Abstract
We recently used in vitro selection to identify many deoxyribozymes that catalyze DNA phosphodiester bond hydrolysis and create 5′-phosphate and 3′-hydroxyl termini. Alternatively, numerous deoxyribozymes have been identified for catalysis of RNA cleavage by 2′-hydroxyl transesterification, forming 2′,3′-cyclic phosphate and 5′-hydroxyl termini. In this study, we investigated the ability of DNA to catalyze RNA cleavage by hydrolysis rather than transesterification, although normally the hydrolysis reaction is substantially disfavored relative to transesterification. Via a series of in vitro selection experiments, we found that reselection of a DNA-hydrolyzing deoxyribozyme leads either to transesterification or hydrolysis, depending on exclusion or inclusion of a stringent selection pressure for hydrolysis. An entirely new selection starting from a random DNA pool, using an all-RNA substrate and imposing the same selection pressure, also leads to RNA hydrolysis. Collectively, these results establish experimentally that small DNA sequences have the catalytic ability to direct a chemical reaction down a disfavored pathway, even when a more favorable mechanism is readily available. Our view of DNA catalysis is therefore expanded beyond merely increasing the rates of reactions that would have occurred more slowly without the catalyst.
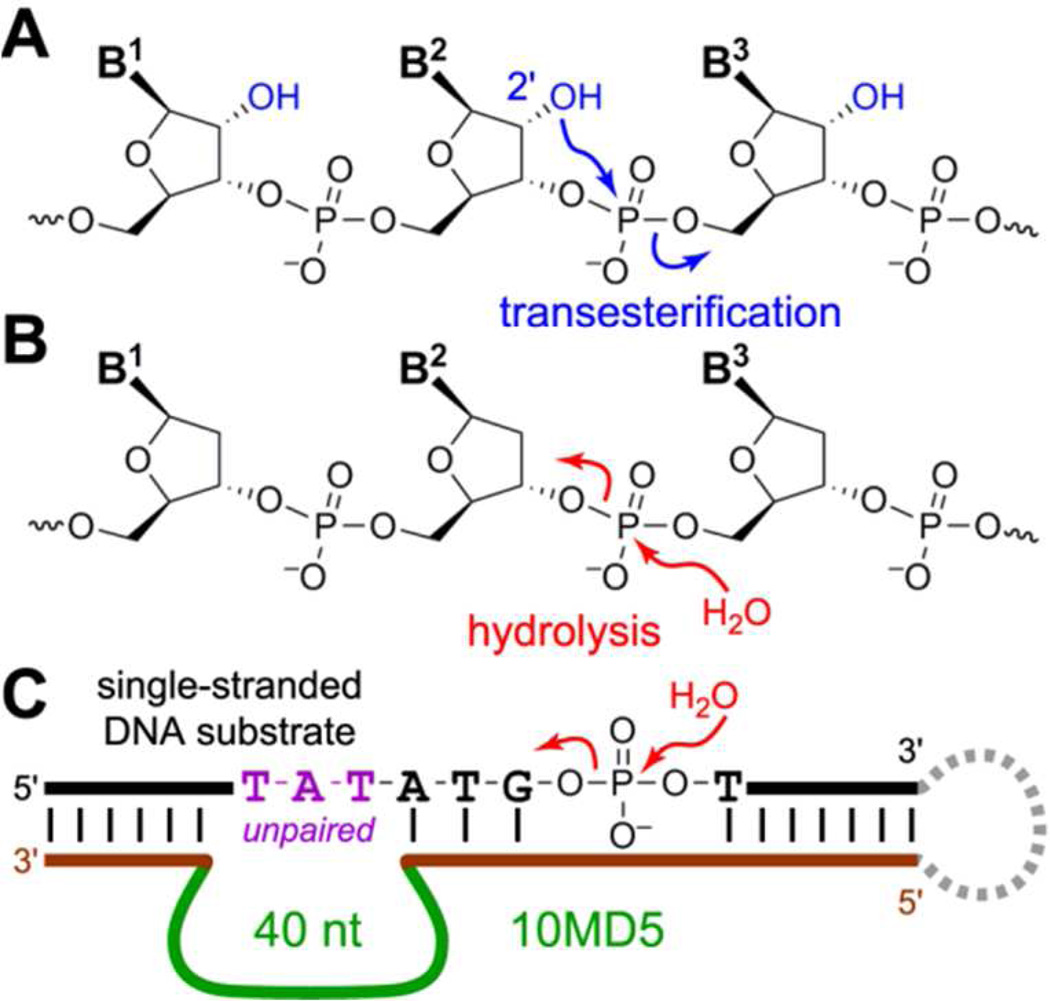
A catalyst is particularly useful when it can direct a reaction down a desired mechanistic pathway that would ordinarily be disfavored relative to one or more competing routes, especially when the inherent bias is strong against the desired pathway. Both natural and artifical proteins can catalyze normally disfavored reactions,1 although for some natural enzymes this may relate to historical contingency rather than mechanistic imperative.2 Nucleic acids (RNA and DNA) can also be catalysts,3,4 but in most known instances the reactions catalyzed by ribozymes and deoxyribozymes are not normally disfavored; the same reaction occurs both with and without the catalyst, merely faster when the catalyst is present. We are interested in understanding catalysis by DNA.5 Here we examined the ability of DNA to catalyze a normally disfavored reaction, RNA cleavage by hydrolysis of the phosphodiester bond, despite the inherent availability of an alternative and much more facile mechanistic pathway in which RNA cleavage by transesterification proceeds with nucleophilic attack of a ribose 2′-hydroxyl group on the neighboring phosphodiester bond (Figure 1A). RNA cleavage by transesterification has been reported many times by both ribozymes and deoxyribozymes.3 The uncatalyzed t1/2 values for transesterification and hydrolysis at near-neutral pH in the absence of divalent metal ions are ~10 years6 and ~30 million years,7 respectively, which is a 105-fold difference. The starting point for our experiments was the 10MD5 deoxyribozyme, which we identified as catalyzing DNA phosphodiester bond hydrolysis with formation of 5′-phosphate and 3′-hydroxyl termini (Figure 1B).8 The original 10MD5-catalyzed DNA hydrolysis reaction has no competing transesterification pathway, because the DNA substrate lacks 2′-hydroxyl groups.
Figure 1.
Oligonucleotide cleavage mechanisms and reaction catalyzed by the 10MD5 deoxyribozyme. (A) RNA cleavage by transesterification at phosphorus. Attack of a ribonucleotide 2′-hydroxyl group at the adjacent phosphodiester linkage leads to 2′,3′-cyclic phosphate and 5′-hydroxyl termini. (B) DNA cleavage by phosphodiester hydrolysis. Attack of a water molecule can form 5′-phosphate + 3′-hydroxyl termini as shown; formation of 3′-phosphate + 5′-hydroxyl is also possible. Competing transesterification cannot occur because no 2′-hydroxyl is present. (C) 10MD5-catalyzed DNA hydrolysis, showing the selection arrangement that enables PAGEshift selection (downward PAGE shift upon substrate cleavage; the dashed loop on the right side enables selection but is dispensable for catalysis). See Figure S1 for details.
10MD5 hydrolyzes its DNA substrate at a specific G^T dinucleotide junction located several nucleotides within the double-stranded region formed by interaction of the deoxyribozyme and substrate (Figure 1C). As a preliminary experiment to examine the effect on 10MD5 catalysis of including a 2′-OH at the scissile phosphodiester linkage, we evaluated the activity of 10MD5 when presented with a substrate that either has only a single ribonucleotide (rG) at the cleavage site or is entirely RNA. 10MD5 retained detectable activity with the rG substrate, albeit with ca. 40-fold lower kobs, but it was not measurably active with the all-RNA substrate (Figure S2). Therefore, introduction of the 2′-OH into the otherwise all-DNA substrate inhibits 10MD5 catalysis, and changing the surrounding nucleotides to RNA is further detrimental.
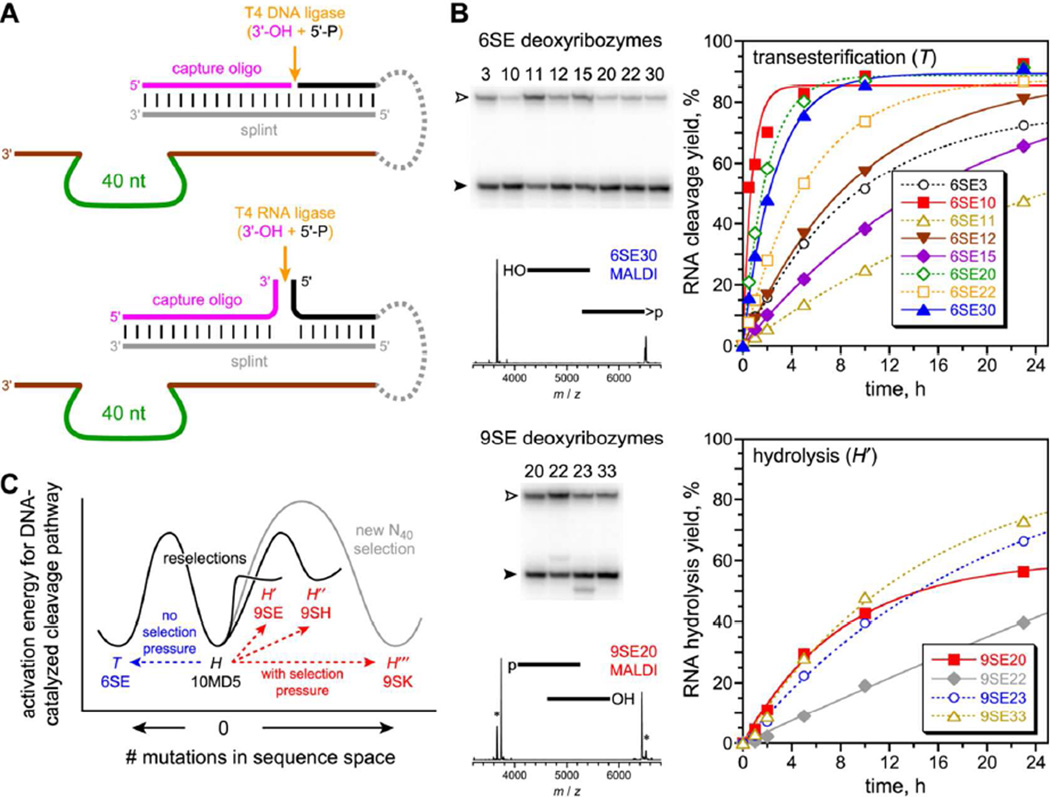
On this basis, we performed several in vitro selection experiments, each of which revealed a different facet of DNA’s ability to catalyze the normally disfavored RNA hydrolysis reaction. Some of these experiments imposed a stringent selection pressure for phosphodiester hydrolysis rather than transesterification, by requiring that the DNA-catalyzed substrate cleavage reaction forms a 5′-phosphate terminus (Figure 2A). This selection pressure either used T4 DNA ligase and an exactly complementary DNA splint, or it used T4 RNA ligase and a DNA splint that leaves several product nucleotides unpaired near the ligation site (see detailed nucleotide sequences in Figure S1).
Figure 2.
10MD5 partial randomization and reselection with exclusion or inclusion of hydrolysis selection pressure. (A) Hydrolysis selection pressure by capture of the 5′-phosphate hydrolysis product using a 3′-hydroxyl capture oligonucleotide, a DNA splint, and T4 DNA or RNA ligase. The 5′-hydroxyl formed by transesterification cannot be captured by this reaction. (B) Selection outcomes, revealed by analysis of individual deoxyribozymes from round 6 (no pressure, top) and round 9 (three additional rounds with pressure, bottom). Open arrowhead = substrate; filled arrowhead = product (t = 24 h). Asterisks denote 9SE signals that arise from transesterification rather than hydrolysis, either as background reaction (for PAGE) or during analysis (for MALDI-MS). See Table S1 for MALDI-MS data and cleavage-site assignments for individual deoxyribozymes. See Figure S3 for selection progressions and Figure S4 for individual deoxyribozyme sequences. See Table S2 kobs values. (C) Schematic model for accessing the competing transesterification (T) and hydrolysis (H, H′...) mechanistic pathways in DNA sequence space, starting from the parent 10MD5 sequence and ending with the deoxyribozymes from each of the four cloned selection rounds. This diagram is intended to assist visualization of the relationships among the various selection experiments and plots activation energy (ΔG‡) versus number of mutations, unlike a conventional energy diagram of free energy (G) versus reaction coordinate. See Supporting Information text for a brief explanation of the components of the diagram.
First, we partially randomized the 10MD5 sequence to the extent of 25%; i.e., each of the 40 catalytic region DNA nucleotides had 75% probability of being the parent nucleotide and 25% probability of being one of the other three nucleotides. This partially randomized 10MD5 pool was subjected to reselection using the original DNA substrate sequence, now containing a single ribonucleotide (rG) at the 10MD5 (r)G^T cleavage site. The selection process relied on simple downward PAGE shift of active deoxyribozyme sequences due to cleavage of the attached oligonucleotide substrate (Figure 1C). After six rounds in which the key selection step used incubation conditions of 70 mM HEPES, pH 7.5, 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, and 150 mM NaCl at 37 °C for 14 h (the same as in the original identification of 10MD5), the pool yield was 54% (Figure S3). Individual deoxyribozymes were cloned, and all eight that were examined were found to catalyze cleavage not by phosphodiester hydrolysis but instead by transesterification using the rG 2′-hydroxyl group as the nucleophile (Figure 2B, top, and Table S1). Their diverse sequences were essentially unrelated to 10MD5 (10–17 mutations, including many mutations within sequence segments found to be conserved in our prior study of 10MD5 variants;9 Figure S4). The overall outcome of this experiment can be depicted schematically as in Figure 2C, which illustrates that transesterification (T) catalyst sequences are readily accessible by reselection from the 10MD5 hydrolysis (H) catalyst as the starting point. Such sequences dominate in the absence of selection pressure to the contrary, consistent with the chemically more facile nature of transesterification compared to phosphodiester hydrolysis.
We resumed the same reselection experiment, now performing three additional rounds with imposition of the hydrolysis selection pressure of Figure 2A, using T4 DNA ligase to capture only those DNA catalyst sequences that cleave by hydrolysis at the rG site. The pool yield initially dropped substantially to undetectable (<0.5%) at round 7 but steadily rose to 20% by round 9 (Figure S3). Four out of four individual deoxyribozymes were observed to catalyze rG^T phosphodiester hydrolysis with 5′-phosphate formation (Figure 2B, bottom and Table S1), despite the presence of the rG 2′-hydroxyl group that could have enabled cleavage by transesterification. These hydrolytic deoxyribozymes have only 4– 9 mutations from 10MD5 and sequence conservation predicted well by our prior study of 10MD5 variants9 (Figure S4). Therefore, the normally favored T outcome in Figure 2C (as observed after six rounds) can be avoided by including the appropriate selection pressure for the normally disfavored H reaction. This outcome is observed even when the H pressure is applied at a relatively late stage of the selection process, indicating that H sequences were present in the population that experienced six rounds with no selection pressure and was therefore dominated by T sequences at that point.
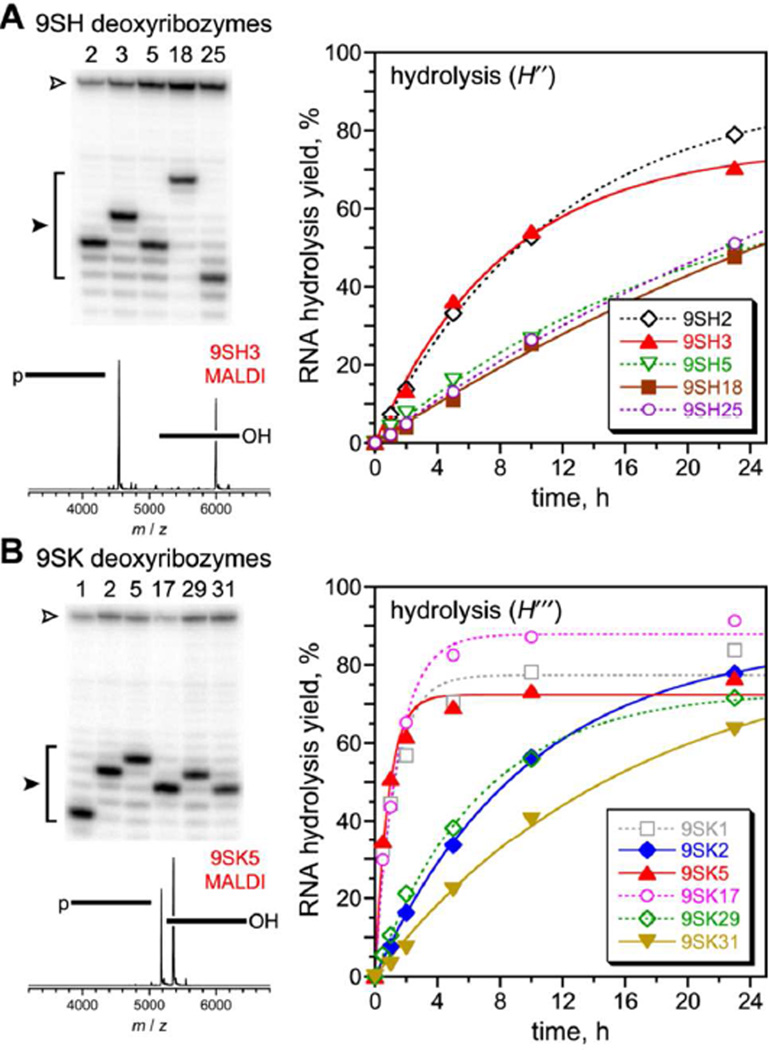
A separate 10MD5 reselection experiment was initiated, now using the all-RNA substrate for which 10MD5 has no measurable cleavage activity, rather than the DNA substrate with a single rG. The hydrolysis selection pressure of Figure 2A was imposed from the outset, now with T4 RNA ligase and a DNA splint that enables capture of the cleavage products from reaction at any of several possible RNA nucleotides. The all-RNA substrate alters the deoxyribozyme-substrate duplex from B-form DNA:DNA to A-form DNA:RNA. Given 10MD5’s inactivity with the all-RNA substrate (Figure S2), it was not known whether accessible 10MD5 variants can cleave the all-RNA substrate by hydrolysis. After nine rounds with the same key incubation conditions as above, the pool yield was 9% (Figure S3). Individual deoxyribozymes were each found to hydrolyze the all-RNA substrate at one of four nearby nucleotide junctions with 5′-phosphate formation (Figure 3A and Table S1). Each of the five new DNA catalyst sequences had 10–14 mutations relative to 10MD5 (Figure S4). Unlike the outcome with the single-rG substrate, here the new DNA sequences did not maintain 10MD5’s conserved motifs, which is sensible because here the substrate cleavage sites have changed; the new deoxyribozymes are essentially unrelated to 10MD5. This outcome indicates that many different hydrolytic catalyst sequences were readily accessible in sequence space, even with the constraint of starting the reselection process with a partially randomized version of the 10MD5 sequence. Without detailed biochemical investigation, we cannot know whether any of these new sequence-unrelated DNA catalysts nevertheless maintain any structural or catalytic features of 10MD5.
Figure 3.
DNA-catalyzed hydrolysis of all-RNA substrates. (A) 10MD5 reselection, seeking hydrolysis at the original cleavage site within the binding arm. (B) Entirely new N40 selection, seeking hydrolysis at the unpaired nucleotides.
Finally, we initiated a new selection experiment using a fully random N40 region, the all-RNA substrate, and the hydrolysis selection pressure (T4 RNA ligase) from the outset. We directed cleavage near a dinucleotide junction located within three unpaired nucleotides of the RNA substrate between the two DNA:RNA binding arms, rather than within the deoxyribozyme-substrate duplex region as for the 10MD5 reselection experiments. After nine rounds with the same key incubation conditions as above, the pool activity reached 28% (Figure S3). Consistent with the outcome of 10MD5 reselection using the all-RNA substrate, four different cleavage sites were used by individual deoxyribozymes (Figure 3B and Table S1), each of which catalyzes RNA hydrolysis with 5′-phosphate formation, and all six new DNA sequences were entirely unrelated to 10MD5 (26–36 nucleotide differences; Figure S4).
The divergent chemical outcomes of the two Figure 2B reselection experiments demonstrate that multiple mechanistic pathways for RNA cleavage by DNA catalysts are accessible depending on the selection pressure (Figure 2C), and the outcome can be controlled by a rather modest number of catalyst sequence differences. Escaping 10MD5’s hydrolysis (H) local minimum in DNA sequence space to access catalysts for the more facile transesterification (T) reaction could, in principle, have been analogous to finding RNA-cleaving hammerhead ribozymes as the simplest solutions in RNA sequence space.10 However, secondary structure calculations (mfold; data not shown) for the eight new transesterification DNA catalysts did not systematically predict any stem-loop structures similar to that of the small 8–17 RNA-cleaving DNA motif, which is thought to be the most common RNA cleavage solution in DNA sequence space.11 If the newly identified transesterification catalysts share any functional relationship with 8–17, then that relationship is not immediately obvious from the folding predictions. Although most of the round 6 pool catalyzed transesterification, a sufficient (albeit small) fraction of the round 6 pool must have been able to catalyze hydrolysis, such that three additional rounds with the hydrolysis selection pressure efficiently redirected the outcome12 to provide hydrolytic deoxyribozymes (i.e., H→H′ rather than H→T), despite the much greater chemical ease of transesterification.
The two selection experiments with the all-RNA substrate (Figure 3) validate that numerous apparently unrelated DNA sequences can catalyze RNA cleavage by hydrolysis rather than transesterification, despite the relative chemical challenge of hydrolysis. The reselection effort that was constrained primarily to survey sequences derived by mutation from 10MD5 nevertheless led readily to new DNA catalyst sequences (Figure 3A). Nevertheless, the successful new N40 selection shows that the partially randomized 10MD5 sequence was not strictly required as the starting point in order to find entirely different DNA catalysts for RNA hydrolysis that function well with the all-RNA substrate (Figure 3B).
RNA cleavage reactions catalyzed by large natural ribozymes such as group I introns,13 group II introns,14 and RNase P15 involve phosphodiester bond cleavage without transesterification by available 2′-hydroxyl groups, and a group I intron ribozyme was evolved to cleave single-stranded DNA.16 However, all of these natural (or naturally derived) ribozymes are an order of magnitude larger than the small artificial deoxyribozymes found in the present study, and notably, all of the small natural RNA-cleaving ribozymes catalyze only transesterification. 17 At the outset of our investigation, this situation left open the possibility that regardless of selection pressure, such small nucleic acid catalysts might be unable to suppress the mechanistic ease of transesterification in order to perform hydrolysis (e.g., evolve H→H′ rather than H→T in Figure 2C, or select for H rather than T from random sequences). Phosphodiester hydrolysis is inherently challenging, 7 and our previous work established that DNA can catalyze this difficult transformation for DNA substrates.8,9,18 The present results expand our understanding to reveal that phosphodiester hydrolysis by DNA catalysts can also be achieved for RNA substrates while readily avoiding the competing and much easier transesterification pathway that uses the 2′-hydroxyl group. This conclusion should invigorate searches for unrelated nucleic acid catalysts that promote a variety of normally disfavored chemical reactions.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants to S.K.S. from the National Institutes of Health (R01GM065966), the Defense Threat Reduction Agency (HDTRA1-09-1-0011), and the National Science Foundation (CHE0842534).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental details and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interests.
REFERENCES
- 1.(a) For example, natural polyketide synthase thioesterase domains form medium-sized rings even though such macrocyclization is normally disfavored relative to formation of smaller rings. Janda KD, Shevlin CG, Lerner RA. Science. 1993;259:490–493. doi: 10.1126/science.8424171. Gouverneur VE, Houk KN, de Pascual-Teresa B, Beno B, Janda KD, Lerner RA. Science. 1993;262:204–208. doi: 10.1126/science.8211138. Cravatt BF, Ashley JA, Janda KD, Boger DL, Lerner RA. J. Am. Chem. Soc. 1994;116:6013–6014. McDunn JE, Dickerson TJ, Janda KD. In: Catalytic Antibodies. Keinan E, editor. Wiley-VCH: Weinheim; 2005. pp. 184–216.
- 2.(a) Mohrig JR, Moerke KA, Cloutier DL, Lane BD, Person EC, Onasch TB. Science. 1995;269:527–529. doi: 10.1126/science.7624773. [DOI] [PubMed] [Google Scholar]; (b) Reynolds KA, Holland KA. Chem. Soc. Rev. 1997;26:337–343. [Google Scholar]
- 3.(a) Doudna JA, Cech TR. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]; (b) Schlosser K, Li Y. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]; (c) Silverman SK. Angew. Chem. Int. Ed. 2010;49:7180–7201. doi: 10.1002/anie.200906345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.This manuscript discusses DNA catalysts as enzymes for which the specific DNA sequence matters critically (analogous to protein enzymes as amino acid sequences), not DNA for templating or stereocontrol applications as discussed in ref. 3c.
- 5.Silverman SK. Acc. Chem. Res. 2009;42:1521–1531. doi: 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Breaker RR. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- 7.Schroeder GK, Lad C, Wyman P, Williams NH, Wolfenden R. Proc. Natl. Acad. Sci. USA. 2006;103:4052–4055. doi: 10.1073/pnas.0510879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra M, Sachdeva A, Silverman SK. Nat. Chem. Biol. 2009;5:718–720. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Xiao Y, Chandra M, Silverman SK. Biochemistry. 2010;49:9630–9637. doi: 10.1021/bi1013672. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xiao Y, Allen EC, Silverman SK. Chem. Commun. 2011;47:1749–1751. doi: 10.1039/c0cc04575f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salehi-Ashtiani K, Szostak JW. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- 11.Schlosser K, Li Y. ChemBioChem. 2010;11:866–879. doi: 10.1002/cbic.200900786. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Silverman SK. Biochemistry. 2005;44:3017–3023. doi: 10.1021/bi0478291. [DOI] [PubMed] [Google Scholar]
- 13.(a) Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]; (b) Cech TR. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 14.Pyle AM. Crit. Rev. Biochem. Mol. Biol. 2010;45:215–232. doi: 10.3109/10409231003796523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]; (b) Walker SC, Engelke DR. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Beaudry AA, Joyce GF. Science. 1992;257:635–641. doi: 10.1126/science.1496376. [DOI] [PubMed] [Google Scholar]; (b) Tsang J, Joyce GF. Biochemistry. 1994;33:5966–5973. doi: 10.1021/bi00185a038. [DOI] [PubMed] [Google Scholar]; (c) Tsang J, Joyce GF. J. Mol. Biol. 1996;262:31–42. doi: 10.1006/jmbi.1996.0496. [DOI] [PubMed] [Google Scholar]
- 17.Cochrane JC, Strobel SA. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 18.(a) Xiao Y, Wehrmann RJ, Ibrahim NA, Silverman SK. Nucleic Acids Res. 2012;40:1778–1786. doi: 10.1093/nar/gkr860. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dokukin V, Silverman SK. Chem. Sci. 2012;3:1707–1714. doi: 10.1039/C2SC01067D. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.