reads with higher specificity compared to other elution techniques [37]. These techniques are analogous to ChIP-seq, which uses antibodies to enrich for chromatin associated with specific proteins.
CHART-seq was used to identify the locations of the Drosophila roX2 lncRNA, which controls dosage compensation [37]. ChIRP-seq was used to identify the genomic localizations of the roX2 lncRNA in Drosophila cells, the telomerase RNA TERC in human cells, and HOTAIR in human cancer cells ectopically expressing HOTAIR [38]. In both cases, the locations of specific histone modifications and/or histone modifying complexes thought to be controlled by the lncRNAs were also probed. These techniques will allow researchers to determine whether mammalian lncRNAs localize to the same regions of the genome whose chromatin state the lncRNAs are thought to control. This information would provide insight into mechanisms by which lncRNAs function as well as allow direct versus indirect targets of the lncRNAs to be determined.
lncRNAs as structural scaffolds to bridge protein complexes
It is becoming apparent that some lncRNAs interact with multiple different proteins; in this way the lncRNAs act as modular scaffolds to bridge protein complexes (Figure 1D). The idea that lncRNAs could function as flexible scaffolds that tether different proteins was first proposed from studies of the yeast telomerase RNA, which was found to have three arms that each binds a distinct protein, all of which are then tethered to the reverse transcriptase protein bound to the central region of the RNA [39, 40]. Following this theme, the lncRNA HOTAIR was found to simultaneously interact with two histone modifying protein complexes [16]. A region in the 5'-end of HOTAIR binds PRC2 and a region near the 3' end of HOTAIR binds LSD1. A model arises in which HOTAIR can recruit both protein complexes to specific chromatin sites resulting in coupled H3K27 methylation (via PRC2) and H3K4 demethylation (via LSD1). More recently, the lncRNA Six3OS was found to bind PRC2 and Eya proteins, the latter of which are protein tyrosine phosphatases that function as transcriptional coregulators [41]. Other examples of lncRNAs proposed to function as molecular scaffolds are 7SK RNA and SRA RNA. 7SK RNA holds together a group of proteins that control the availability of active P-TEFb [42], while SRA RNA can bring together proteins functioning as transcriptional coactivators, RNA helicases, and gene insulators [43]. It seems likely that many known lncRNAs will be found to act as flexible, modular protein-interaction scaffolds in the near future.
lncRNAs controlling nuclear organization
The localization of genes to domains or bodies within the nucleus (e.g. nuclear speckles, paraspeckles, and perinucleolar compartment) has been found to correlate with the transcriptional activity of the genes [44]. For example, genes that localize to nuclear speckles are generally active, although the cause of this phenomenon is not fully understood. Several lncRNAs have been found associated with distinct nuclear bodies and potentially control the formation of these structures, as well as regulate the transcriptional state of associated mRNA genes (Figure 1E).
The lncRNA Kcnq1ot1 is involved in silencing genes in the Kcnq1 region of the paternal genome [12]. Interestingly, when Kcnq1ot1 RNA was expressed from an episome, it targeted the episome to the perinucleolar compartment [45], which is known to contain highly compact heterochromatin [44]. A separate study showed that the Kcnq1 domain of one of the parental chromosomes is often associated with the nucleolar compartment [36]. These observations indicate that the Kcnq1ot1 RNA both recruits the Kcnq1 domain on the paternal chromosome to the perinucleolar compartment and also causes silencing of transcription in the this domain; however, whether there is a causal relationship between these two phenomena remains unknown.
Two other lncRNAs found associated with nuclear bodies are NEAT1 (nuclear paraspeckle assembly transcript 1, also known as nuclear enriched abundant transcript 1 and MENepsilon/beta) and MALAT-1 (metastasis-associated lung adenocarcinoma transcript 1, also known as NEAT2) [46]. MALAT-1 was initially identified as being abundant in a variety of tumors [47, 48], and has been shown to transcriptionally up-regulate genes involved in cell motility, potentially contributing to metastasis [49]. MALAT-1 was found to localize to nuclear speckles (also known as interchromatin granule clusters) [46], and more recently to recruit growth-control genes to nuclear speckles where they become transcriptionally active [50]. Depletion of MALAT-1 in cells did not lead to loss of the nuclear speckles themselves, suggesting MALAT-1 is not be required for their maintenance [51]. Recently, several labs knocked MALAT-1 out in mice and found no obvious phenotypes, and no effect on the formation of nuclear speckles [52–54]. Knockout did affect transcription of several genes that neighbor the MALAT-1 gene in the genome, suggesting that the MALAT-1 lncRNA or transcription of the MALAT-1 gene acts in cis to control transcription of neighboring genes [54].
Although it is not yet known whether NEAT1 controls transcription, this lncRNA does control the formation of paraspeckles, a distinct class of nuclear bodies that retain certain mRNAs in the nucleus [55]. NEAT1 was initially discovered in a search for transcripts that are abundant in the nuclei of human cells, and localization studies determined that it was associated with paraspeckles [46]. Paraspeckles disappeared when NEAT1 was knocked down and the number of paraspeckles increased when NEAT1 was over-expressed, leading to the conclusion that NEAT1 plays an essential role as an architectural component of paraspeckles [56–59]. Moreover, knockdown of NEAT1 resulted in loss of structured and A to I edited mRNAs from paraspeckles [57]. Live cell imaging showed that paraspeckle formation and maintenance required ongoing transcription of NEAT1 [60]. Knockout of the NEAT1 gene in mice eliminated paraspeckles, however the mice showed no other apparent phenotypes [61]. Hence, NEAT1 is the first example of an lncRNA that plays an active role in establishing a nuclear body; this precedent will likely lead to the discovery of other ncRNAs that function to control nuclear structure thereby regulating chromatin and transcription.
Future perspective
If the past several years are any indication, the discovery of new lncRNA regulators of gene expression will continue to advance at a rapid pace. This is likely to be fueled in part by the decreasing cost and increasing availability of high-throughput sequencing, which is arguably the simplest means to identify new transcripts and new lncRNA/protein interactions in a diversity of biological systems. The major challenge will remain the functional characterization of these novel lncRNAs and lncRNA/protein complexes. Knock-down studies have been extremely useful, however, it is difficult to delineate direct and indirect effects on gene expression due to reducing the level of an lncRNA. Therefore, additional mechanistic and structural studies will be required, which may well involve the development of new techniques. In addition, it will be important to better characterize known lncRNA regulators of transcription in order to understand their mechanisms of action. As one example, how lncRNA regulators of chromatin control the activity of histone modifying complexes and recruit them to specific regions of the genome remains a pressing question in the field. Lastly, the vast majority of functional studies of lncRNA regulators of gene expression have been performed with cultured cells. A limited number of mouse knock-out studies suggest that conclusions about the functions of lncRNAs in cultured cells might not hold true in vivo [17, 52–54]. Future studies are likely to uncover new modes of regulation, modify existing models, and better develop our current understanding of how lncRNAs function to control gene expression.
Executive summary.
lncRNAs controlling chromatin structure
lncRNAs interact with histone modifying complexes to direct the placement of histone modifications on specific regions of the genome, which in turn controls transcription.
Chromatin modifying lncRNAs function in a diversity of biological systems, including disease states and during normal cellular development and differentiation.
lncRNAs can direct the placement of both activating marks (e.g. HOTTIP and DBE-T) and repressive marks (e.g. HOTAIR and ANRIL).
The mechanism(s) by which lncRNAs target histone modifying complexes to specific regions of chromatin remains unknown.
lncRNAs controlling transcription factors
These lncRNAs regulate transcription at sites removed from their own site of transcription by binding to and controlling the activity of protein transcriptional regulators.
Some lncRNAs bind to gene-specific transcriptional activators in response to a cellular signal and repress the activator's activity; examples include PANDA and Gas5.
Some lncRNAs bind to and regulate proteins that function in the general transcription reaction; examples include 7SK binding P-TEFb and B2 or Alu RNA binding Pol II.
Determining the locations of lncRNA occupancy on the genome
New techniques have been developed to localize lncRNAs on the genome.;
The ChIRP and CHART techniques use antisense oligonucleotides against lncRNAs to enrich for the chromatin associated with the lncRNAs in crosslinked cells.
Chromatin enriched by ChIRP and CHART can be deep-sequenced to obtain a global view of where lncRNAs are localized on the genome.
These techniques will allow researchers to correlate regions bound by an lncRNA with regions at which the lncRNA affects chromatin and/or transcription, thereby shedding light on direct versus indirect targets of the lncRNA.
lncRNAs as structural scaffolds to bridge protein complexes
lncRNAs can act as scaffolds to tether different proteins and protein complexes together.
The functional role of the lncRNA scaffold differs depending on the system. For example, in the case of HOTAIR the scaffold coordinates histone modifications, whereas the 7SK RNA scaffold controls the availability and activity of P-TEFb.
lncRNAs controlling nuclear organization
lncRNAs are implicated in establishing nuclear bodies to which genes with different transcriptional states localize.
The mechanistic interplay between transcriptional activity, sub-nuclear localization, and lncRNAs such as Kcnq1ot1, MALAT-1 and NEAT1 are ongoing topics of study.
In the future, it will be important to determine whether lncRNAs that control transcript levels also control localization to nuclear bodies, and to determine the causal relationship between localization and transcriptional activity.
References
- 1.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 2010;20(2):142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cabianca DS, Casa V, Bodega B, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. **These studies identified an epigenetic switch controlled by an lncRNA (DBE-T), which provides a mechanistic understanding of why a specific chromosomal deletion results in Facioscapulohumeral muscular dystrophy.
- 4.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kogo R, Shimamura T, Mimori K, et al. Long non-coding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 8.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4(11):e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotake Y, Nakagawa T, Kitagawa K, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yap KL, Li S, Munoz-Cabello AM, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. *These experiments provide mechanistic insight into the interplay between a chromatin modifying complex (PRC1) interacting with an lncRNA (ANRIL) and/or a histone.
- 11.Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr. Opin. Genet. Dev. 2012;22(2):62–71. doi: 10.1016/j.gde.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4(5):277–286. [PubMed] [Google Scholar]
- 13. Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137(15):2493–2499. doi: 10.1242/dev.048181. * This study found that Kcnq1ot1 - in addition to controlling transcription via repressive histone modifications - also mediates DNA methylation at the Kncq1 locus by binding to DNMT1.
- 14. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. *This paper describes an lncRNA (HOTTIP) that activates transcription of genes within its own locus by recruiting chromatin modifying complexes to create a higher-order, looped chromosome structure.
- 15.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7(5):e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcr. 2010;1(3):165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. * High density promoter arrays of cell-cycle genes identified many lncRNAs whose expression changes with disease state or the cell cycle. One lncRNA, PANDA, was found to control transcription of pro-apoptotic genes by binding the transcription factor NF-YA.
- 23.Lanz RB, McKenna NJ, Onate SA, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97(1):17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 24.Foulds CE, Tsimelzon A, Long W, et al. Research resource: expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol. Endocrinol. 2010;24(5):1090–1105. doi: 10.1210/me.2009-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caretti G, Schiltz RL, Dilworth FJ, et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell. 2006;11(4):547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24(22):2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatchell EC, Colley SM, Beveridge DJ, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol. Cell. 2006;22(5):657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Downes M, Xie W, et al. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15(9):1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010;17(7):815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobhian B, Laguette N, Yatim A, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010;38(3):439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariner PD, Walters RD, Espinoza CA, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29(4):499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA. 2009;106(14):5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA represses TFIIH phosphorylation of RNA polymerase II. Transcr. 2011;2(1):45–49. doi: 10.4161/trns.2.1.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagano T, Mitchell JA, Sanz LA, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 35.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32(4):623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 36.Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 37.Simon MD, Wang CI, Kharchenko PV, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108(51):20497–20507. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. *This paper, along with reference 37, each describe methods to determine the locations of lncRNA occupancy across eukaryotic genomes that couple antisense oligo affinity purification of lncRNAs and associated chromatin from crosslinked cells with high throughput sequencing of recovered DNA.
- 39.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. U S A. 2004;101(27):10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zappulla DC, Cech TR. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 2006:71217–71224. doi: 10.1101/sqb.2006.71.011. [DOI] [PubMed] [Google Scholar]
- 41.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;632 doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He N, Jahchan NS, Hong E, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29(5):588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colley SM, Leedman PJ. Steroid Receptor RNA Activator - A nuclear receptor coregulator with multiple partners: Insights and challenges. Biochimie. 2011;93(11):1966–1972. doi: 10.1016/j.biochi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27(8):295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohammad F, Pandey RR, Nagano T, et al. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol. Cell. Biol. 2008;28(11):3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;839 doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 48.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 49.Tano K, Mizuno R, Okada T, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Yang L, Lin C, Liu W, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eissmann M, Gutschner T, Hammerle M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8) doi: 10.4161/rna.21089. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Rep. 2012;2(1):111–123. doi: 10.1016/j.celrep.2012.06.003. *Knockout of MALAT-1 in mice showed that it is not necessary for pre- and post-natal development and is not involved in general transcriptional regulation, but instead might function in cis to control transcription of neighboring genes.
- 55.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186(5):637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. **This paper provided compelling evidence that NEAT1 localizes to paraspeckles and is required for their formation.
- 57.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35(4):467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106(8):2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011;13(1):95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]