Abstract
An O18-labeling assisted LC-MS method was designed for unambiguous assignment of aspartyl/isoaspartyl products produced by Asn deamidation and Asp isomerization. By preparing the acid- and base–catalyzed deamidation standards in H2O18, isomer-specific mass tags were introduced to aspartyl- and isoaspartyl-containing peptides, which could be easily distinguished by mass spectrometry (MS). In contrast to the traditional ways to assign the isomers based on their elution order in reverse phase HPLC, the new method is more reliable and universal. Furthermore, the new method can be applied to the entire protein digest, and is therefore more time- and cost-effective compared with existing methods that use synthetic aspartyl- and isoaspartyl-containing peptide standards. Finally, since the identification of isomers in the new method only relies on LC-MS analysis, it can be easily implemented using the most basic and inexpensive MS instrumentation, thus providing an attractive alternative to tandem MS-based approaches. The feasibility of this new method is demonstrated using a model peptide as well as the entire digest of human serum transferrin.
INTRODUCTION
Deamidation of asparaginyl (Asn) and isomerization of aspartyl residues (Asp) in proteins are the most frequent non-enzymatic post-translational modifications (PTMs) in vivo.1 Under physiological conditions, both deamidation and isomerization proceed through formation of a succinimide intermediate, and lead to a mixture of aspartyl (Asp) and isoaspartyl residues (isoAsp).2,3 Asn deamidation introduces an additional negative charge on the protein surface; furthermore, formation of isoAsp residue elongates the protein backbone by inserting an extra methylene group, resulting in the β-peptide linkage. Thus, these modifications affect protein conformation,4 function,5, 6 activity,7, 8 stability,9 aggregation10 and even immune responses.11, 12In vivo, the level of isoAsp is controlled by protein L-isoaspartyl O-methyltransferase (PIMT), which catalyzes the conversion of isoAsp to Asp.13 The lack of such a mechanism in vitro leads to the accumulation of isoAsp with time as the Asn deamidation and Asp isomerization can also occur during protein production and long term storage. It is especially important for protein pharmaceuticals, where the effect of isoAsp formation has been widely examined.14 Therefore, close monitoring of Asn or Asp degradation products in protein pharmaceuticals is highly desirable and necessary.15
Traditional ways of detection and localizing isoAsp residues in stressed protein involve chemical hydrolysis and proteolysis. Edman sequencing is useful for identifying the formation of isoAsp residue, because these reactions stop at the iso-peptide bond.16 PIMT is also frequently applied, since it leads to the methylation of isoAsp residues as an intermediate step. Due to the longer retention time of methylated isoaspartyl-containing peptide compared to its aspartyl counterpart on RP-HPLC analysis, valid assignment can be made.17 Furthermore, the byproduct of the methylation reaction, S-Adenosyl-L-homocysteine (SAH), has been used to achieve the global analysis of the isoAsp residue content in protein samples, which has already been commercialized under the name IsoQuant.18, 19 The utility of endoproteinase Asp-N for the differentiation of aspartyl- and isoaspartyl-containing peptides has also been explored (taking advantage of the selective cleavage of aspartyl-containing peptides but not their isoaspartyl counterparts).20, 21 Detection of deamidated peptides by MS is relatively straightforward, since a mass shift of +0.984 Da occurs upon each deamidation event, which can be readily detected by MS with high resolution. However, the differentiation between aspartyl- and isoaspartyl-containing peptides from Asn deamidation or Asp isomerization presents a significant challenge for MS due to their identical mass. Even when separated by HPLC, unambiguous identification of the two isomers can be difficult using LC-MS analysis alone. In fact, most of the assignments were made based on either the ca. 3:1 relative peak intensity ratio or the elution order of the aspartyl- and isoaspartyl-containing peptides (isoaspartyl-containing peptides are believed to be more hydrophilic than their aspartyl counterparts due to the greater acidity at the side-chain, therefore have shorter retention time during RP-HPLC analysis). However, as have been reported by other researchers,21, 22, 23 the elution order is strongly dependant on the chromatographic conditions as well as the properties of the peptides. In fact, the inverted elution order (isoaspartyl-containing peptides have longer retention time than aspartyl-containing peptides) has been consistently observed in peptides with Asp/isoAsp residues located at the non-acetylated N-terminus.22,24 We have also observed examples with inverted elution order in this work (vide infra). Clearly, accurate assignment of aspartyl- and isoaspartyl-containing peptides cannot be achieved by elution order alone. Likewise, assignment of aspartyl- and isoaspartyl-containing peptides based on relative peak intensity ratio (ca. 1:3) is not always reliable. Since these peptides usually eluted at different time, their relative ionization efficiencies can be modulated differently by mobile phase and/or co-eluting species, and may not reflect their fractional concentrations in solution. Furthermore, the 1:3 ratio had been established based on the analysis of products of deamidation of short, unstructured peptides, while in proteins this may vary due to influence of the higher order structure.25,26
Other approaches to aspartyl- and isoaspartyl-containing peptides differentiation were mainly focused on applying different tandem mass spectrometry techniques. Several successful examples were reported by using different collision-activated dissociation (CAD) techniques, where the differentiation was mainly based on the specific reporter ions27 or the relative fragment ion intensity ratios.28 However, these approaches are highly dependent on the peptide sequence and very sensitive to experimental conditions. A much more successful strategy is the application of electron-based fragmentation techniques (electron capture dissociation, ECD, and electron transfer dissociation, ETD), which generate a pair of reporter ions (c·+57 and z-57) that are unique to isoaspartyl-containing peptides,29 and another pair of reporter ions (z-44 and (M+nH)(n-1)+·-60) that are unique to aspartyl-containing peptides.30, 31 This approach has already been applied in a high-throughput manner to achieve proteome-scale identification and quantitation of isoaspartyl residues in biological samples.32, 33 However, this technique is limited to ECD-capable instruments, and sometimes can be hampered due to the low abundance of the reporter ions.21
Recently, a number of O18-labeling based strategies were developed to facilitate the Asn deamidation or Asp isomerization studies.34, 35 Most of them were taking advantage of the increased mass increment after deamidation due to O18 incorporation (from 1 Da to 3 Da). Particularly, by using H2O18 for sample storage and protein digestion, in vitro and in vivo occurring deamidation processes can be readily differentiated.36, 37 However, none of these methods have explored the ability of using O18-labeling for aspartyl- and isoaspartyl-containing peptides differentiation and assignment. In this work, we demonstrate the possibility of using O18-labeling and LC-MS method for unambiguous differentiation of aspartyl- and isoaspartyl-containing peptides. Taking advantages of different deamidation mechanisms in acidic and basic environments, isomer-specific mass tags were introduced to O18-labeled aspartyl- and isoaspartyl-containing peptides, which could be easily distinguished by MS. Feasibility of this new method is demonstrated using a synthesized peptide EWSVNSVGK, followed by successful aspartyl- and isoaspartyl-containing peptides assignment for at least six peptides containing Asn residue, and two peptides containing Asp-Gly motif, from the tryptic digestion of an 80 kDa non-glycosylated form of human serum transferrin (hTf), which is a part of a number of biopharmaceutical products that are currently under development.15, 38, 39, 40, 41
MATERIALS AND METHODS
Non-glycosylated human serum transferrin (hTf) was provided by Prof. Anne B. Mason (University of Vermont College of Medicine, Burlington, VT); Peptides EWSVNSVGK were custom synthesized by Peptide 2.0 (Chantilly, VA, USA); Proteomic-grade trypsin and H2O18 (97% purity) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). All other chemicals and solvents used in this work were of analytical grade or higher.
The synthesized peptide (~0.4 mg) was dissolved in 20 μL of DMSO and 178 μL of H2O18, and then split into two equal volumes. Acid-catalyzed deamidation and base-catalyzed deamidation were initiated by the addition of 1 μL of TFA (pH ca. 1) and 1 μL of ammonium hydroxide (pH ca. 10) respectively. Both solutions were incubated at 50 °C, and sampled over 48 hours by taking an aliquot every 12 hours. The extent of deamidation was monitored by LC-MS analysis.
Deamidation of hTf and consequent identification of deamidated sites and aspartyl- and isoaspartyl-containing peptides assignment were carried out as follows. A solution containing 100 μg of hTf in 200 μL of 0.1 M ammonium bicarbonate (pH 8.0) was reduced with 10 mM of DTT at 50 °C for 20 min, and followed by alkylation of free sulfhydrals with 20 mM of iodoacetic acid (50 °C for 20 min in the dark). After removing the excess of iodoacetic acid by a spin concentrator with a molecular weight cutoff of 10,000 Da, the digestion was initiated by addition of trypsin at a 15:1 substrate to enzyme ratio and followed by incubation at 37 °C for 4 hours. The tryptic peptides were recovered by spinning down the digestion solution (50 μL of 100 μM ammonium bicarbonate buffer was added to wash the filter and recovered and combined with the digestion solution). After drying the digestion products under a stream of nitrogen, the acid- and base-catalyzed deamidation was carried out using the procedure outlined above (24 hours of incubation).
The O18-labeled deamidation standards were analyzed by LC/MS/MS using an LC Packings Ultimate (Dionex/Thermo Fisher Scientific, Sunnyvale, CA) nano-HPLC system coupled with a QStar-XL (AB SCIEX, Toronto, Canada) hybrid quadrupole/time-of-flight mass spectrometer. The samples were first loaded onto a trap column (C18 PepMap 100, 5 μm, 100 Å, 300 μm i.d. × 1 mm) at a relatively high flow rate (30 μL/min) for preconcentration and cleaning. Then the peptides were resolved using a C18 column (Acclaim PepMap 100 C18, 3 μm, 75 μm i.d. × 15 cm) at a flow rate of 0.2 μL/min. For the model peptide, the gradient was as follows: 0-20% solvent B in 6 min, 20-26% solvent B in 40 min, 26-80% solvent B in 3 min, 80% solvent B in 2 min, followed by 0% solvent B in 10 min. For the tryptic digests of hTf, the following gradient was used: 0-16% solvent B in 6 min, 16-24% solvent B in 40 min, 24-36% solvent B in 30 min, 36-80% solvent B in 4 min, 80% solvent B in 5 min, followed by 0% solvent B in 15 min. The mobile phase A was water with 0.1% formic acid. Mobile phase B was acetonitrile with 0.1% formic acid.
The ECD MS/MS experiments were carried out using an Agilent 1100 HPLC system (Agilent Technologies, Santa Clara, CA) coupled with a SolariX 7 (Bruker Daltonics, Billerica, MA) Fourier transform ion cyclotron resonance mass spectrometer. The O18-labeled deamidation standards were separated by RP-HPLC, and the produced ions were selected and subjected to ECD MS/MS to produce fragment ions.
RESULTS AND DISCUSSION
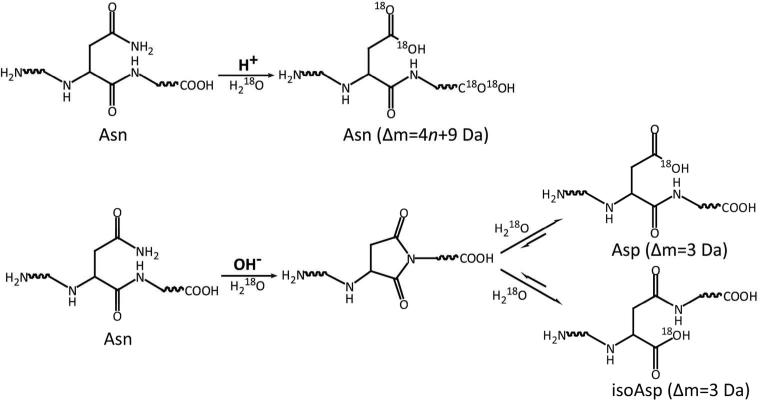
The mechanisms of Asn deamidation under both basic conditions and acidic conditions have been extensively studied.2 Unlike deamidation under basic conditions, which generates both aspartyl- and isoaspartyl-containing peptides, acid-catalyzed deamidation only leads to formation of aspartyl-containing peptides. By preparing the forced deamidation standards in H2O18 under different conditions, different levels of O18 incorporation in aspartyl- and isoaspartyl-containing peptides can be achieved, which can be exploited for unambiguous isomer differentiation (Scheme 1). As previously reported,42 acid-catalyzed labeling and deamidation result in O18 incorporation in all carboxylic groups, leading to a maximum mass increment of 4n + 9 Da (4 Da from each acidic residues, 4 Da from C-terminus, and 5 Da from deamidation-formed Asp residue), where n is the number of acidic residues and carboxymethylated Cys residues in this peptide. On the other hand, base-catalyzed deamidation only leads to a mass increment of 3 Da (1 Da from deamidation and 2 Da from incorporation of one O18 atom), because only one O18 atom is incorporated during the hydrolysis of succinimide intermediate. Indeed, due to the much lower rate of Asp/isoAsp isomerization under basic condition, the formation and subsequent hydrolysis of succinimide intermediate for the second time is limited within the experimental time-scale.
Scheme 1.
Preparation of O18-labeled deamidation standards
Under basic conditions, Asn deamidation in proteins occurs at a much lower rate compared to deamidation of short unstructured peptides due to the protection afforded by the higher order structures. Therefore, both acid- and base-catalyzed deamidation standards were prepared using peptides as starting material. Some variations in the deamidation rate under basic conditions were still observed, since this process is influenced by the sequence of the peptide, especially the neighboring residue on the C-terminal side of Asn. Particularly, deamidation occurs much faster in Asn followed by a polar residue with a relatively small side chain (Gly, Ser, Thr and Asp), compared to that followed by a hydrophobic residue with a bulky side chain (Val, Leu and Ile).43 In order to accelerate the deamidation process, elevated pH (1% NH4OH, pH ~ 10) and temperature (50 °C) were applied. In contrast, the deamidation rate of Asn under acidic conditions is barely affected by the peptide sequence. By incubating the model peptide EWSVNSVGK in H2O18 at 50 °C and in the presence of 1% TFA, the deamidation process was monitored by LC/MS analysis overtime. After 24 hours’ incubation, nearly 15% of the peptides were in aspartyl form, and the isotopic peak with maximum O18 incorporation could be readily detected (see Supplementary Material). It is probably worth to mention that the peptide cleavage is a side reaction during acid-catalyzed deamidation, although significant peptide cleavage was only observed at Asp-Pro site (the most susceptible site for acid cleavage) for 24 hours’ incubation. Thus, the acid- and base-catalyzed deamidation standards were both prepared by 24 hours’ incubation using the conditions described above.
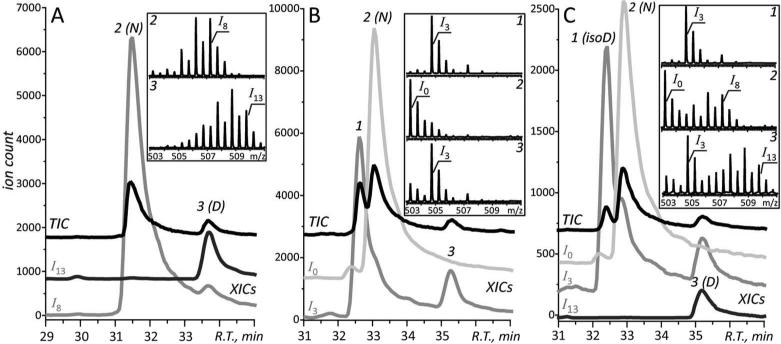
Forced deamidation of the model peptide EWSVNSVGK was carried out in H2O18 under both acidic and basic conditions using the procedure described above. Subsequently, the O18-labeled deamidation standards were analyzed by LC-MS. As previously reported,42, 44 oxygen atom exchange occurs between carboxylic groups and solvent at low pH, leading to a maximum mass increment of 8 Da (I8) for peptide EWSVNSVGK (Figure 1A). In addition, the hydrolysis of the amide group in Asn residue under acidic conditions leads to an additional mass increase of up to 5 Da (the I13 species). Indeed, only one deamidation product with a maximum mass increase of 13 Da (designated as peak 3 in the chromatogram shown in Figure 1A) is observed under acidic conditions. In contrast, the base-catalyzed deamidation leads to the formation of two indistinguishable deamidation products (designated as peaks 1 and 3 in the chromatogram shown in Figure 1B), with a maximum mass increase of 3 Da (the I3 species) for both. The analysis of the mixture of acid- and base-catalyzed deamidation standards reveals co-elution of species I3 and I13 (peak 3), which can be unambiguously assigned to aspartyl-containing peptide. The other chromatographic peak for species I3 (peak 1) can be assigned to isoaspartyl-containing peptide (Figure 1C). The corresponding mass spectra also allow the unambiguous differentiation between the two isomers to be made.
Figure 1.
LC-MS analysis of the O18-labeled acid-catalyzed (A) and base-catalyzed (B) deamidation standards generated using model peptide EWSVNSVGK, and their mixture (C). The total ion chromatogram (TIC) is shown in black trace, and the extracted ion chromatogram (XIC) of ionic species I0, I3, I8, and I13 are shown in grey trace, respectively. The elution peaks are labeled with numbers, and the corresponding mass spectra are shown in the insets. N: Asn; D: Asp; isoD: isoAsp.
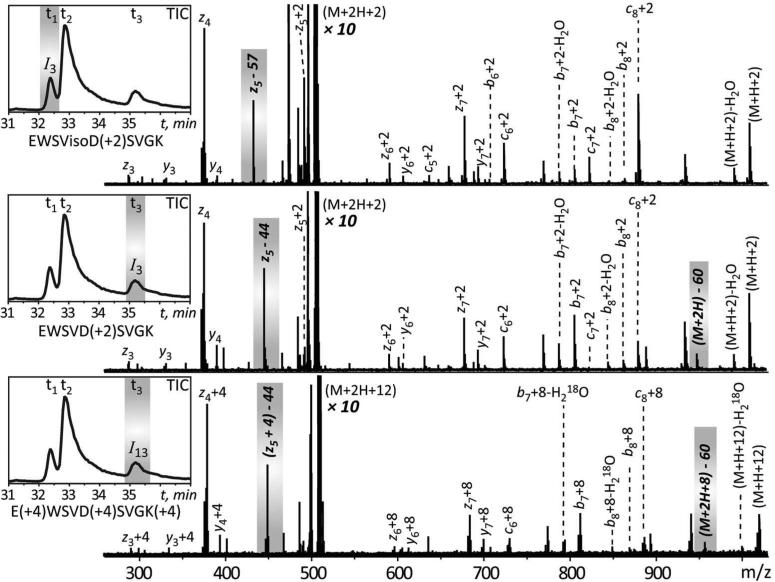
To validate this approach, a mixture of acid- and base-catalyzed deamidation standards prepared from model peptide EWSVNSVGK was further analyzed by LC-MS/MS using ECD to produce fragment ions. ECD MS/MS analysis allows a distinction to be made between aspartyl- and isoaspartyl-containing peptides, since it can generate a pair of reporter ions (cr·+58 and zk-r-57) that are unique to isoaspartyl-containing peptides, and another pair of ions (zk-r-44 and (M+nH-60)(n-1)+·) that are unique to aspartyl-containing peptides (r is the position of Asp/isoAsp residue from the N terminus and k is the total number of residues in the peptide). Both ion I3 and ion I13 (elution at t1 and t3) were selected for ECD MS/MS analysis (Figure 2). The reporter ion (z5-57) was only observed from the fragmentation of ion I3 at elution time t1, which confirmed the identity of this species as base-catalyzed isoaspartyl-containing peptide. On the other hand, both (z5-44) and (M+2H-60)+· fragment ions were observed in the ECD spectrum of species I3 acquired at elution time t3, but not from the fragmentation of species I3 at elution time t1, which confirmed the identity of this species as base-catalyzed aspartyl-containing peptide. In addition, the corresponding ions (z5+4)-44 and (M+2H+8)-60 with additional incorporation of O18 atoms were observed in the fragmentation of ion I13, which confirmed the identity of this species as acid-catalyzed aspartyl-containing peptide. Despite the O18 incorporation in the carboxylic group of the Asp residue, the reporter ions did not carry any O18 atom due to the consequent loss of O18 atoms in the form of either CO2 (44 Da) or C2H4O2 (60 Da) during the fragmentation. These results are in excellent agreement with the proposed mechanism29 for the neutral loss involving Asp residues during ECD fragmentation. The ECD MS/MS analysis clearly validates the O18-labeling approach to identification of aspartyl- and isoaspartyl-containing peptides in deamidated peptides.
Figure 2.
The ECD MS/MS analysis of O18-labeled deamidation standards generated using model peptide EWSVNSVGK. The inset in each panel represents the total ion chromatogram (TIC), and the ion selected for each ECD MS/MS analysis is highlighted by the grey area. The signature ions for aspartyl- and isoaspartyl-containing peptides are also highlighted by the grey area. The number in brackets after the amino acid residues indicates the mass increment in this residue due to O18 incorporation.
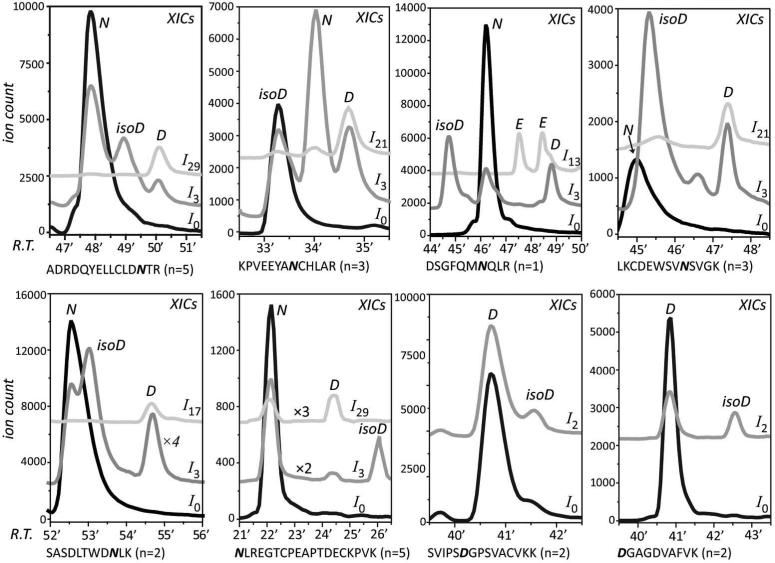
Following validation of the new methodology with ECD MS/MS, it was applied to detect and identify deamidation products of an 80 kDa plasma protein hTf, whose ability to traverse physiological barriers and be internalized by malignant cells is attracting significant attention in the biopharmaceutical sector. Deamidation of protein pharmaceuticals is always a big concern during the protein production and storage. A general procedure to monitor the deamidation in proteins is by detecting the deamidated peptides in the protein digest (mostly by trypsin) mixtures. However, as has been reported before,23 the traditional overnight tryptic digestion can introduce false positives (artificial deamidation, especially in Asn-Gly motif). Therefore, in this study, a shortened digestion time (4 hours) was used to minimize the digestion-induced deamidation. In addition, an extra step of removing alkylation reagent as well as an increased enzyme to substrate ratio was applied in order to improve the digestion efficiency. The tryptic digests were further subjected to acid- and base-catalyzed deamidation in H2O18, and followed by LC/MS analysis. Successful assignment of aspartyl- and isoaspartyl-containing peptides was achieved for six Asn-containing peptides and two Asp-Gly motif containing peptides from hTf. Based on the number of acidic residues in the peptide (n), a specific ion (I4n+9) was extracted together with ion I3. As discussed earlier, the co-eluting species I4n+9 and I3 indicated the presence of aspartyl-containing peptide, while the other elution peak of ion I3 indicated the presence of isoaspartyl-containing peptide. In the case of peptide DSGFQMNQLR, due to the presence of two Gln residues which are also targets of acid-catalyzed deamidation, three peaks of ion I13 eluting at different times were observed. Nevertheless, even in this case the assignment of aspartyl-containing peptide can still be made based on the co-elution of one of such peaks with one of the elution peaks of ion I3. By subtracting, the other two peaks of ion I13 can be assigned to either one of the Gln-deamidated peptides. Furthermore, by extracting ion I2, the isoaspartyl-containing peptide produced by Asp isomerization can be easily detected. Since Asp isomerization occurs much slower than Asn deamidation under basic conditions, it was only observed in peptides with an Asp-Gly motif, which is the most susceptible isomerization site.
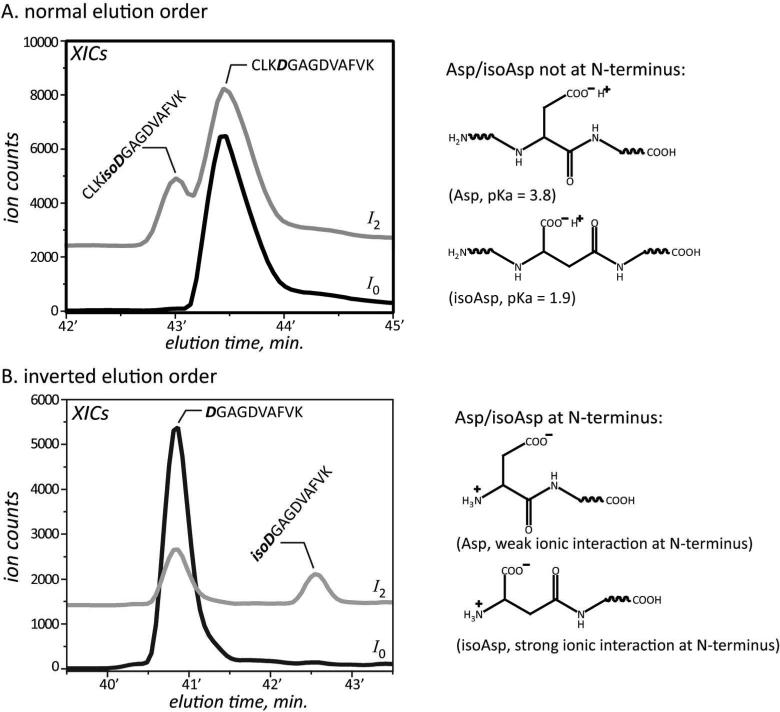
The elution order of these aspartyl- and isoaspartyl-containing peptides was mostly in agreement with the general observation that isoaspartyl-containing peptides elute earlier than aspartyl-containing peptides due to the greater acidity at the side-chain. However, the inverted elution order did occur in the case of peptides NLREGTCPEAPTDECKPVK and DGAGDVAFVK, where both the Asn and Asp-Gly were localized at the N-terminus. It is possible that the more favorable interaction between the N-terminal amine group and the carboxyl group of isoAsp residue could increase the overall hydrophobicity of the peptide, leading to longer elution time (Figure 4). Indeed, another peptide CLKDGAGDVAFVK, as an extended version of peptide DGAGDVAFVK with three more residues at the N-terminal side of Asp, exhibited ‘normal’ elution order vis-a-vis aspartyl- and isoaspartyl-containing peptides. This could be explained by the unfavorable ionic interaction at N-terminus. Furthermore, peptide SVIPSDGPSVACVKK also exhibited inverted elution order after Asp isomerization, probably due to the hydrogen bonding between the side-chains of Ser and isoAsp residues. These results suggest the unreliability of assigning the aspartyl- and isoaspartyl-containing peptides based on the elution order. Finally, formation of isoaspartyl-containing peptide was generally observed to be more efficient than the formation of aspartyl-containing peptide during the base-catalyzed deamidation of tryptic peptides, which could also be used to support the assignment.
Figure 4.
Examples of peptides exhibited both normal and inverted elution order for their aspartyl- and isoaspartyl-forms, and the possible explanations. The XICs of ionic species I0 and I2 are shown in black and grey, respectively.
CONCLUSIONS
A new method based on O18-labeling and LC-MS detection was developed for unambiguous assignment of aspartyl- and isoaspartyl-containing peptides produced by Asn deamidation and Asp isomerization. By preparing the acid- and base–catalyzed forced deamidation standards in H2O18, specific mass tags were introduced to aspartyl- and isoaspartyl-containing peptides, which could be easily distinguished by MS. Compared to the traditional method where the assignment of the isomers is based on the assumption of elution orders, this new method is more accurate and reliable. Furthermore, since the method can be applied to the entire digests of a protein, multiple assignments can be made simultaneously. The procedure is considerably more cost-effective and faster than the use of synthetic aspartyl- and isoaspartyl-containing peptide standards. The new method also offers an attractive alternative to the direct ECD MS/MS analysis of deamidation products, which relies on detection of reporter ions and, therefore, may not produce a conclusive answer if the fragment ion intensity is insufficient. Furthermore, isomer identification can be carried out with the new methodology using a generic and inexpensive LC-MS system without tandem capabilities, such as single quadrupole MS.
Supplementary Material
Figure 3.
The aspartyl- and isoaspartyl-containing peptides assignment from the tryptic digestion of hTf. The number in brackets after the peptide sequence indicates the total number of acidic residues and carboxymethylated Cys residues in this peptide. The XICs of ionic species I0, I3 and I4n+9 are shown in black, dark grey and light grey, respectively. N: Asn; D: Asp; isoD: isoAsp; E: Glu.
ACKNOWLEDGMENTS
This work was supported by Grant R01 GM061666 from the National Institutes of Health. Fourier transform ion cyclotron resonance mass spectrometer was acquired using a grant CHE-0923329 from the National Science Foundation under the Major Research Instrumentation program. The authors are grateful to Prof. Anne B. Mason (University of Vermont College of Medicine, Burlington, VT) for providing the transferrin samples and to Dr. Cedric E. Bobst and Mr. Guanbo Wang (UMass-Amherst) for assistance with ECD experiments.
REFERENCES
- 1.Geiger T, Clarke S. Deamidation, Isomerization, and Racemization at Asparaginyl and Aspartyl Residues in Peptides - Succinimide-Linked Reactions That Contribute to Protein-Degradation. J Biol Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 2.Clarke S. Propensity for Spontaneous Succinimide Formation from Aspartyl and Asparaginyl Residues in Cellular Proteins. Int J Pept Prot Res. 1987;30:808–821. doi: 10.1111/j.1399-3011.1987.tb03390.x. [DOI] [PubMed] [Google Scholar]
- 3.Reissner KJ, Aswad DW. Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious signals? Cell Mol Life Sci. 2003;60:1281–1295. doi: 10.1007/s00018-003-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noguchi S. Structural Changes Induced by the Deamidation and Isomerization of Asparagine Revealed by the Crystal Structure of Ustilago sphaerogena Ribonuclease U2B. Biopolymers. 2010;93:1003–1010. doi: 10.1002/bip.21514. [DOI] [PubMed] [Google Scholar]
- 5.Curnis F, Longhi R, Crippa L, Cattaneo A, Dondossola E, Bachi A, Corti A. Spontaneous formation of L-isoaspartate and gain of function in fibronectin. J Biol Chem. 2006;281:36466–36476. doi: 10.1074/jbc.M604812200. [DOI] [PubMed] [Google Scholar]
- 6.Charache S, Fox J, Mccurdy P, Kazazian H, Winslow R. Post-Synthetic Deamidation of Hemoglobin Providence (Beta-82 Lys-]Asn Asp) and Its Effect on Oxygen-Transport. J Clin Invest. 1977;59:652–658. doi: 10.1172/JCI108683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman AR, Ichhpurani AK, Brown DM, Hillman RM, Krabill LF, Martin RA, Zurcherneely HA, Guido DM. Degradation of Growth-Hormone Releasing-Factor Analogs in Neutral Aqueous-Solution Is Related to Deamidation of Asparagine Residues - Replacement of Asparagine Residues by Serine Stabilizes. Int J Pept Prot Res. 1991;37:14–20. doi: 10.1111/j.1399-3011.1991.tb00727.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang LH, Li JR, Wroblewski VJ, Beals JM, Riggin RM. In vivo deamidation characterization of monoclonal antibody by LC/MS/MS. Anal Chem. 2005;77:1432–1439. doi: 10.1021/ac0494174. [DOI] [PubMed] [Google Scholar]
- 9.Solstad T, Flatmark T. Microheterogeneity of recombinant human phenylalanine hydroxylase as a result of nonenzymatic deamidations of labile amide containing amino acids - Effects on catalytic and stability properties. Eur J Biochem. 2000;267:6302–6310. doi: 10.1046/j.1432-1327.2000.01715.x. [DOI] [PubMed] [Google Scholar]
- 10.Nilsson MR, Driscoll M, Raleigh DP. Low levels of asparagine deamidation can have a dramatic effect on aggregation of amyloidogenic peptides: Implications for the study of amyloid formation. Protein Science. 2002;11:342–349. doi: 10.1110/ps.48702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mamula MJ, Gee RJ, Elliott JI, Sette A, Southwood S, Jones PJ, Blier PR. Isoaspartyl post-translational modification triggers autoimmune responses to self-proteins. Journal of Biological Chemistry. 1999;274:22321–22327. doi: 10.1074/jbc.274.32.22321. [DOI] [PubMed] [Google Scholar]
- 12.Doyle HA, Gee RJ, Mamula MJ. Altered immunogenicity of isoaspartate containing proteins. Autoimmunity. 2007;40:131–137. doi: 10.1080/08916930601165180. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu T, Matsuoka Y, Shirasawa T. Biological significance of isoaspartate and its repair system. Biol Pharm Bull. 2005;28:1590–1596. doi: 10.1248/bpb.28.1590. [DOI] [PubMed] [Google Scholar]
- 14.Aswad DW, Paranandi MV, Schurter BT. Isoaspartate in peptides and proteins: formation, significance, and analysis. J Pharmaceut Biomed. 2000;21:1129–1136. doi: 10.1016/s0731-7085(99)00230-7. [DOI] [PubMed] [Google Scholar]
- 15.Kaltashov IA, Bobst CE, Abzalimov RR, Wang GB, Baykal B, Wang SH. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol Adv. 2012;30:210–222. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edman P. Method for Determination of the Amino Acid Sequence in Peptides. Acta Chem Scand. 1950;4:283–293. [Google Scholar]
- 17.Paranandi MV, Guzzetta AW, Hancock WS, Aswad DW. Deamidation and Isoaspartate Formation during in-Vitro Aging of Recombinant Tissue-Plasminogen Activator. J Biol Chem. 1994;269:243–253. [PubMed] [Google Scholar]
- 18.Schurter BT, Aswad DW. Analysis of isoaspartate in peptides and proteins without the use of radioisotopes. Anal Biochem. 2000;282:227–231. doi: 10.1006/abio.2000.4601. [DOI] [PubMed] [Google Scholar]
- 19.Johnson BA, Aswad DW. Optimal Conditions for the Use of Protein L-Isoaspartyl Methyltransferase in Assessing the Isoaspartate Content of Peptides and Proteins. Analytical Biochemistry. 1991;192:384–391. doi: 10.1016/0003-2697(91)90553-6. [DOI] [PubMed] [Google Scholar]
- 20.Kameoka D, Ueda T, Imoto T. A method for the detection of asparagine deamidation and aspartate isomerization of proteins by MALDI/TOF-mass spectrometry using endoproteinase Asp-N. J Biochem. 2003;134:129–135. doi: 10.1093/jb/mvg120. [DOI] [PubMed] [Google Scholar]
- 21.Ni WQ, Dai SJ, Karger BL, Zhou ZH. S. Analysis of Isoaspartic Acid by Selective Proteolysis with Asp-N and Electron Transfer Dissociation Mass Spectrometry. Anal Chem. 2010;82:7485–7491. doi: 10.1021/ac101806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter D, Pipkorn R, Lehmann WD. Separation of peptide isomers and conformers by ultra performance liquid chromatography. J Sep Sc. 2009;32:1111–1119. doi: 10.1002/jssc.200800691. [DOI] [PubMed] [Google Scholar]
- 23.Krokhin OV, Antonovici M, Ens W, Wilkins JA, Standing KG. Deamidation of -Asn-Gly-sequences during sample preparation for proteomics: Consequences for MALDI and HPLC-MALDI analysis. Anal Chem. 2006;78:6645–6650. doi: 10.1021/ac061017o. [DOI] [PubMed] [Google Scholar]
- 24.Sargaeva NP, Goloborodko AA, O'Connor PB, Moskovets E, Gorshkov MV. Sequence-specific predictive chromatography to assist mass spectrometric analysis of asparagine deamidation and aspartate isomerization in peptides. Electrophoresis. 2011;32:1962–1969. doi: 10.1002/elps.201000507. [DOI] [PubMed] [Google Scholar]
- 25.Harris RJ, Kabakoff B, Macchi FD, Shen FJ, Kwong M, Andya JD, Shire SJ, Bjork N, Totpal K, Chen AB. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J Chromatogr B. 2001;752:233–245. doi: 10.1016/s0378-4347(00)00548-x. [DOI] [PubMed] [Google Scholar]
- 26.Kossiakoff AA. Tertiary Structure Is a Principal Determinant to Protein Deamidation. Science. 1988;240:191–194. doi: 10.1126/science.3353715. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez LJ, Shimizu T, Satomi Y, Betancourt L, Besada V, Padron G, Orlando R, Shirasawa T, Shimonishi Y, Takao T. Differentiating alpha- and beta-aspartic acids by electrospray ionization and low-energy tandem mass spectrometry. Rapid Commun Mass Sp. 2000;14:2092–2102. doi: 10.1002/1097-0231(20001130)14:22<2092::AID-RCM137>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann WD, Schlosser A, Erben G, Pipkorn R, Bossemeyer D, Kinzel V. Analysis of isoaspartate in peptides by electrospray tandem mass spectrometry. Protein Science. 2000;9:2260–2268. doi: 10.1110/ps.9.11.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cournoyer JJ, Pittman JL, Ivleva VB, Fallows E, Waskell L, Costello CE, O'Connor PB. Deamidation: Differentiation of aspartyl from isoaspartyl products in peptides by electron capture dissociation. Protein Sci. 2005;14:452–463. doi: 10.1110/ps.041062905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sargaeva NP, Lin C, O'Connor PB. Identification of Aspartic and Isoaspartic Acid Residues in Amyloid beta Peptides, Including A beta 1-42, Using Electron-Ion Reactions. Anal Chem. 2009;81:9778–9786. doi: 10.1021/ac901677t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cournoyer JJ, Lin C, O'Connor PB. Detecting deamidation products in proteins by electron capture dissociation. Anal Chem. 2006;78:1264–1271. doi: 10.1021/ac051691q. [DOI] [PubMed] [Google Scholar]
- 32.Yang HQ, Fung EYM, Zubarev AR, Zubarev RA. Toward Proteome-Scale Identification and Quantification of Isoaspartyl Residues in Biological Samples. Journal of Proteome Research. 2009;8:4615–4621. doi: 10.1021/pr900428m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang HQ, Lyutvinskiy Y, Soininen H, Zubarev RA. Alzheimer's Disease and Mild Cognitive Impairment are Associated with Elevated Levels of Isoaspartyl Residues in Blood Plasma Proteins. Journal of Alzheimers Disease. 2011;27:113–118. doi: 10.3233/JAD-2011-110626. [DOI] [PubMed] [Google Scholar]
- 34.Xiao G, Bondarenko PV, Jacob J, Chu GC, Chelius D. O-18 labeling method for identification and quantification of succinimide in proteins. Anal Chem. 2007;79:2714–2721. doi: 10.1021/ac0617870. [DOI] [PubMed] [Google Scholar]
- 35.Terashima I, Koga A, Nagai H. Identification of deamidation and isomerization sites on pharmaceutical recombinant antibody using (H2O)-O-18. Anal Biochem. 2007;368:49–60. doi: 10.1016/j.ab.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 36.Li XJ, Cournoyer JJ, Lin C, O'Cormora PB. Use of O-18 labels to monitor deamidation during protein and peptide sample processing. J Am Soc Mass Spectr. 2008;19:855–864. doi: 10.1016/j.jasms.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaza-Bulseco G, Li BQ, Bulseco A, Liu HC. Method to Differentiate Asn Deamidation That Occurred Prior to and during Sample Preparation of a Monoclonal Antibody. Anal Chem. 2008;80:9491–9498. doi: 10.1021/ac801617u. [DOI] [PubMed] [Google Scholar]
- 38.Kaltashov IA, C. E. B., Zhang M, Leverence R, Gumerov DR. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biochim. Biophys. Acta. 2012;1820:417–426. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bobst CE, Wang S, Shen W-C, Kaltashov IA. Mass spectrometry study of a transferrin-based protein drug reveals the key role of protein aggregation for successful oral delivery. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13544–13548. doi: 10.1073/pnas.1206924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen SN, Bobst CE, Kaltashov IA. Mass spectrometry-guided optimization and characterization of a biologically active transferrin-lysozyme model drug conjugate. Mol. Pharm. 2013;10:1988–2007. doi: 10.1021/mp400026y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luck AN, Mason AB. Structure and dynamics of drug carriers and their interaction with cellular receptors: Focus on serum transferrin. Advanced Drug Delivery Reviews. 2013 doi: 10.1016/j.addr.2012.11.001. in press (doi 10.1016/j.addr.2012.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang SH, Bobst CE, Kaltashov IA. Pitfalls in Protein Quantitation Using Acid-Catalyzed O(18) Labeling: Hydrolysis-Driven Deamidation. Anal. Chem. 2011;83:7227–7232. doi: 10.1021/ac201657u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson NE, Robinson AB. Molecular clocks. Proc. Natl. Acad. Sci. U. S. A. 2001;98:944–949. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niles R, Witkowska HE, Allen S, Hall SC, Fisher SJ, Hardt M. Acid-Catalyzed Oxygen-18 Labeling of Peptides. Anal. Chem. 2009;81:2804–2809. doi: 10.1021/ac802484d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.