SUMMARY
The ability of a bacterial pathogen to monitor available carbon sources in host tissues provides a clear fitness advantage. In the group A streptococcus (GAS), the virulence regulator Mga contains homology to phosphotransferase system (PTS) regulatory domains (PRDs) found in sugar operon regulators. Here we show that Mga was phosphorylated in vitro by the PTS components EI/HPr at conserved PRD histidines. A ∆ptsI (EI-deficient) GAS mutant exhibited decreased Mga activity. However, PTS-mediated phosphorylation inhibited Mga-dependent transcription of emm in vitro. Using alanine (unphosphorylated) and aspartate (phosphomimetic) mutations of PRD histidines, we establish that a doubly phosphorylated PRD1 phosphomimetic (D/DMga4) is completely inactive in vivo, shutting down expression of the Mga regulon. Although D/DMga4 is still able to bind DNA in vitro, homo-multimerization of Mga is disrupted and the protein is unable to activate trancription. PTS- mediated regulation of Mga activity appears to be important for pathogenesis, as bacteria expressing either nonphosphorylated (A/A) or phosphomimetic (D/D) PRD1 Mga mutants were attenuated in a model of GAS invasive skin disease. Thus, PTS-mediated phosphorylation of Mga may allow the bacteria to modulate virulence gene expression in response to carbohydrate status. Furthermore, PRD-containing virulence regulators (PCVRs) appear to be widespread in Gram-positive pathogens.
INTRODUCTION
Pathogens that infect humans must successfully gain access to essential nutrients in order to grow and persist in their host during infection. The ability to rapidly sense and respond to preferred carbon sources provides a significant selective growth advantage. Not surprisingly, bacteria have evolved pathways that sense the presence of preferred sugars, repress the utilization of non-preferred substrates, and regulate sugar metabolism based on this information flow (Deutscher et al., 2006, Gorke & Stulke, 2008). Given the importance of this process, one would predict that pathogenesis is linked to the nutrient supply of the bacteria in the host. In fact, genomic studies on many pathogens have found that genes encoding proteins involved in carbohydrate uptake, utilization, and metabolic regulation are up regulated in vivo and contribute to the disease process (Cho & Caparon, 2005, Iyer & Camilli, 2007, Moyrand et al., 2007, Munoz-Elias & McKinney, 2005, Rollenhagen & Bumann, 2006, Son et al., 2007). Although these connections have been appreciated for some time, the molecular mechanisms that allow regulatory interplay between virulence and sugar metabolism remain unclear.
The phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS) is a conserved multiprotein pathway found in many eubacteria that couples the transport of sugars across the cytoplasmic membrane with their phosphorylation (Deutscher et al., 2006). Importantly, the PTS also serves as a signal transduction system that monitors carbohydrate utilization based on the flux of phosphate through the pathway (Fig. 1A). The phosphorelay provides a flow of phosphate from PEP generated during glycolysis to the EI protein, and then to a catalytic histidine (His15) of HPr (Fig. 1A). HPr-His∼P in turn phosphorylates one of several sugar-specific EIIA proteins, which transfers the phosphate to its cognate EIIB, then ultimately phosphorylates the incoming sugar transported by EIIC. In Gram-positive organisms, HPr can also be phophorylated by ATP on a specific serine (Ser46) via HPr kinase/P-Ser-HPr phosphorylase (HPrK/P). HPr–Ser∼P is then able to interact with carbon catabolite protein A (CcpA) and bind to catabolite responsive elements (cre) sites in promoter regions and either repress or activate transcription of targets. Hence, when a favorable carbon source is present, carbon catabolite repression (CCR) is mediated by CcpA to prevent metabolism of an inferior sugar (Stulke & Hillen, 2000, Deutscher et al., 2006, Gorke & Stulke, 2008). However, in the absence of a rapidly metabolizable sugar, HPr–His∼P may begin to accumulate. Both HPr–His∼P and EIIB∼P are also capable of phosphorylating conserved histidines within PTS regulatory domains (PRDs) of transcriptional antiterminators (e.g., LicT) and activators, (e.g., MtlR, LevR) thereby modulating their ability to regulate the expression of alternative sugar operons (Deutscher et al., 2006). Thus, the PTS provides a sophisticated sensory pathway for monitoring the metabolism of carbohydrates in Gram-positive bacterial pathogens.
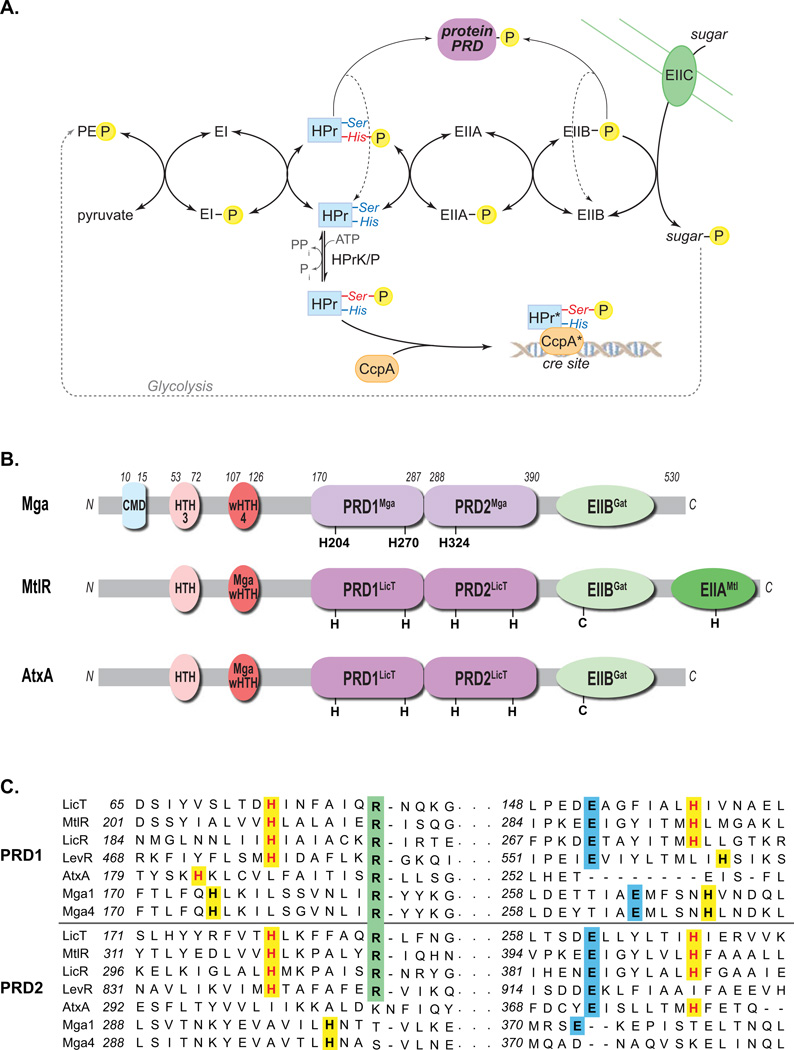
Figure 1. PTS pathway and alignment of Mga with PRD-containing regulators.
(A) The phosphotransferase system (PTS) in Gram-positive bacteria couples the phosphorylation and import of sugars. The general cytoplasmic enzymes (EI and HPr) and sugar-specific EII components form a phosphorelay to transfer phosphate from phosphoenol pyruvate (PEP) produced by glycolysis to the incoming sugar. Carbon catabolite repression (CCR) results from HPrK/P phosphorylation of serine 46 of HPr, which then complexes with CcpA to repress target promoters via cre sites. HPr-His∼P and EIIB∼P can also phosphorylate PTS regulatory domains (PRDs) of sugar regulators, thereby modulating their activity. (B) Domain alignment of GAS Mga with B. subtilis MtlR and B. anthracis AtxA. EII (green), PRD (purple), DNA-binding (red) and conserved (blue) domains are indicated with conserved histidines (H) and cysteines (C). PRDMga and PRDLicT refer to Pfam 08270 and Pfam 00874, respectively. (C) Amino acid sequence alignment of the PRDs from B. subtilis LicT, LicR, LevR, G. stearothermophilus MtlR, B. anthracis AtxA, and GAS Mga (M1 and M4 serotypes), highlighting the regions containing the conserved histidine (highlighted yellow), arginine (highlighted green), and aspartate (highlighted blue) residues. Histidine residues previously shown to be phosphorylated are colored red. (Supplemental Figure S1 contains the full alignment.)
The group A streptococcus (GAS) or Streptococcus pyogenes is a significant human-restricted pathogen causing a wide array of acute diseases (pharyngitis, impetigo, necrotizing fasciitis, streptococcal toxic shock syndrome) and autoimmune sequelae (rheumatic fever) in its host, resulting in over half a million deaths worldwide each year (Cunningham, 2000, Carapetis et al., 2005). A hallmark of GAS is the ability to successfully colonize and adapt to many different tissue sites in the human host. Both ex vivo and in vivo studies have firmly established that GAS exhibit significant changes in its transcriptome during infection (Cho & Caparon, 2005, Graham et al., 2005, Graham et al., 2006, Sumby et al., 2006, Virtaneva et al., 2005). Given that GAS are fastidious fermentative organisms that preferentially utilize sugars as carbon sources, it is not surprising that carbohydrate uptake (PTS & ABC transporters), metabolic operons, and CCR genes are induced in vivo and are required for full virulence in models of GAS infection (Kinkel & McIver, 2008, Loughman & Caparon, 2006, Shelburne et al., 2008c, Shelburne et al., 2008a, Shelburne et al., 2009, Shelburne et al., 2008b). Therefore, GAS appears to depend upon carbohydrate uptake systems such as the PTS for their ability to survive in the host and elicit disease.
In addition to classical two-component signal transduction systems (TCS) (Kreikemeyer et al., 2003), GAS utilize ‘stand-alone’ regulators representing transcription factors controlling virulence gene regulons by sensory components that have yet to be fully defined (McIver, 2009). The multiple virulence gene regulator (Mga) was the first such stand-alone regulatory network described in GAS and allows the pathogen to adapt and flourish in host environments favourable for growth (Hondorp & McIver, 2007). The gene encoding Mga (mga) has been found in all sequenced GAS genomes and strains tested, exhibiting two divergent alleles (mga-1, mga-2) that correlate with different tissues sites of infection (Bessen et al., 2005). Mga is critical for multiple pathogenic phenotypes, including biofilm formation, growth in whole blood, resistance to phagocytosis, and optimal virulence (Cho & Caparon, 2005, Graham et al., 2005, Hondorp & McIver, 2007, Virtaneva et al., 2005).
Mga strongly activates the transcription of a number of established GAS virulence genes during the exponential phase of growth (carbohydrate rich) and in conditions conducive to growth (optimal temperature, available iron). These “core” Mga-regulated genes encode mostly multifunctional surface molecules important for adherence to host tissues and evasion of the host immune responses, including M protein (emm), M-like proteins (arp), C5a peptidase (scpA), collagen-like protein (scp1, scpA), fibronectin-binding proteins (fba, sof), and the secreted inhibitor of complement (sic) (Hondorp & McIver, 2007). Transcriptomic analyses in several GAS serotypes found that Mga regulates over 10% of the genome, including sugar metabolism operons and ccpA (Ribardo & McIver, 2006) and synthesis of the Mga-regulated M protein can be influenced by specific sugars such as glucose (Ribardo & McIver, 2006, Pine & Reeves, 1978). CcpA was recently found to regulate mga expression via an upstream cre site (Almengor et al., 2007). Furthermore, Mga regulon expression was shown to peak during the acute phase of infection in a primate model of GAS pharyngitis, directly correlating with carbohydrate utilization genes (Virtaneva et al., 2005). Taken together, these findings suggest that Mga activity is linked to sugar metabolism; however, it is not clear how Mga is able to monitor the carbohydrate status.
The 62 kDa Mga protein (Fig. 1B) possesses two helix-turn-helix (HTH-3 and HTH-4) motifs and a conserved region (CMD) towards its amino terminus that are involved in DNA-binding activity, with the winged HTH domain (HTH-4) being absolutely required for binding to all Mga-binding sites tested thus far (McIver & Myles, 2002, Vahling, 2006). We have recently shown that the conserved carboxy-terminal region of Mga, containing a PTS EIIB-like domain, is important for oligomerization as well as regulon gene activation in vivo (Hondorp et al., 2012). In this study, an in silico analysis comparing Mga to proteins of known structure in the Protein Database (PDB) revealed two potential PTS regulatory domains (PRDs) in the central region of Mga. Inactivation of the PTS (∆ptsI) in an M4 GAS background led to alteration in Mga regulon expression. We also found that the conserved histidines within the PRDs of Mga are phosphorylated by the PTS, leading to a defect in protein oligomerization, altered gene expression and attenuation of virulence in a mouse model of GAS infection. We propose that as GAS encounters different sugar availabilities in the host environment, PTS-mediated phosphorylation of Mga serves to modulate protein activity (both positively and negatively) to regulate the Mga regulon, thereby linking Mga regulation of virulence directly to the sugar status of the cell.
RESULTS
Mga shares homology to PRD-containing regulators
To identify structural homologs to domains within Mga, we previously undertook an in silico analysis to compare Mga to proteins of known structure in the SCOP database (Andreeva et al., 2004, Deutscher et al., 2005, Hondorp & McIver, 2007). The central region of Mga was predicted to have strong structural homology to PTS regulatory domains (PRDs) in the B. subtilis antiterminator LicT, the only PRD-containing protein for which a structure had been determined (Deutscher et al., 2005, Hondorp & McIver, 2007). Interestingly, subsequent Pfam analysis indicates that the putative PRDs of Mga may possess a unique but related form of PRD, termed “PRD_Mga” (PF08270, here called PRDMga), distinct from the classic PRD of sugar-specific antiterminators and activators (PF00874, here called PRDLicT); however, both domains are members of the PRD clan (CL0166). To further investigate these findings, domain analysis of Mga (from serotypes M4 and M1T1) was performed using the Protein Homology/analogY Recognition Engine v2.0 (Phyre2, http://www.sbg.bio.ic.ac.uk/phyre2) algorithm and the current worldwide Protein Data Bank (wwPDB, http://www.wwpdb.org/) of protein structures (Kelley & Sternberg, 2009). The resulting domain prediction of Mga most closely resembles that of the mannose operon activator MtlR from Geobacillus stearothermophilus, which is modulated via phosphorylation of conserved histidines within its PRD domains (Deutscher et al., 2006). A pair of PRDs with strong homology to PRDLicT (termed PRDMga) is located between the N-terminal DNA-binding HTH domains and the C-terminal PTS EIIBGat-like domain (Fig. 1B). MtlR possesses an additional EIIA-like domain that appears to be missing from Mga. The closest overall protein structure identified by Phyre2 with similarity to Mga was the recently solved crystal structure for an Enterococcus faecalis Mga-like transcriptional activator EF3013 (PDB 3SQN), however, nothing is known about the biological function of this protein (Osipiuk, 2011). The Bacillus anthracis AtxA virulence regulator, a long-established homolog of Mga, also shares a similar predicted domain structure (Fig. 1B) (Hammerstrom et al., 2011, Tsvetanova et al., 2007).
The amino acid sequences of putative PRDs from divergent alleles of Mga (GAS serotypes M1T1 [mga-1] and M4 [mga-2]) were aligned with the PRD-containing regulators LicT (B. subtilis), MtlR (G. stearothermophilus), LicR (B. subtilis), LevR (B. subtilis), and AtxA (B. anthracis) using ClustalW (Fig. 1C). As evidenced by the sugar regulators (LicT, MtlR, LicR and LevR), classic PRDs are characterized by an essential histidine as well as a conserved arginine spaced 7 residues downstream, a strongly conserved glutamate at amino acid 57, and often a conserved histidine at residue 63 (Fig. 1C and S1) (Stulke et al., 1998). One or both of the conserved histidines (Fig 1C, highlighted yellow) serve as sites of PTS phosphorylation. The histidines within the B. anthracis AtxA PRDs do not align exactly with those of the sugar regulator paradigm (Fig. 1C), yet are reported to be phosphorylated (Tsvetanova et al., 2007). Both Mga alleles possess three histidine residues (2 in PRD1 and 1 in PRD2) that align similarly to those of AtxA (Fig. 1C) and are conserved among Mga proteins from all sequenced GAS genomes (Fig. S2). Thus, Mga has the potential to be a PRD-containing virulence regulator that might interact with the PTS to sense carbohydrate availability and utilization in the GAS cell.
A ∆ptsI PTS mutant alters Mga-dependent virulence gene regulation in M4 GAS
Genomic components of the PTS are highly conserved in low G+C Gram-positive bacteria (Deutscher et al., 2006), particularly genes for the ‘general’ PTS proteins HPr (ptsH) and EI (ptsI) encoded in the ptsHI operon. To assess the role of the PTS in Mga-dependent regulation, an EI mutant (ΔptsI) was constructed in the M4 strain GA40634. Wild-type ptsI was replaced with an in-frame deletion (ΔptsI) containing a non-polar aad9 spectinomycin resistance cassette (Lukomski et al., 2000) in the GA40634 genome. The resulting ∆ptsI mutant (GA40634ΔptsI) was verified by PCR (data not shown) and qRT-PCR (Fig. 2C). Although wild-type GA40634 had a slightly increased lag phase compared to the mutant in rich THY media, growth kinetics were comparable (Fig. 2A). GA40634∆ptsI also grew at the same rate as the parental GA40634 in low glucose C medium, except the mutant reached a slightly lower overall yield (Fig. 2A). Introduction of a ptsI complementing plasmid into the ∆ptsI mutant was problematic, possibly due to a detrimental effect of over expressing ptsI in GAS. To address this issue, multiple independent ∆ptsI mutants were generated in M4 GA40634, M1T1 MGAS5005, M1T1 5448, and M1T1 5448-AP, and all exhibited identitical PTS-related growth defects (Fig. 2 and data not shown).
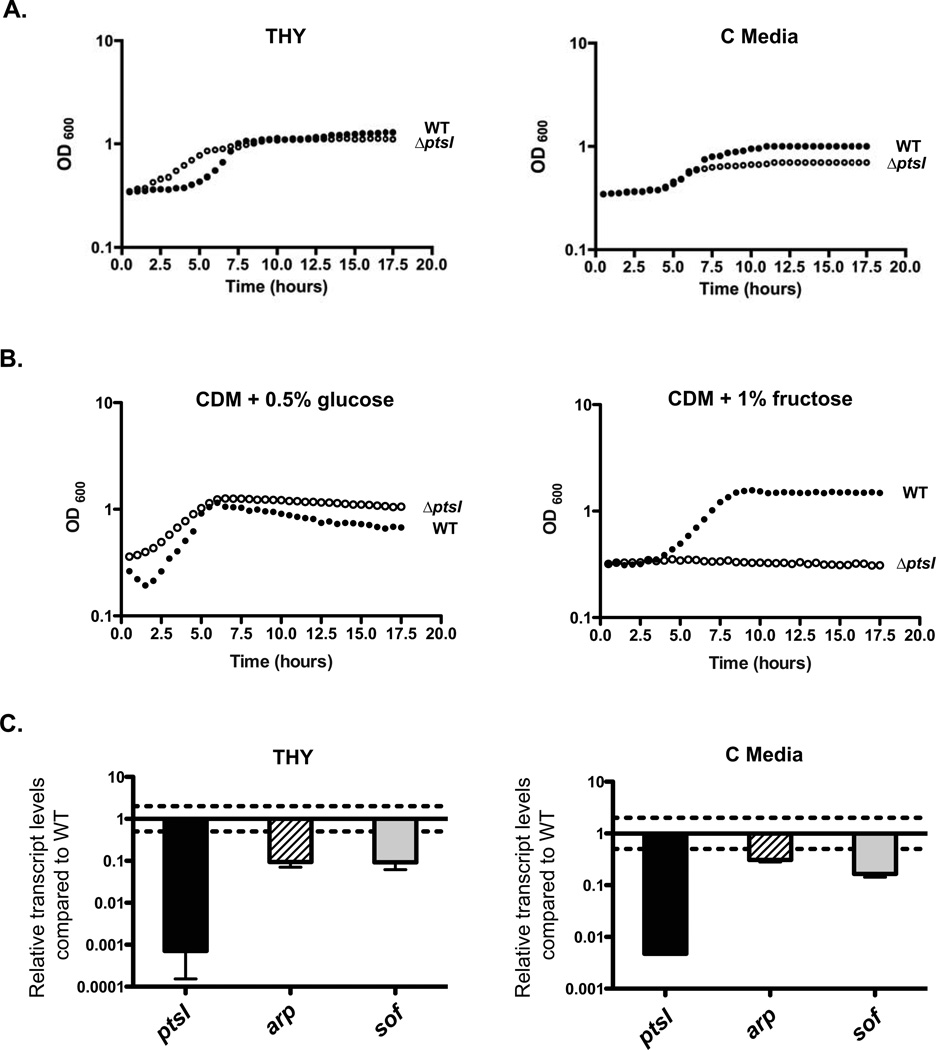
Figure 2. A ∆ptsI mutant of GAS is altered in PTS-dependent growth and Mga regulon expression.
(A) Growth curves of wild type GA40634 (closed circles) and GA40634∆ptsI (open circles) in THY or C medium as indicated. Data are representative of three independent experiments. (B) Growth curves of wild type GA40634 (closed circles) and GA40634∆ptsI (open circles) in CDM supplemented with either 0.5% (v/v) glucose or 1% (v/v) fructose as indicated. Data are representative of three independent experiments. (C) Transcript levels of ptsI (black), arp (striped), and sof (grey) were measured by qRT-PCR from late logartithmic phase cultures grown in THY and C medium for GA40634 ∆ptsI compared to GA40634. Two-fold differences in expression (dashed line) were considered significant. Standard error was determined from three biological replicates.
Carbohydrate-specific phenotypes of the wild-type GA40634 and the GA40634∆ptsI mutant were analyzed by growth assays in chemically defined media (CDM) supplemented with various carbohydrates serving as the sole carbon source. In CDM containing 0.5% glucose, GA40634ΔptsI showed a comparable growth rate to GA40634, except with higher yields and no significant lag phase (Fig. 2B). In contrast, GA40634∆ptsI was unable to grow when the PTS sugars fructose (Fig. 2B), lactose, sucrose, galactose, trehalose, and mannose were tested (data not shown). This mutant phenotype was identical to that found for independent ∆ptsI mutants generated in three different M1T1 GAS strains MGAS5005, 5448, and 5448-AP (Gera and McIver, in submission). These data suggest that GA40634∆ptsI lacks a functional PTS.
To investigate whether the GA40634∆ptsI PTS-defective mutant affects the Mga virulence regulon, qRT-PCR was performed on mRNA isolated from wild type and the ΔptsI mutant at late logarhithmic phase of growth in both THY (rich) and C (low glucose) media, probing for the Mga-regulated genes arp (M-like IgA receptor protein) and sof (serum opacity factor; Fn-binding protein). Relative transcript levels of GA40634∆ptsI compared to wild-type GA40634 were determined, such that full activity gives a ratio of 1.0 and a difference of greater than 2-fold was considered significant (Fig. 2C, dotted lines). As expected, ptsI transcript levels were reduced 2 to 3 logs in the mutant (Fig. 2C). In addition, both arp and sof transcript levels were significantly reduced (3- to 10-fold) in the ∆ptsI mutant compared to wild type, indicating that a functional PTS influences Mga-dependent virulence gene expression during logarithmic phase growth in GAS.
The PTS directly phosphorylates conserved PRD histidines in Mga and inhibits activity in vitro
To determine whether Mga could be phosphorylated by the PTS, the pathway was reconstituted in vitro. Mga (mga-2), EI (ptsI), and HPr (ptsH) from a serotype M4 GAS were expressed as His-tagged fusion proteins (Mga4-His6, His6-EI, and His6-HPr) in E. coli and purified. The radiolabeled [32P]-PEP phosphodonor (*PEP) was synthesized from [γ-32P]-ATP using purified E. coli PEP carboxykinase (pepck). Protein phosphorylation was then assessed by monitoring the transfer of radiolabel from *PEP following separation by SDS-PAGE and phosphorimager analysis (Fig. 3A). As expected, EI alone was phosphorylated by *PEP, whereas HPr required EI in order to be phosphorylated (Fig. 3A, lanes 1–3). Mga4 phosphorylation was dependent on the presence of all the components of the PTS pathway (Fig. 3, lanes 4–6). In contrast, heat-denatured (HD) Mga4 was not phosphorylated (Fig. 3A, lane 7), indicating that the native fold of Mga is likely critical for PTS-mediated phosphotransfer. To verify that phosphorylation targeted Mga proteins in general, the divergent M1T1 Mga1 (mga-1) was analyzed and was also phosphorylated in a PTS-dependent manner (Fig. 3B, lane 10). Phosphorylation was also observed for a truncated Mga4 (∆139Mga4-His6) that retains both PRD domains, but lacks the C-terminal EIIB-like domain (Fig. 1B; Fig. 3B, lane 9). Thus, in vitro the general proteins of the GAS PTS can directly phosphorylate Mga containing predicted PRD domains.
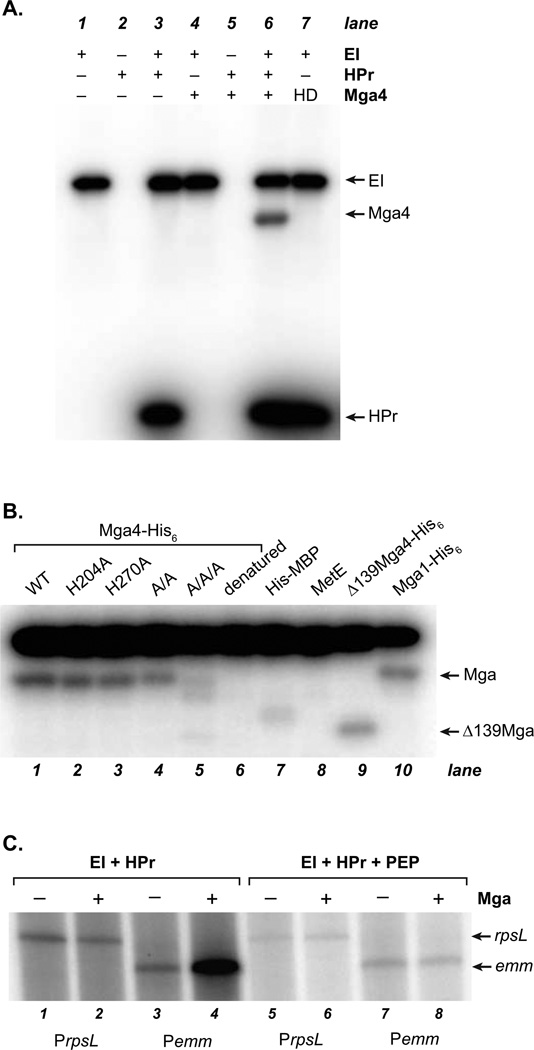
Figure 3. In vitro phosphorylation and inactivation of Mga by the PTS.
(A, B) Phosphorylation was assessed by incubation of [32P]-PEP with purified His6-EI, His6-HPr, and Mga4-His6. Reactions were then subjected to SDS-PAGE and phosphorimager analysis. (A) Proteins included in each reaction are indicated; HD refers to heat-denatured Mga4-His6(B) Wild type and various PRD Mga4-His6 mutants were analyzed, alongside heat-denatured Mga4-His6, His6-MBP, MetE, ∆139Mga4-His6, and Mga1-His6 controls as indicated. (C) The ability of Mga1-His6 to activate transcription of the constitutive PrpsL and Mga-regulated Pemm promoters was assessed in vitro immediately following phosphorylation reactions (containing or lacking PEP). Products of the in vitro transcription assays were then separated on a 6% sequencing acrylamide gel and subjected to phosphorimager analysis.
To determine whether conserved histidines within the PRDs of Mga were the target of PTS phosphorylation, Mga4-His6 mutants were first constructed where each of the three conserved histidines or both of the conserved PRD1 histidines were replaced by an alanine (H204A, H270A, H324A, PRD1 A/A) to prevent phosphorylation. The mutant proteins were purified from E. coli and assayed for in vitro phosphorylation by HPr-His∼P (Fig. 3B). Neither the single alanine mutations in the PRD1 histidines (H204A, H270A) nor the double mutation in both PRD1 histidines (A/A) appeared to prevent in vitro phosphorylation, although a modest decrease may be observed for the A/A mutation compared to wild type (Fig. 3B, lanes 2–4). This suggested that all three conserved PRD histidines might serve as targets for PTS-specific phosphorylation of Mga in vitro. Therefore, a triple Mga mutant replacing all three PRD histidines with alanines (A/A/A) was generated and assayed for PTS phosphorylation. The A/A/A Mga showed a dramatic loss of phosphorylation compared to wild type (Fig. 3B, lanes 1, 5). A very faint band could be seen at the Mga position with similar intensity to that of a His-MBP control (Fig. 3B, lane 7). Because this was not observed with an untagged protein of similar size (cobalamin-independent methionine synthase, MetE, lane 8), we suspect that the His tag might be weakly phosphorylated by GAS HPr in this system. Nevertheless, the lack of significant A/A/A phosphorylation indicates that the conserved Mga PRD histidines are likely the target of PTS-mediated phosphorylation by HPr-His∼P.
To assess whether PTS phosphorylation of Mga altered its activity, an in vitro transcription assay of Mga-dependent activation was utilized. Wild-type Mga1-His6 was subjected to in vitro PTS phosphorylation either in the presence or absence of PEP and tested for the ability to activate transcription from a constitutive PrpsL control promoter and the Mga-regulated Pemm promoter (Fig. 3C). As expected, Mga had no effect on transcription from the control PrpsL promoter regardless of PTS phosphorylation (Fig. 3C, lanes 1–2, 5–6). In the absence of phosphorylation in vitro (no PEP), Mga was able to significantly activate transcription from the Mga-dependent Pemm promoter (Fig. 3C, lanes 3–4). However, PTS phosphorylation (with PEP) of the same wild-type Mga1-His6 resulted in loss of Pemm activation in vitro. Identical results were observed using wild-type Mga4-His6 (data not shown). Thus, PTS phosphorylation appears to inhibit the ability of Mga to activate virulence gene expression in vitro.
PTS phosphorylation of PRD1 impacts Mga activity in vivo
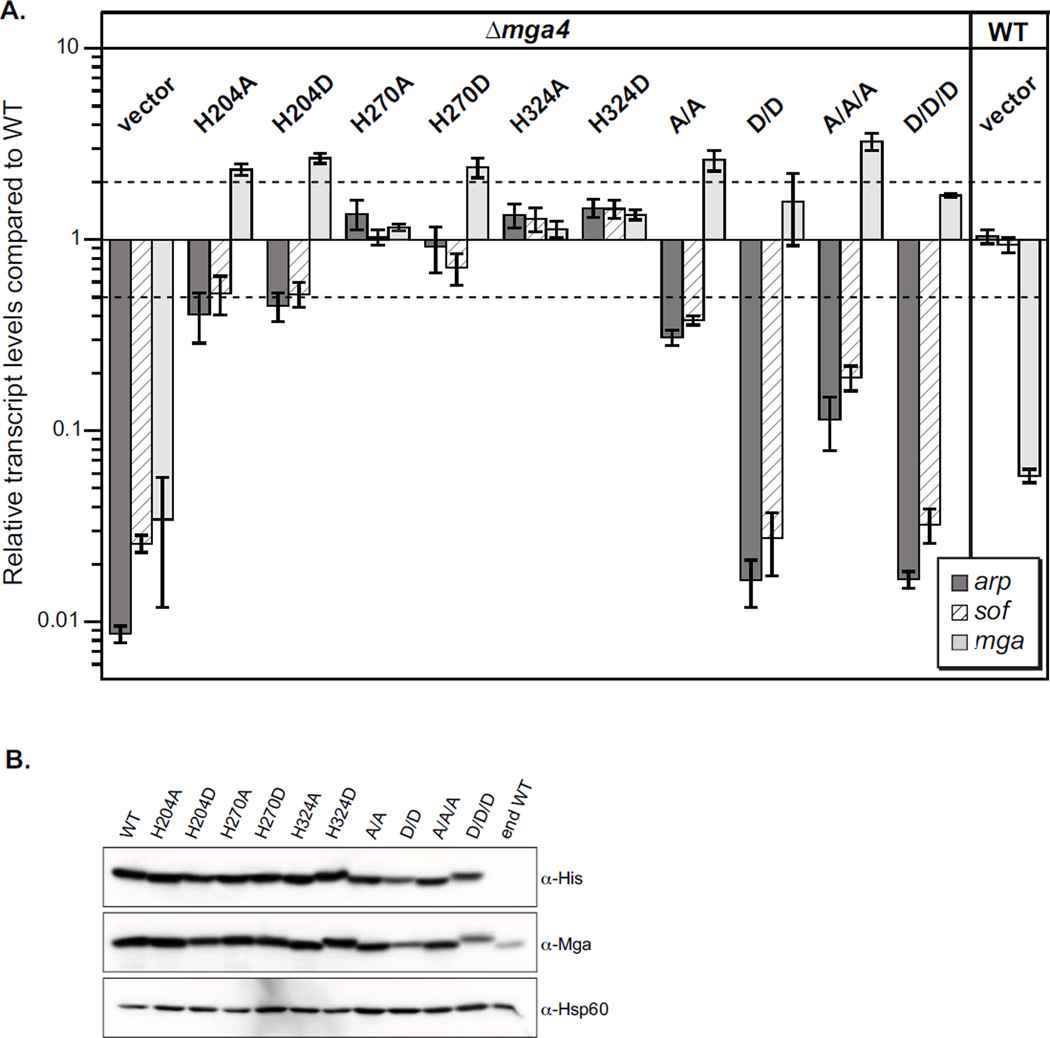
We next sought to explore PTS-mediated phosphorylation of Mga in GAS. A second series of mutants was generated whereby each PRD histidine was replaced by an aspartate (H204D, H270D, and H324D) to mimic the phosphorylated residue. The corresponding double PRD1 H204/H270 mutants (D/D) and triple H204/H270/H324 mutants of both PRD1 and PRD2 (D/D/D) were also constructed. The mutant proteins were then tested for their ability to influence Mga-regulated gene expression in vivo. Plasmids expressing wild type and mutant mga4 alleles from the native promoter (Pmga4) were transformed into an M4 ∆mga GAS background (KSM547.4) and Mga-regulated expression of arp and sof was assayed by qRT-PCR (Fig. 4A). Relative transcript levels of ∆mga4 GAS expressing the mutant alleles compared to a ∆mga GAS expressing the wild-type mga allele was determined, such that full activity gives a ratio of 1.0 and a difference of greater than 2-fold was considered significant (Fig. 4A, dotted lines). All proteins were stably expressed in GAS at levels above that of the fully active endogenous Mga4 (end WT) produced from the isogenic wild-type strain (Fig. 4B). The vector control in the ∆mga4 GAS background showed significantly reduced arp and sof levels (dark & striped bars) compared to the same strain expressing wild-type Mga (Fig. 4A, lane 1). However, none of the single alanine (non-phosphorylated) or aspartate (phosphomimetic) substitutions of the conserved histidine residues significantly altered Mga-dependent arp and sof expression (Fig. 4A). The PRD1 double A/A Mga mutant exhibited a modest reduction in sof and arp expression; in contrast, the phosphomimetic double D/D Mga mutant was almost completely inactive compared to wild type (Fig. 4A). The PRD1/PRD2 triple A/A/A mutation also showed an ca. 9-fold reduction in Mga activity, but not to the level of the PRD1/PRD2 D/D/D protein, which was similar to PRD1 D/D Mga4 (Fig. 4A). Comparable results were observed for PRD mutations in a divergent M1 Mga (mga-1 in strain SF370), although decreased levels of the D/D protein were observed (Fig. S3). Similar to our recent results (Hondorp et al., 2012), inactive Mga mutations did not appear to impact mga transcript levels (Fig. 4A, light grey bars), suggesting that mga may not be autoregulated in this plasmid-based system. Taken together with our in vitro data (Fig. 3), phosphorylation of Mga at both PRD1 histidines appears to inactivate the protein in vivo, shutting down expression of the Mga regulon. These findings suggest that direct phosphorylation of both PRD1 histidines leads to an inactive Mga. In addition, the inability to phosphorylate all three conserved residues (A/A/A) appears to also reduce Mga activity.
Figure 4. The conserved PRD1 histidines are important for M4 Mga activity in vivo.
(A)In vivo Mga activity was assessed by real-time RT-PCR analysis of arp (dark grey), sof (striped) and mga (light grey) mRNA. Transcript levels that were greater than 2-fold different compared to wild type (dotted lines) were considered significant. Single mutations in Mga are indicated, along with H204A/H270A (A/A), H204D/H270D (D/D), H204A/H270A/H324A (A/A/A), H204D/H270D/H324D (D/D/D), and an empty vector (vector) in the mga4-inactivated GAS strain KSM547.4. An isogenic wild type M4 strain GA40634 (WT) with vector was also assayed to show endogenous Mga4 (end WT) activity. (B) Protein levels in cell lysates were determined by immunoblotting with α-His and α-Mga4 antibodies; the α-Hsp60 antibody was employed as a loading control.
Mga PRD1 phosphomimetics do not prevent DNA binding
Mutations that target the primary winged HTH domain (wHTH-4) of Mga and block DNA binding result in the inability to activate virulence gene expression (McIver & Myles, 2002). Thus, one mechanism by which PTS phosphorylation could inactivate Mga is by preventing the binding of target promoters Parp and Psof. Hence, it was important to assess Mga-DNA binding in vitro. Because purified Mga is difficult to extensively manipulate in vitro (Hondorp et al., 2012) and the phospho-histidine bond is notoriously labile, we sought to determine whether the more stable, but inactive phosphomimetic D/D Mga retained the ability to bind cognate DNA.
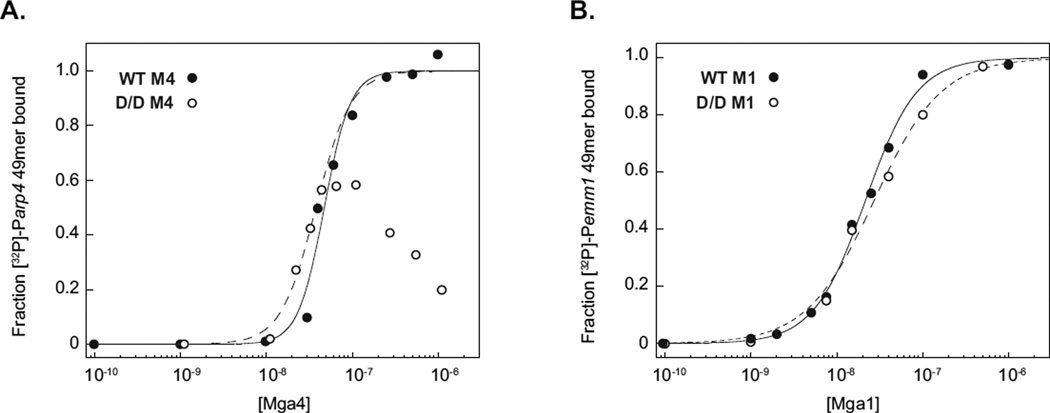
The PRD1 D/D Mga4-His6 mutant was expressed in E. coli and purified using similar methods to those developed for the wild-type protein. Interestingly, D/D Mga4-His6 levels were considerably lower than those of the recombinant wild type, resulting in a mutant protein that was more difficult to isolate and ∼30% pure based on coomassie staining of an SDS-PAGE gel. Nevertheless, Mga-DNA binding could be analyzed using filter-binding assays, which do not require a completely pure system as long as the protein concentration is known. Mutant concentrations were estimated from densitometric analysis of an immunoblot in which serial dilutions of D/D Mga4-His6 were compared with the wild-type protein at a known concentration. Binding was then assessed with a 49-mer double-stranded oligonucleotide that encompasses the 44-bp Parp Mga binding site with some flanking sequences (Hondorp et al., 2012). Varying concentrations of purified wild type and D/D Mga4-His6 were incubated with the [32P]-labeled Parp 49mer and then filtered through nitrocellulose. The amount of radiolabeled DNA bound to Mga was quantitated by densitometry and plotted to determine the concentration at half-maximal binding or apparent Kd (Kd,app, Fig. 5A). Wild-type Mga4-His6 bound to the Parp 49mer with a Kd,app of ca. 47 nM (Fig. 5A), which is consistent with our previous finding of 51 nM in this system (Hondorp et al., 2012). D/D Mga4-His6 was found to bind Parp similarly to the wild type at low protein concentrations (≤ 45 nM), but exhibited a decrease in binding at higher concentrations (Fig. 5A), perhaps due to increased aggregation of the isolated D/D protein. Analogous experiments with the Mga1-His6 protein analyzed under conditions that more closely resemble those used in the electrophoretic mobility shift assays indicate that the wild-type and D/D PRD1 mutant bind to the target emm promoter target (Pemm1) 49mer with comparable affinities (Fig. 5B, Kd,app 22 and 27 nM for wild type and D/D Mga1-His6, respectively). The deviation in binding profile observed in the M4 system compared to M1 might hint at differences in the mechanism of binding upon phosphorylation of Mga or may simply be an artifact of the system; further studies will be needed to address whether PRD1 phosphorylation might impact the mode of Mga-DNA interactions. Nevertheless, phosphorylation of the Mga PRD1 histidines does not appear to preclude DNA binding per se. These results are similar to those of a carboxy-terminal truncation mutant of Mga (∆139), which is monomeric and not active in vivo, but is still able to bind DNA in vitro with an affinity comparable to the full length protein (Hondorp et al., 2012).
Figure 5. The PRD1 D/D Mga phosphomimetic mutant is able to bind target DNA in vitro.
Filter-binding assays were employed to analyze protein-DNA associations for M4 (A) and M1 (B) wild type (closed circles) and PRD1 D/D phosphomimetic (open circles) Mga-His6 proteins. Proteins were incubated with [32P]-labeled 49-bp double-stranded oligonucleotides containing the Mga-binding sites of the arp(A) or emm(B) promoters and then filtered through nitrocellulose. The protein-bound radiolabel was quantified by phosphorimager analyses and the calculated fraction of DNA bound was plotted versus Mga protein concentrations.
Homomeric interactions are impaired for the Mga PRD1 phosphomimetic
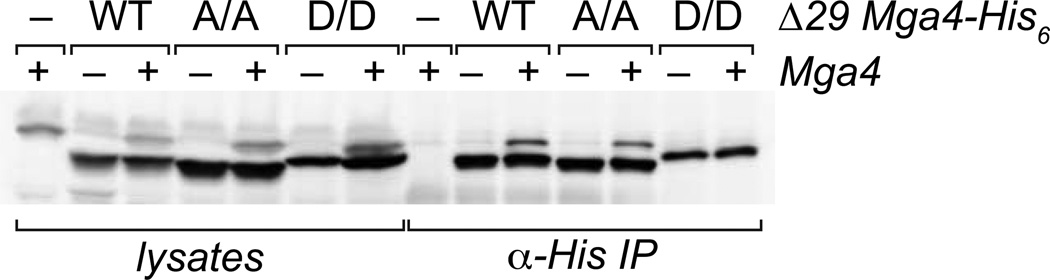
For many Gram-positive PRD-containing PTS sugar regulators, PRD phosphorylation controls activity through altering the dimerization of the regulator (Deutscher et al., 2006). We recently demonstrated that Mga forms multimers in solution and this process appears necessary for Mga-dependent virulence gene activation, but not for binding to the regulated promoters (Hondorp et al., 2012). To ascertain whether PRD1 phosphorylation controls activity of Mga activity by altering regulator multimerization, co-immunoprecipitation (co-IP) experiments were performed based on our published studies (Hondorp et al., 2012). We employed a 29 residue C-terminal truncated version of Mga (∆29Mga4-His6), which does not impact in vivo activity or disrupt oligomerization but is distinguishable from the full-length protein by SDS-PAGE analysis. His-tagged wild type and PRD1 double mutants of ∆29Mga4-His6 proteins were expressed from a plasmid in a M4 GA40634 GAS background, which expresses the endogenous untagged Mga4 protein (Fig. 6 lysates). Immunoprecipitation using an α-His antibody was performed with lysates from exponentially growing cells. Western blot analysis with an α-Mga4 antibody that recognizes both tagged and native Mga4 indicates that the truncated wild-type ∆29Mga4-His6 and the PRD1 A/A ∆29Mga4-His6 double mutant are able to interact with endogenous full-length native Mga4 and co-immunoprecipitate, whereas the inactive phosphomimetic PRD1 D/D ∆29Mga4-His6 does not (Fig. 6). The PRD1 D/D ∆29Mga4-His6 protein migrates as a slightly larger protein on SDS-PAGE compared to ∆29Mga4-His6, but is still distinguishable from the larger native Mga4 (Fig. 6, D/D lysates). These data strongly suggest that PRD1 phosphorylation inhibits Mga oligomerization and results in an inactive Mga molecule.
Figure 6. PRD1 phosphomimetic inhibits Mga oligomerization.
Co-immunoprecipitation of Mga4 with PRD1 mutant proteins. Wild type (WT), PRD1 A/A (A/A), and PRD1 D/D phosphomimetic (D/D) versions of the His-tagged 29-residue truncated Mga4 (∆29Mga4-His6) were expressed in isogenic M4 GAS strains containing (+) or lacking (−) endogenous Mga4 (GA40634 and KSM547.4, respectively). His-tagged ∆29 proteins from exponentially growing GAS cell lysates were immunoprecipitated with an α-His antibody, and coimmunoprecipitation of endogenous Mga4 was assessed by SDS-PAGE and immunoblot analysis with an α-Mga4 antibody (right). Western blot analysis of whole cell lysates prior to immunoprecipitation is shown alongside for comparison (left).
PTS phosphorylation of Mga PRD1 is important for GAS virulence in vivo
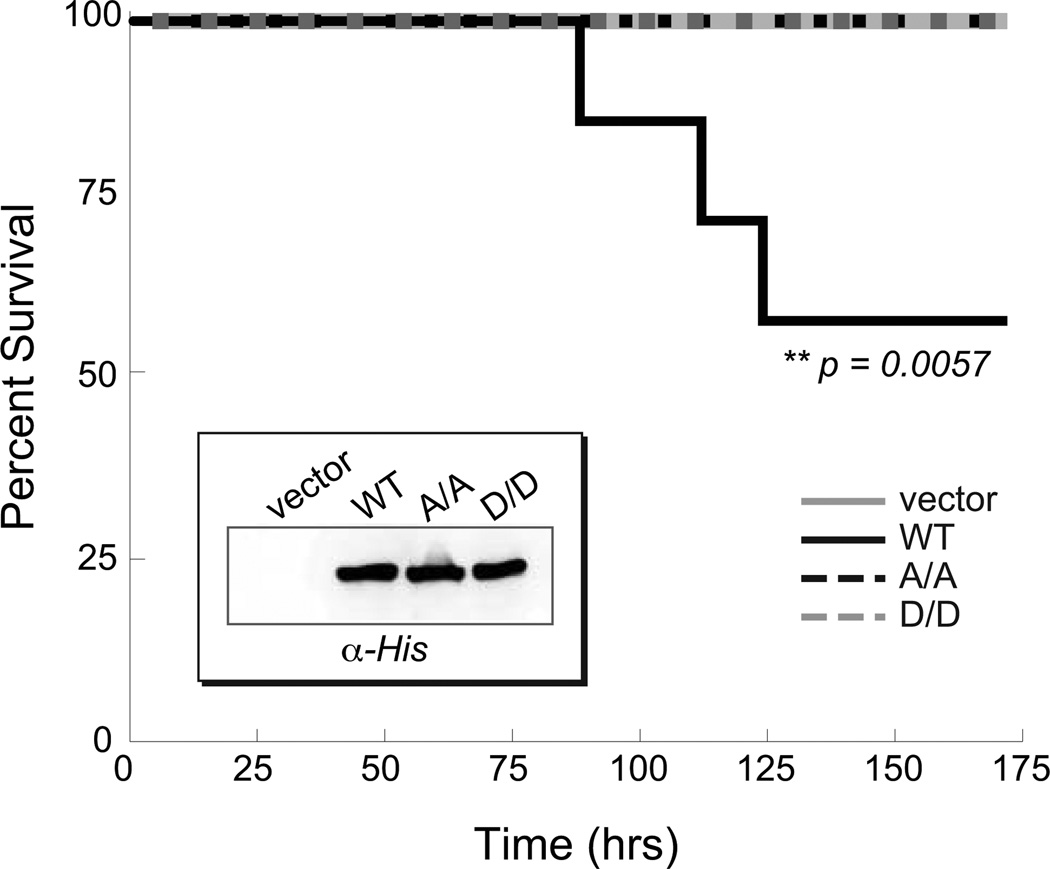
Phosphorylation of Mga PRD1 appears to strongly influence Mga activity under laboratory conditions; however, it was important to determine whether this link to the PTS and carbohydrate availability is significant during GAS infection. To test this, the ability of the PRD mutants to complement virulence attenuation in a ∆mga GAS mutant was assessed in a subcutaneous mouse model of infection. Because M4 GA40634 GAS does not exhibit virulence in mouse models of subcutaneous skin infection and functional results with M1 Mga were similar to M4 (Fig. S3), the M1T1 MGAS5005 strain was employed. Vector alone, wild type Mga1-His6, the PRD1 D/D phosphomimetic allele, and the PRD1 A/A mutant allele were expressed from native Pmga1 in the ∆mga KSM165-L.5005 background. All Mga proteins were expressed at comparable levels as determined by western blot analysis using an α−His antibody (Fig. 7, inset). Analysis of antibiotic resistance patterns of GAS isolated from mice spleen and blood showed that plasmids were retained in vivo over the time course of the experiment (data not shown). Expression of wild-type Mga1-His6 complements the virulence defect of the ∆mga strain compared to the vector alone control (Fig. 7, WT) resulting in significant mortality over 7 days. Interestingly, both the phosphomimetic Mga1-His6 PRD1 D/D mutant and the non-phosphorylated Mga1-His6 PRD1 A/A mutants were attenuated for virulence compared to wild type (Fig. 7) and did not exhibit any lethality at the doses used. It appears that either preventing phosphorylation of Mga (A/A) or constitutive phosphorylation (D/D) severely impacts the course of infection, indicating that the ability to modulate Mga activity via PTS-mediated phosphorylation of PRD1 may play a critical role in GAS virulence gene regulation and pathogenesis.
Figure 7. PTS phosphorylation of PRD1 is important for GAS virulence.
Survival curve for CD-1 mice infected subcutaneously with an M1T1 MGAS5005 Mga mutant (KSM165-L.5005) containing an empty vector (light grey solid, 1.4 × 108 cfu) or Mga1-His6 proteins (WT, black solid, 1.5 × 108 cfu; A/A, black dashed, 2.1 × 108 cfu; D/D, light grey dashed, 2.0 × 108 cfu) expressed from the native Pmga1 promoter. Data shown represent 8–10 mice for each strain. Significance was determined using Kaplan-Meier survival analysis and log rank test. Western blot analysis of GAS cell lysates is shown (inset).
DISCUSSION
There is an increasingly appreciated connection between bacterial pathogenesis and sugar availability during infection; however, the molecular mechanisms that might allow this interplay are not well understood. Our discovery that the Mga stand-alone regulator possesses homology to PRD domains found in sugar-specific regulators suggested a mechanism by which Mga might link the PTS to global virulence regulation in GAS (Deutscher et al., 2005, Hondorp & McIver, 2007). The results presented here support this hypothesis and demonstrate that Mga activity may be regulated by PTS-mediated phosphorylation of conserved histidines, providing a mechanism whereby the pathogen could modulate expression of Mga-regulated virulence genes in response to environmental sugar status.
Mga activity is directly modulated by PTS phosphorylation
Mga was phosphorylated and inactivated by the general components of the PTS in vitro (Fig. 3). While the stoichiometry of phosphorylation cannot be determined from these experiments due to technical issues related to handling purified Mga, it is clear that EI/HPr are able to phosphorylate Mga in vitro. Moreover, EI/HPr-mediated phosphorylation of Mga appears to be specific to the PRD histidines within the folded protein, because mutant proteins lacking the conserved histidines or heat-denatured wild type were not phosphorylated. For PRD-containing sugar regulators, the functional impact of PTS phosphorylation can be quite variable, with phosphorylation often showing antagonistic effects depending on which domain is targeted (Deutscher et al., 2006). While all three PRD histidines of Mga appear to be phosphorylated in vitro (Fig. 3B), phosphorylation of PRD1 seems to have the most significant functional consequences for GAS in vivo. Real-time RT-PCR analysis of a doubly phosphorylated PRD1 phosphomimetic protein (D/D Mga4) showed complete loss of activity regardless of the phosphorylation status of PRD2 (Fig. 4), indicating that PRD1-mediated inactivation of Mga is dominant. Yet interestingly, the data also suggest that phosphorylation of PRD2 might enhance Mga activity. When phosphorylation of PRD1 is prevented (in the A/A PRD1 mutant), the detrimental effect of not being able to phosphorylate His324 in PRD2 is evidenced by a 2- to 3-fold loss in activity for the A/A/A mutant compared to the A/A background (Fig. 4). Moreover, in the GA40634 ∆ptsI mutant, the lack of a functional PTS was found to result in decreased Mga activity (Fig. 2), suggesting that the PTS can also serve a stimulatory role. Although real-time RT-PCR analyses indicate that single mutations in PRD2 do not affect Mga activity, it is quite likely that this is an artifact of the multi-copy plasmid-based system employed for these experiments. Even though Mga levels are significantly higher than when endogenously expressed, equivalent activity is observed (Fig. 4), which suggests that another factor may be limiting and maximal activity is capped under these conditions. Therefore, even if preventing PRD2 phosphorylation in the H324A mutant decreases Mga activity, protein concentrations may be high enough to compensate such that expression of the Mga regulon by the H324A mutant appears similar to that of the wild-type protein. Furthermore, because Mga activity is already saturated in this system, the H324D phosphomimetic would not be expected to exhibit increased activity. Thus enhancement of Mga activity by PRD2 phosphorylation may only be observed in the compromised A/A background (Fig. 4) or in the ∆ptsI mutant where mga is in single copy (Fig. 2).
In order to examine the mechanism by which PRD1 phosphorylation might inactivate Mga, we focused on analyzing the inactive His204Asp/His270Asp (D/D) phosphomimetic protein. Previous studies suggest that the activity of PRD-containing sugar regulators is modulated by the negative charge introduced by phosphorylation, and analyses of stable phosphomimetics has proven valuable for characterizing the effects of phosphorylation, as the phospho-histidine bond is exceptionally labile (Deutscher et al., 2006). We found that wild type and non-phosphorylated PRD1 Mga (A/A) are able to form multimers in cell lysates and co-immunoprecipitate, whereas the inactive D/D phosphomimetic appears to be defective for oligomerization and remains predominantly monomeric (Fig. 6). Control of protein activity via modulation of oligomerization and nucleic acid binding affinity are themes common to PRD-containing sugar regulators. For the well-characterized LicT antiterminator, dimerization is a part of the activation mechanism by which significant conformational changes induce stabilization of the N-terminal RNA binding domain upon phosphorylation of PRD2 (Tortosa et al., 2001). Phosphorylation-induced activation and inactivation of the MtlR sugar regulator are directly correlated with increases and decreases in DNA binding affinity, respectively (Henstra et al., 2000). Thus a straightforward mechanism predicts that phosphorylation of Mga PRD1 prevents protein oligomerization, which weakens DNA binding and impedes transcriptional activation. Yet interestingly, we found that D/D Mga is still able to bind promoter DNA in vitro. These results are similar to those obtained for a truncated version of Mga (∆139 Mga) that is missing the C-terminal EIIBGat-like domain (Hondorp et al., 2012). The ∆139 Mga protein lacks activity in vivo and is impaired in multimerization, but still binds DNA in vitro, suggesting that the mechanism by which Mga activates transcription is more complex than simply binding to cognate promoters and multimerization likely plays an additional key role.
Although structural information is lacking for GAS Mga, the crystal structure of a putative E. faecalis homolog EF3103 (PDB# 3SQN) was recently solved (Osipiuk, 2011). There are no data concerning its biological function, but the domain structure is identical to that predicted for GAS Mga: an N-terminal wHTH domain followed by two PRDs and a C-terminal EIIB-like domain. However, the PRD histidines are missing in the E. faecalis homolog (data not shown), and indeed, it is not apparent from the structure how PRD phosphorylation would influence oligomerization and activity. Hence there is a need to understand the structural basis for Mga activities in order to further assess the means by which phosphorylation may modulate activity in GAS.
PTS regulation of the Mga virulence regulon during infection
The importance of being able to regulate Mga function for pathogenesis is highlighted by our in vivo results. Both blocking PTS-mediated inactivation of Mga via PRD1 alanine substitutions as well as phosphomimetic Mga PRD1 mutations (constitutive inactivation) led to attenuation of GAS virulence in a mouse model of disseminating subcutaneous skin infection (Fig. 7). While it is not surprising that the inactive D/D Mga mutant would behave similarly to the ∆mga strain, attenuation of virulence by the active A/A mutant suggests that not only is Mga activity critical to GAS pathogenesis, but that it is also important for the bacteria to be able to shut down the Mga regulon during an infection. Therefore, the ability to link control of the Mga regulon to the PTS, and thus sugar availability, appears to be critical during the course of disease, and likely reflects the requirement for proper temporal expression of Mga-regulated virulence determinants.
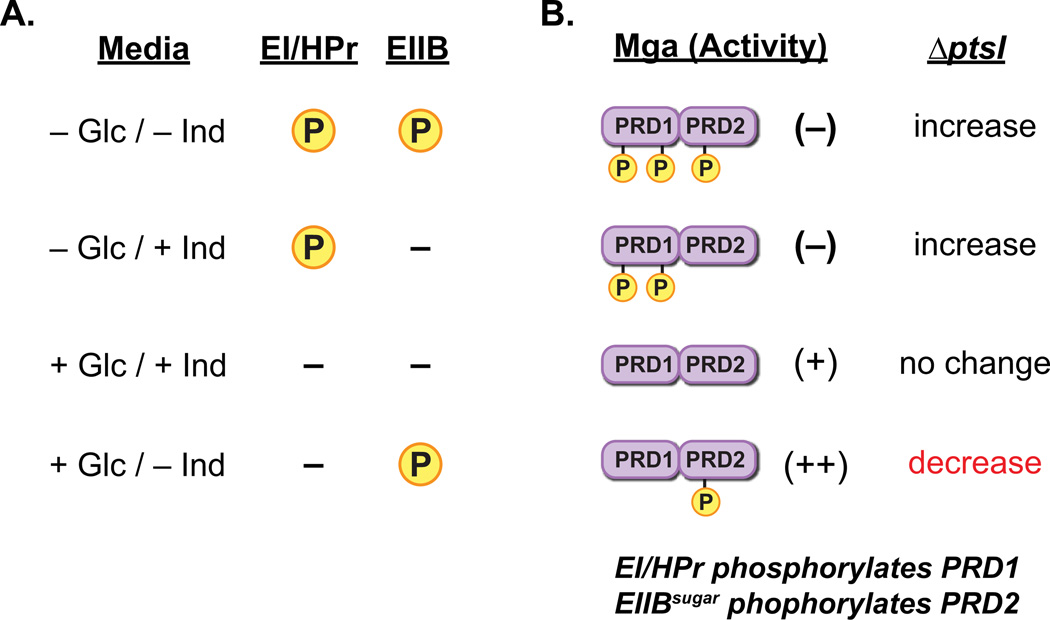
In general, phosphorylation of PRD-containing sugar regulators by an inducer-specific EIIBsugar protein inactivates the regulator while phosphorylation by EI/HPr enhances activity (Deutscher et al., 2006). In this study, we found that EI/HPr phosphorylates and inactivates Mga in vitro (Fig. 3). Yet, because EI/HPr is known to be able to phosphorylate PRDs in vitro when an EIIBsugar is the in vivo phosphodoner (Deutscher et al., 2006), the specific roles of the PTS protein(s) modulating Mga activity in vivo are uncertain. However, the results of the ∆ptsI mutant combined with the known physiology of GAS allows for speculation. Assuming that non-phosphorylated Mga is active, the PRD1-phosphorylated protein is inactive (and dominant) and the PRD2-phosphorylated species has enhanced activity, the impacts on Mga function can be proposed under various growth scenarios (Fig. 8A). Based on this model, the observed decrease in Mga activity in the ∆ptsI mutant compared to the isogenic wild type (Fig 2) would only be expected to occur in the presence of glucose and absence of inducer if EI/HPr inactivates Mga and an EIIBsugar enhances activity. In both THYB and C media, cells are growing in the presence of glucose, which argues that EI/HPr inactivates Mga via phosphorylation of PRD1 and EIIBsugar phosphorylation of PRD2 enhances activity (Fig. 8AB). Hence, EI/HPr and EIIBsugar may modulate Mga activity in an opposite manner to that which is common for PRD-containing sugar regulators. However, this mechanism of control would make sense given the biological role of Mga in pathogenesis. Niches where preferred PTS sugars are available appear to present a desirable setting for GAS colonization such that expression of the Mga regulon would be highly beneficial. Under these favorable growth conditions, ready phosphorylation of incoming sugars would limit the availability of HPr-His∼P to phosphorylate and inactivate Mga; furthermore, buildup of serine-phosphorylated HPr may act in concert with CcpA to increase mga expression (Almengor et al., 2007). In contrast, carbohydrate-poor conditions may provide a signal for GAS dissemination in host tissues mediated by down regulation of Mga-regulated adherence factors. A lack of preferred PTS sugars would lead to the accumulation of HPr-His∼P, which could phosphorylate PRD1 of Mga, thereby precluding the formation of active multimers and shutting down expression of the Mga regulon (Fig. 8B). Thus while these results allow us to speculate on the nuances of the mechanism in GAS, the details of PTS-mediated control of Mga in vivo will need to be further explored. Nevertheless, this study clearly shows that the PTS plays direct role in regulation of Mga activity and virulence of the pathogen.
Figure 8. Model for PTS/Mga interactions in vivo.
(A) Predicted availability of phosphate for transfer to Mga by either EI/Hpr or a sugar-specific EIIB component when growing in the presence or absence of a preferred sugar source (i.e., glucose) and/or an EIIB-specific inducer sugar. In the absence of glucose, EI/Hpr can phosphorylate PRD domains, whereas it cannot in its presence. Phosphorylation of PRD domains by a cognate EIIB protein would only occur in the absence of inducer. (B) Proposed role of PTS-mediated Mga phosphorylation on activity based upon sugar source in (A). In the absence of glucose, Mga would be inactivated through phosphorylation of PRD1. Inactivation of PTS (∆ptsI) would be expected to increase Mga-regulated expression. With both glucose and inducer present, Mga is not phosphorylated at either PRD and is active. Loss of PTS (∆ptsI) would have no effect. In the presence of glucose only (THY, C media, or CDM + glucose only), phosphorylation of Mga PRD2 by inducer-specific EIIB leads to enhancement of activity. In this case, loss of PTS ((∆ptsI) would result in a decrease in Mga regulon expression.
A novel family of PRD-containing virulence regulators (PCVR) in Gram-positive pathogens
Mga, along with B. anthracis AtxA, appear to represent the paradigms for a growing family of PRD-containing virulence regulators (PCVRs) broadly distributed in Gram-positive pathogens. Homologs of Mga can be found in the genome sequences of most pathogenic streptococci, as well as enterococci and Listeria spp. and many other Gram-positive pathogens. Phyre analysis indicates that they possess a similar domain structure with two internal PRDMga domains, a carboxy-terminal EIIBGat-like domain, and amino-terminal DNA-binding motifs (data not shown). Alignment of streptococcal Mga PRDs reveals that His204 is conserved within PRD1, however, His270 is only found in GAS Mga (Fig. S4). Interestingly, His324 is present within most Mga homologs and a second PRD2 histidine appears to be conserved among all streptococcal Mga proteins except those found in GAS. This suggests that while PTS-mediated control of Mga is likely a conserved means of linking virulence to metabolism, the mechanistic details may vary between pathogens. Such customization is not unprecedented for PRD-containing regulators. For example, phosphorylation of PRD2 was reported for B. subtilis GlcT but not the S. carnosus homolog, and G. stearothemophilus MtlR PRD1 is phosphorylated in vitro, but the B. subtilis protein is not (Henstra et al., 2000, Joyet et al., 2010, Knezevic et al., 2000, Schmalisch et al., 2003).
Phyre analyses also indicate that GAS RofA-like proteins (RALPs) have predicted domain structures that are similar to Mga including PRDs (data not shown) and therefore may be classified as PCVRs. The RALPs represent another important family of stand-alone virulence regulators in GAS and other pathogenic streptococci that affect host cell attachment and avoidance of host cell damage as the pathogen transitions from exponential to stationary phase growth (McIver, 2009). Interestingly, several RALPs are known to directly impact the expression of mga in the GAS cell (Roberts & Scott, 2007, Siemens et al., 2012). Therefore, it is distinctly possible that direct interaction with the PTS also impacts the activity of the RALPs and future studies will be needed to assess this hypothesis.
The B. anthracis virulence regulator AtxA, which has homology to Mga, also appears to be a PCVR. Moreover, phosphorylation of AtxA PRD histidines was reported to impact transcriptional activity in a manner dependent on an intact PTS (Tsvetanova et al., 2007). Thus Mga and AtxA represent the archetypes for this growing class of proteins and reveal how pathogens appear to have developed an efficient means for linking pathogenesis to the metabolic state of the cell.
EXPERIMENTAL PROCEDURES
The plasmids used in this study are described in Table 1. General DNA manipulations, construction of plasmids, and tables detailing primers and supplemental plasmids (Table S1 and Table S2, respectively) can be found in the supporting materials.
Table 1.
Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pJRS525 | broad host range cloning vector; aad9 (Spr) | (McIver & Scott, 1997) |
| pProExHTb | expression vector with N-terminal 6x His; bla (Apr) | Invitrogen |
| pKSM357 | his6-ptsI; bla (Apr) | this study |
| pKSM637 | luc under PtrxT in pJRS525; aad9 (Spr) | (Gold, 2011) |
| pBlue∆ptsI | ∆ptsI in pBluescriptII KS-; bla (Apr) | this study |
| pKSM645 | ∆ptsI with non-polar aad9 (Spr) | this study |
| pKSM712 | his6-ptsH; bla (Apr) | (Hondorp et al., 2012) |
| pMga1-His | WT mga1 (SF370) with C-terminal His6; bla (Apr) | (Hondorp et al., 2012) |
| pKSM801 | WT mga4 (GA40634) with C-terminal His6; bla (Apr) | (Hondorp et al., 2012) |
| pKSM807 | WT mga1 under native Pmga1; aad9 (Spr) | (Hondorp et al., 2012) |
| pKSM808 | WT mga4-his6 (GA40634) under native Pmga4; aad9 (Spr) | (Hondorp et al., 2012) |
| pKSM809 | WT mga1-his6 under native Pmga1; aad9 (Spr) | (Hondorp et al., 2012) |
| pKSM819 | ∆29mga4-his6 under PrpsL; aad9 (Spr) | (Hondorp et al., 2012) |
| pKSM822 | ∆29mga4-his6 under native Pmga4; aad9 (Spr) | (Hondorp et al., 2012) |
| pKSM824 | H324A mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM825 | H324D mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM832 | H324A mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM833 | H324D mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM836 | H204A mga1 with C-terminal His6; bla (Apr) | this study |
| pKSM837 | H204D mga1 with C-terminal His6; bla (Apr) | this study |
| pKSM838 | H270A mga1 with C-terminal His6; bla (Apr) | this study |
| pKSM839 | H270D mga1 with C-terminal His6; bla (Apr) | this study |
| pKSM841 | H204A/H270A mga1 with C-terminal His6; bla (Apr) | this study |
| pKSM842 | H204D/H270D mga1 with C-terminal His6; bla (Apr) | this study |
| pKSM849 | H204A/H270A mga1-his6 under native Pmga1; aad9 (Spr) | this study |
| pKSM850 | H204D/H270D mga1-his6 under native Pmga1; aad9 (Spr) | this study |
| pKSM857 | H204D mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM858 | H270A mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM859 | H270D mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM860 | H204A mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM861 | H204A/H270A mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM862 | H204D/H270D mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM863 | H204A/H270A/H324A mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM864 | H204D/H270D/H324D mga4 with C-terminal His6; bla (Apr) | this study |
| pKSM865 | H204A mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM866 | H204D mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM867 | H270A mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM868 | H270D mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM869 | H204A/H270A mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM870 | H204D/H270D mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM871 | H204A/H270A/H324A mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM872 | H204D/H270D/H324D mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM876 | D/D ∆29mga4-his6 under native Pmga4; aad9 (Spr) | this study |
| pKSM877 | A/A ∆29mga4-his6 under constitutive PrpsL; aad9 (Spr) | this study |
| pKSM879 | his6-pepck; bla (Apr) | this study |
Bacterial Strains and Media
GA40634 is a clinical isolate of the GAS M4 serotype and SF370 is a sequenced M1 strain. KSM547.4 and KSM165-L are isogenic strains of GA40634 and SF370, respectively, that contain an insertion in the mga gene (Ribardo & McIver, 2006, Almengor & McIver, 2004). GAS strain KSM165-L.5005 contains a similar suicide-based inactivation of mga in the M1T1 MGAS5005 strain and was constructed as described for KSM165-L (Almengor & McIver, 2004). Escherichia coli strain DH5α was used for cloning and proteins were expressed in either BL21[DE3]-Gold, JM109, or C41[DE3](Miroux & Walker, 1996), as indicated. GAS were grown in Todd-Hewitt medium containing 0.2% yeast extract (THY) and growth was monitored with a Klett-Summerson colorimeter equipped with an A filter. E. coli were grown in Luria-Bertani broth or ZYP-5052 (Studier & Moffatt, 1986) for protein expression. Antibiotics were utilized at the following concentrations: 100 µg/ml ampicillin, 100 µg/ml spectinomycin, and 50 µg/ml and 300 µg/ml kanamycin for E. coli and GAS, respectively.
Construction of the PTS mutant GA40634∆ptsI in GAS
PCR SOE-ing was used to delete the ptsI gene as previously described (Kinkel & McIver, 2008). Briefly, the primers ptsI-1a and ptsI-1b (Table S1) were used to amplify from MGAS5005 genomic DNA (gDNA) an 819 bp upstream region containing 578 bp of ptsI, and a BglII restriction site. Primers ptsI-2a and ptsI-2b (Table S1) were used to amplify an 812 bp region containing the 3’ of ptsI with BglII ends and a 10 bp overlap with the first fragment at the 5’ end. These fragments were combined as template DNA for PCR SOE-ing with ptsI-3a and ptsI-3b (Table S1) to generate the ptsI deletion. The resulting product was blunt end ligated into pBluescript II KS- to create pBlue∆ptsI. The non-polar aad9 spectinomycin resistance cassette was amplified from pSL60-1 using primers aad9L2-bglII and aad9R2-bglII (Table S1), digested with BglII, and ligated into BglII-digested pBlue∆ptsI. The resulting XbaI/XhoI ΔptsI∷aad9 fragment from pBlue∆ptsI was ligated into XbaI/XhoI-digested pCRK to yield pKSM645. A ∆ptsI mutant (GA40634∆ptsI) was constructed in GA40634 by temperature sensitive allelic exchange with pKSM645 as previously described (Perez-Casal et al., 1993). GAS ptsI mutants were screened for sensitivity to kanamycin and verified by PCR on genomic DNA for internal, external and junction regions. Multiple independent ∆ptsI mutations were generated in GA40634, M1T1 MGAS5005, M1T1 5448, and M1T1 5448-AP, and all exhibited identitical PTS-related growth defects (data not shown).
Growth analyses
Growth of GAS in CDM supplemented with a single carbhohydrate were assayed as follows: overnight cultures of GAS in THY (10 ml) were adjusted to OD600 of 0.2 in saline and inoculated into CDM with appropriate carbohydrates added (0.5% glucose or 1% for all other sugars), and 50 µl aliquots were added to each well of a 24-well microplate (Corning/Costar) followed by sealing with plate tape. Growth was followed for 12–17 hours at 37°C using a FLUOstar Omega microplate spectrophotometer (BMG), with measurements taken at 30 min intervals.
Protein Purifications
Mga-His6 proteins and His6-PEPCK were purified from C41[DE3] E. coli containing the appropriate plasmid (pKSM801, pMga1-His6, pKSM804, pKSM860, pKSM858, pKSM861, pKSM863, pKSM862, pKSM842, and pKSM879 for Mga4-His6, Mga1-His6, ∆139Mga4-His6, H204A Mga4-His6, H270A Mga4-His6, A/A Mga4-His6, A/A/A Mga4-His6, D/D Mga4-His6, D/D Mga1-His6, and His6-PEPCK, respectively); His6-HPr and His6-EI were purified from BL21[DE3]-Gold containing pKSM712 and JM109 containing pKSM357, respectively. Cultures were grown as previously described (Hondorp et al., 2012), with the following modifications. Cultures (12 L) with strains expressing alanine mutant Mga4-His6 proteins were grown at 37°C with rotary shaking at 250 rpm for 8 hours and then moved to room temperature for 16 hours while shaking at 225 rpm; the D/D Mga4-His6 culture (24 L) was grown similarly, except that the room temperature incubation was extended to 40 hours. E. coli expressing D/D Mga1-His6 (9 L) was cultured at 30°C for 50 hours, while His6-HPr, His6-EI, and His6-PEPCK expressing strains (1.5 L) were grown at 37°C for 24 hours.
All Mga-His6 proteins were then purified and prepared as previously described (Hondorp et al., 2012). Protein concentrations were estimated based on the absorbance at 280 nm using calculated extinction coefficients (Mga4-His6 ε280 ∼ 61,730 M−1 cm−1; Mga1-His6 ε280 ∼ 59,170 M−1 cm-1; Δ139Mga4-His6 ε280 ∼ 56,610 M−1 cm−1 (Hondorp et al., 2012)), except for D/D Mga-His6 proteins, which were only approximately 30% pure as judged by coomassie staining of an SDS-PAGE gel. Hence D/D Mga-His6 concentrations were estimated by western blot analysis with an α-Mga4 or α-Mga1 antibody, comparing serial dilutions of the mutant protein to that of the wild-type following densitometric analysis.
His6-HPr, His6-EI, and His6-PEPCK were similarly purified, except that the column was washed with 30 ml 0% buffer B (500 mM imidazole, 20 mM NaPi, 500 mM NaCl, pH 7.4), followed by 25 ml 6% B (for His6-EI only); His-tagged proteins were then eluted with 25 ml 100% B. For His6-HPr and His6-EI, fractions containing purified protein were combined and dialyzed against 4 L of 50 mM Tris, pH 7.4 at 4°C overnight. For His6-PEPCK, EDTA was added to the combined fractions to a final concentration of 5 mM and then dialyzed against 4 L of 1 mM EDTA, 100 mM KCl, 20 mM Tris, pH 5.5 at 4°C. Proteins were then concentrated using Amicon ultra-15 concentrators (10 kDa MWCO) and judged to be > 97% pure by coomassie staining of an SDS-PAGE gel. Protein concentrations were estimated from the absorbance at 280 nm using the calculated extinction coefficients (ɛ280(His6-HPr) = 6,400 M−1 cm−1, ɛ280(His6-EI) = 34,660 M−1 cm−1, and ɛ280(His6-PEPCK) = 78,090 M−1 cm−1).
Preparation of [32P]-PEP
Radiolabeled PEP was synthesized enzymatically using the method of Matoo and Waygood (Mattoo & Waygood, 1983). Briefly, a 1-ml reaction containing 100 µM oxaloacetic acid (Sigma), 50 µCi [γ−32P]-ATP (Perkin Elmer), 0.1 µM His6-PEPCK, 10 mM MgCl2, and 50 mM HEPES, pH 7.5 was incubated for 15 to 20 min at room temperature. The mixture was diluted 10-fold with water and then loaded onto a 2-ml AG-1-X8-bicarbonate column (BioRad). Reaction components were eluted using 5 ml of 0.3, 0.4, 0.6, and 0.7 M triethylammonium bicarbonate buffer, pH 8.5 (Sigma). The 0.6 and 0.7 M fractions containing purified PEP were combined and dried. Solid PEP was then dissolved in 100 µl 50 mM HEPES, pH 7.5 and stored at −20°C.
In vitro PTS phosphorylation
Reactions (20 µl) were prepared containing 250 nM His6-EI, 1 µM His6-HPr, 5 µM substrate protein (Mga-His6, His-MBP, or MetE), and [32P]-PEP in 10 mM MgCl2, 50 mM HEPES, pH 7.5. For the heat-denatured sample, Mga in buffer was heated at 99°C for 10 min, cooled on ice, and then His6-EI and His6-HPr were added. The reactions were initiated by the addition of [32P]-PEP (1 µl, ∼750 kcpm) and incubated at 37°C. After 20 min, SDS loading buffer (pH 7.4) was added and the mixture was placed on ice. Samples were loaded on a 10% SDS-PAGE gel and run at 20 mA for 1 hr, 40 min. The gel was briefly dried for approximately 5 min and then exposed to a phosphorimager cassette.
In vitro transcription assay after PTS phosphorylation
The phosphorylation reaction described above was modified to contain 1 µM DNA template (PrpsL or Pemm), 1 µM Mga4-His6, and 50 mM MgCl2. The PrpsL and Pemm templates were described previously (Almengor et al., 2006). Phosphorylation reactions were initiated by the addition of PEP (25 mM, Sigma) and incubated for 20 min at 37°C. The subsequent in vitro transcription assay was performed as described previously (Opdyke et al., 2001). Purified GAS RNA polymerase (10 µg) was mixed with purified GAS σ70 (10 µg) and incubated on ice for at least 10 min. The resulting holoenzyme (12 µg) was then added to the phosphorylation reaction and incubated at 37°C for 10 min. 1 µL NTP mix (1 µL each 10 mM ATP, GTP, CTP, [γ]32P UTP and dH2O) was added and incubated for 5 min at 37°C. 1 µL cold UTP was added and incubated for 5 min at 37°C. The reaction was then stopped with 12.5 µL Stop buffer (80% formamide, 12.5% 0.5% bromophenol blue, 20 mM EDTA, in 1X TBE). Reactions were denatured for 5 min at 80°C, then spun briefly. Reactions were run on a 6% sequencing acrylamide gel alongside a sequencing reaction to determine the size of transcripts following phosphorimager analysis.
Western blot analysis
GAS lysates were prepared as previously described using the PlyC bacteriophage lysin (Hondorp et al., 2012). Western blots were performed with the following antibody dilutions: rabbit α-Mga4 polyclonal antibody 1:1,000 or 1:2,000; rabbit α-Mga1 polyclonal antibody conjugated to HRP 1:1,000; mouse α-His monoclonal antibody (Novagen) 1:1,000; mouse α-Hsp60 monoclonal antibody (StressGen Biotechnologies Corp.) 1:2,500.
Real-time RT-PCR
GAS transcript levels were analyzed by real-time RT-PCR as previously described (Hondorp et al., 2012). Briefly, RNA was purified from at least three biological replicates of cultures grown to late exponential phase using a Triton X-100/chloroform extraction method (Sung et al., 2003). Contaminating DNA was removed by DNase treatment and verified by PCR analysis. Isolated RNA (25 ng) was added to a SYBR Green Master Mix (Applied Biosystems) containing 250 nM of each primer and 3 U multiscribe reverse transcriptase (Applied Biosystems). Each sample was assessed in triplicate and processed on a Lightcycler 480 real-time PCR system (Roche). Transcript levels in mutant strains were compared to the wild type, relative to gyrA expression.
Filter-Binding Assays
Mga-DNA binding was measured using filter-binding assays as previously described (Hondorp et al., 2012), with the following modifications. Mga4 binding was assayed with 0.1 nM [32P]-Parp 49mer in 1X BB (100 mM NaCl, 5 mM MgCl2, 50 mM Na-HEPES, pH 7.5), while Mga1 binding was assayed with 1.7 nM [32P]-Pemm 49mer in buffer containing 25 µg/ml BSA, 0.6 mM DTT, 1 mM EDTA, 60 mM KCl, 5 mM MgCl2, and 12 mM Na-HEPES, pH 7.5, which is similar to the conditions used previously for electrophoretic mobility shift assays (McIver et al., 1995). The double-stranded 49-bp oligomer containing the Mga-binding site of the Pemm1 promoter was produced by annealing the PAGE-purified MBS-Pemm1 and MBS-Pemm1-RC single-stranded oligomers (Table S1) as previously described (Hondorp et al., 2012); the concentration of the [32P]-labeled duplex was determined spectrophotometrically using an ɛ260 of 758,212 M−1 (calculated at http://biophysics.idtdna.com/UVSpectrum.html). The average band intensities obtained from phosphorimager analyses were plotted versus the protein concentrations using Kaleidagraph (Synergy Software) and then analyzed by the Hill equation: I = (Imax [Mga]n) /(Kd,appn + [Mga]n), where I represents the average intensity, Imax the maximum intensity, Kd,app is the apparent dissociation constant for half maximal binding, and n provides an estimate of the Hill coefficient. In order to provide a comparison of binding on the same plot, the fractional binding was then estimated by calculating I/Imax, which was plotted versus protein concentrations. For D/D Mga4-His6, the Imax for wild-type Mga4-His6 was used since binding of the two proteins was assayed together and band intensities of the mutant protein consistently decreased at high protein concentrations.
Co-immunoprecipitations
Co-immunoprecipitation experiments of GAS strains (KSM547.4 and GA40634 containing pJRS525, pKSM819, pKSM877, pKSM876) were performed as described (Hondorp et al., 2012). Briefly, cells from 12.5-ml cultures of GAS in late exponential growth were washed, resuspended in 185 µl PBS, and lysed by incubating 15 min on ice following the addition of 250 U PlyC and 20 U TURBO DNaseI (Applied Biosystems). Lysates were clarified by centrifugation at 16,000 × g for 10 min at 4°C and 40-µl samples were removed, added to SDS-PAGE loading buffer, and heat denatured. Lysates (150 µl) were diluted 2-fold with PBS containing 1X complete EDTA-free protease inhibitors (Roche) and incubated with 40 µl of Protein A agarose (50% slurry equilibrated in 0.5% BSA, CalBiochem) for 15 min with gentle rocking. The supernatant was removed and added to 10 µl α-His monoclonal antibody (Novagen). After gently rocking for 1 hr at 4°C, 40 µl Protein A agarose (50% slurry equilibrated in PBS) was added. Samples were gently rocked for 1 hr at 4°C and then the resin was pelleted and washed with 500 µl PBS. Samples were resuspended in 40 µl of SDS-PAGE loading buffer containing 10% SDS and heat denatured for 10 min at 95°C. Supernatant proteins were separated by SDS-PAGE and analyzed by immunoblotting with α-Mga4 (1:1,000) and the Clean-Blot IP detection reagent (Pierce, 1:200), which only detects native antibodies. Blots were developed using SuperSignal West Femto chemiluminescent reagents (Pierce) and visualized with a Fuji LAS3000 CCD scanner.
Mouse invasive skin infection model
All animal work was performed in AAALAC-accredited ABSL-2 facilities at the University of Maryland following IACUC-approved protocols for humane treatment of animal subjects (R-09–74) in accordance with guidelines set up by the Office of Laboratory Animal Welfare (OLAW) at NIH, Public Health Service, and the Guide for the Care and Use of Laboratory Animals. The department of Laboratory Animal Resources at U. Maryland provided animal husbandry and veterinary services. Every effort to limit distress and pain to animals were taken. Mice were monitored for signs of moribund state and euthanized immediately using CO2 asphyxiation consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
An overnight culture (5 ml) was used to inoculate 75 ml of THY and incubated static at 37°C until late-logarithmic phase. KSM165-L.5005 Mga mutant containing pKSM637 (empty vector), pKSM809 (WT mga-1), pKSM849 (Mga1 PRD1 A/A), and pKSM850 (Mga1 PRD1 D/D) were vortexed for 5 min, centrifuged for 20 min at 7,500 × g at 4°C and the pellet resuspended in 3 ml of saline (Table S1). Approximately 2 × 109 colony forming units (CFU)/ml each, as determined by microscope counts and verified by plating for viable colonies, were used to infect mice as previously described (Schrager et al., 1996). Briefly, anesthetized 6 to 7-week old female CD-1 mice (Charles River Laboratories) were depilated for an ∼2 cm2 area of their haunch with Nair (Carter Products, New York, NY) and 100 µl of the cell suspension (∼ 2 × 108 CFU/mouse) was injected subcutaneously with 1.4 × 108 cfu (KSM165-L.5005/pKSM637), 1.5 × 108 cfu (KSM165-L.5005/pKSM809), 2.1 × 108 cfu (KSM165-L.5005/pKSM849), and 2.0 × 108 cfu (KSM165-L.5005/pKSM850). Mice were monitored twice daily for seven (7) days and were euthanized by CO2 asphyxiation upon signs of systemic morbidity (hunching, lethargy, hind leg paralysis). Survival data was assessed by Kaplan-Meier survival analysis and tested for significance by log rank test. Data shown represent 8–10 mice for each strain. Significance was determined using Kaplan-Meier survival analysis and log rank test.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Cheryl Vahling for construction of the His-EI and His-HPr plasmids used in this study, and Yan Wang for her efforts. We are grateful to Daniel Nelson (U. Maryland Institute for Bioscience and Biotechnology Research) for supplying the PlyC phage lysin. We thank Vincent Lee (University of Maryland) for critical review of this manuscript and for providing the His-MBP. MetE was kindly provided by Rowena Matthews (University of Michigan). This work was supported by a postdoctoral fellowship from the American Heart Association (E.R.H., GRNT0825477E) and a grant from the NIH National Institute of Allergy and Infectious Diseases (K.S.M., AI47928). The authors have no conflicts of interest to disclose.
REFERENCES
- Almengor AC, Kinkel TL, Day SJ, McIver KS. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the group A streptococcus. J Bacteriol. 2007;189:8405–8416. doi: 10.1128/JB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, McIver KS. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J Bacteriol. 2004;186:7847–7857. doi: 10.1128/JB.186.23.7847-7857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, Walters MS, McIver KS. Mga is sufficient to activate transcription in vitro of sof/sfbX and other Mga-regulated virulence genes in the group A streptococcus. J Bacteriol. 2006;188:2038–2047. doi: 10.1128/JB.188.6.2038-2047.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen DE, Manoharan A, Luo F, Wertz JE, Robinson DA. Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes. J Bacteriol. 2005;187:4163–4172. doi: 10.1128/JB.187.12.4163-4172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, McDonald M, Wilson NJ. Acute rheumatic fever. Lancet. 2005;366:155–168. doi: 10.1016/S0140-6736(05)66874-2. [DOI] [PubMed] [Google Scholar]
- Cho KH, Caparon MG. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol Microbiol. 2005;57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. P-Ser-HPr--a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim Biophys Acta. 2005;1754:118–125. doi: 10.1016/j.bbapap.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Gold KM. Cell Biology and Molecular Genetics. University of Maryland: College Park; 2011. Characterization of the TrxSR two-component signal transduction system of Streptococcus pyogenes and its role in virulence regulation; p. 163. [Google Scholar]
- Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Microbiol. Nat. Rev. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Barry WT, Gowen BB, Johnson CR, Wright FA, Musser JM. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Gardner DJ, Long RD, Welty DM, Barry WT, Johnson CA, Parkins LD, Wright FA, Musser JM. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerstrom TG, Roh JH, Nikonowicz EP, Koehler TM. Bacillus anthracis virulence regulator AtxA: oligomeric state, function and CO(2) -signalling. Mol Microbiol. 2011;82:634–647. doi: 10.1111/j.1365-2958.2011.07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henstra SA, Duurkens RH, Robillard GT. Multiple phosphorylation events regulate the activity of the mannitol transcriptional regulator MtlR of the Bacillus stearothermophilus phosphoenolpyruvate-dependent mannitol phosphotransferase system. J Biol Chem. 2000;275:7037–7044. doi: 10.1074/jbc.275.10.7037. [DOI] [PubMed] [Google Scholar]
- Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- Hondorp ER, S.C. H, Hempstead AD, Hause LL, Beckett DM, McIver KS. Characterization of the group A streptococcus Mga virulence regulator reveals a role for the C-terminal region in oligomerization and transcriptional activation. Mol Microbiol. 2012;83:953–967. doi: 10.1111/j.1365-2958.2012.07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Camilli A. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol Microbiol. 2007;66:1–13. doi: 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyet P, Derkaoui M, Poncet S, Deutscher J. Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR. Mol Microbiol. 2010;76:1279–1294. doi: 10.1111/j.1365-2958.2010.07175.x. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kinkel TL, McIver KS. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect Immun. 2008;76:3451–3463. doi: 10.1128/IAI.00343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic I, Bachem S, Sickmann A, Meyer HE, Stulke J, Hengstenberg W. Regulation of the glucose-specific phosphotransferase system (PTS) of Staphylococcus carnosus by the antiterminator protein GlcT. Microbiology. 2000;146(Pt 9):2333–2342. doi: 10.1099/00221287-146-9-2333. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Loughman JA, Caparon MG. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 2006;25:5414–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo RL, Waygood EB. An enzymatic method for [32P]phosphoenolpyruvate synthesis. Anal Biochem. 1983;128:245–249. doi: 10.1016/0003-2697(83)90372-x. [DOI] [PubMed] [Google Scholar]
- McIver KS. Stand-alone response regulators controlling global virulence networks in streptococcus pyogenes. Contributions to Microbiology. 2009;16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- McIver KS, Heath AS, Green BD, Scott JR. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver KS, Myles RL. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol Microbiol. 2002;43:1591–1602. doi: 10.1046/j.1365-2958.2002.02849.x. [DOI] [PubMed] [Google Scholar]
- McIver KS, Scott JR. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol. 2007;64:771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Scott JR, Moran CP. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol Microbiol. 2001;42:495–502. doi: 10.1046/j.1365-2958.2001.02657.x. [DOI] [PubMed] [Google Scholar]
- Osipiuk J, R. Wu, Jedrzejczak R, Moy S, Joachimiak A. Structure for the Putative Mga family transcriptional regulator from Enterococcus faecalis. Protein Data Bank (PDB); 2011. M. C. f. S. G. (MCSG) (ed) [Google Scholar]
- Perez-Casal J, Price JA, Maguin E, Scott JR. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- Pine L, Reeves MW. Regulation of the synthesis of M protein by sugars, Todd Hewitt broth, and horse serum, in growing cells of Streptococcus pyogenes. Microbios. 1978;21:185–212. [PubMed] [Google Scholar]
- Ribardo DA, McIver KS. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol Microbiol. 2006;62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Scott JR. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol Microbiol. 2007;66:1506–1522. doi: 10.1111/j.1365-2958.2007.06015.x. [DOI] [PubMed] [Google Scholar]
- Rollenhagen C, Bumann D. Salmonella enterica highly expressed genes are disease specific. Infect Immun. 2006;74:1649–1660. doi: 10.1128/IAI.74.3.1649-1660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalisch MH, Bachem S, Stulke J. Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation. Elucidation of the phosphorylation chain leading to inactivation of GlcT. J Biol Chem. 2003;278:51108–51115. doi: 10.1074/jbc.M309972200. [DOI] [PubMed] [Google Scholar]
- Schrager HM, Rheinwald JG, Wessels MR. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J Clin Invest. 1996;98:1954–1958. doi: 10.1172/JCI118998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2008a;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Keith DB, Davenport MT, Beres SB, Carroll RK, Musser JM. Contribution of AmyA, an extracellular alpha-glucan degrading enzyme, to group A streptococcal host-pathogen interaction. Mol Microbiol. 2009;74:159–174. doi: 10.1111/j.1365-2958.2009.06858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Keith DB, Davenport MT, Horstmann N, Brennan RG, Musser JM. Molecular characterization of group A Streptococcus maltodextrin catabolism and its role in pharyngitis. Mol Microbiol. 2008b;69:436–452. doi: 10.1111/j.1365-2958.2008.06290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, Davenport MT, Keith DB, Musser JM. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008c;16:318–325. doi: 10.1016/j.tim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens N, Fiedler T, Normann J, Klein J, Munch R, Patenge N, Kreikemeyer B.Effects of the ERES Pathogenicity Region Regulator Ralp3 on Streptococcus pyogenes serotype M49 Virulence Factor Expression 2012J Bacteriol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MS, Matthews WJ, Jr., Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Stulke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD--a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- Stulke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathogens. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung K, Khan SA, Nawaz MS, Khan AA. A simple and efficient Triton X-100 boiling and chloroform extraction method of RNA isolation from Gram-positive and Gram-negative bacteria. FEMS Microbiol Lett. 2003;229:97–101. doi: 10.1016/S0378-1097(03)00791-2. [DOI] [PubMed] [Google Scholar]
- Tortosa P, Declerck N, Dutartre H, Lindner C, Deutscher J, Coq DLe. Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY. Mol Microbiol. 2001;41:1381–1393. doi: 10.1046/j.1365-2958.2001.02608.x. [DOI] [PubMed] [Google Scholar]
- Tsvetanova B, Wilson AC, Bongiorni C, Chiang C, Hoch JA, Perego M. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol Microbiol. 2007;63:644–655. doi: 10.1111/j.1365-2958.2006.05543.x. [DOI] [PubMed] [Google Scholar]
- Vahling CA. Mol Microbiol. Vol. 63. Dallas: Univesity of Texas Southwestern Medical Center; 2006. Functional Domains in the Multigene Regulator of the Group A Streptococcus; p. 185. [Google Scholar]
- Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, Parkins LD, Romero RA, Corn GJ, Gardner DJ, Bailey JR, Parnell MJ, Musser JM. Longitudinal analysis of the group A streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci U S A. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.