Abstract
Background
Azacitidine and decitabine are two hypomethylating agents approved by the Food and Drug Administration for the treatment of patients with myelodysplastic syndrome (MDS). The efficacy of one agent post failure of the other is unknown.
Methods
Fourteen patients with MDS post azacitidine failure/lack of response/intolerance were treated with decitabine.
Results
Overall 3 patients achieved a complete remission and 1 patient had hematologic improvement, for an overall response rate of 28%. Of the responders 1 stopped prior 5-azacitidine due to disease progression, 2 for no response and l for severe skin toxicity. Grade 3-4 drug related side-effects were minimal. Global methylation studies in patient samples showed decrease of methylation after treatment with decitabine. As in our previous studies, there was no difference in hypomethylation between responders and non-responders.
Conclusions
We conclude that clinically significant responses with decitabine can be seen in patients post azacitidine failure without significant toxicity.
Keywords: azacitidine failure, decitabine, myelodysplastic syndrome
INTRODUCTION
The cytidine analogs modified in position 5 of the pyrimidine ring, such as 5-azacitidine (azacitidine) and 5-aza-2′ deoxycytidine (decitabine), are potent inhibitors of deoxyribonucleic acid (DNA) methylation. Both agents are approved by the Food and Drug administration (FDA) for treatment of myelodysplastic syndromes (MDS). The efficacy of azacitidine and decitabine as anti-neoplastic agents is two-fold: at high concentrations they act as cytotoxic agents; at lower concentrations they induce hypomethylation,1, 2 leading to effects that are distinct from immediate cytotoxicity.
Both azacitidine and decitabine form covalent complexes with DNA methyl transferase 1 (DNMT1) and deplete the cell of functional DNA methylating activity. Incorporation of decitabine and azacitidine into DNA is required for their DNA hypomethylating activity.3 Decitabine being a deoxyribonucleoside, is incorporated only into DNA; azacitidine, a ribonucleoside, is predominantly incorporated into ribonucleic acid (RNA), and to a lesser extent into DNA. Their incorporation into DNA results in profound inhibition of DNMT1 activity. Functional consequences of incorporation of azacitidine into RNA include disassembly of polyribosomes and inhibition of protein production. Decitabine is a more potent hypomethylator in vitro.2 Based on these differences between azacitidine and decitabine, it is conceivable that patients with MDS post failure on one of these agents may respond to the other agent. We are conducting a phase II trial of decitabine in patients with MDS and failure on azacitidine (resistance or intolerance). Here we report on the first 14 patients treated as part of interim analysis. We also evaluated methylation studies (global and individual genes) in samples obtained from these patients at multiple time points.
PATIENTS AND METHODS
Study Group
All patients fulfilling the French, American and British (FAB) and/or World Health Organization (WHO) criteria for MDS and with ≥5% blasts in the bone marrow or International Prognostic Scoring System (IPSS)4 risk intermediate or high were eligible. No prior combination chemotherapy or high-dose cytarabine (≥ 1 g/m2) was permitted. Prior biologic therapies, targeted therapies, or single agent chemotherapy were allowed. Patients must have been treated with azacitidine and failed that treatment. Failure was defined as 1) no response after at least three cycles of azacitidine, 2) progression of disease on azacitidine or 3) grade 3-4 non-hematologic toxicity. Other eligibility criteria included performance status ≤2 (according to Eastern Cooperative Oncology Group scale), normal organ functions including creatinine ≤2 mg/dL bilirubin ≤2 mg/dL and a wash-out period of two weeks from prior therapy.
Treatment
Decitabine was administered at 20 mg/m2 intravenously (IV) over one hour daily times five days. Courses of decitabine were given every four weeks, at least in the first three courses, regardless of the counts, as long as (1) there were no significant myelosuppressive, life-threatening complications with a particular course, such as pneumonia, severe infection or bleeding, or severe organ damage, and (2) there was evidence of persistent disease. For patients achieving complete remission (CR), subsequent courses were started after recovery of counts (granulocytes ≥ 0.75 × 109/L and platelets ≥ 50 × 109/L).
Dose modifications were permitted for grade 3/4 non-hematological toxicity and for sustained low counts (granulocytes <0.5 × 109/L or a platelet count <30 × 109/L) for more than two consecutive weeks in the previous cycle (only applicable for patients with response to therapy). Use of hematopoietic growth factors was permitted according to institutional guidelines . In the absence of treatment delays due to adverse events, treatment was continued until relapse or lack of response after ≥ three courses.
Samples
Bone marrow and/or peripheral blood cells were collected from consenting patients according to institutional guidelines and IRB approved protocol and mononuclear cells were isolated. Genomic DNA was isolated using DNA STAT-60 reagent (Tel Test Inc. Diagnostics, Friendswood, TX) according to the manufacturer's instructions.
Bisulfite modification of DNA: Bisulfite induces deamination of unmethylated cytosines, converting unmethylated CpG sites to UpG without modifying methylated sites. Bisulfite treatment of genomic DNA was done according to published methods.5 Bisulfite treated DNA was purified with a Wizard miniprep Column (Promega Co., San Diego, CA), desulfonated with 0.3 mol/L NaOH at 25°C for five minutes, precipitated with ammonium acetate and ethanol, and resuspended in 20 μL distilled water.
Pyrosequencing: To study gene-related methylation in MDS samples, we used the pyrosequencing method.6 Polymerase chain reactions (PCRs) were carried in 50 μL reactions, including 2 μL bisulfite-treated DNA, 2 mmol/L MgCl2, 0.25 mmol/L deoxynucleotide triphosphate, 2.5 unit Taq polymerase, 16 mmol/L (NH4)2SO4, 67 mmol/L Tris-HCl (pH 8.8), 1 mmol/L 2-mercaptoethanol, and 100 nmol/L primers. Two-step PCRs were done. In the first step, we used 100 nmol/L forward primer and reverse primer, 100 nmol/L of 20-bp universal sequence connected to the 5′ end of the reverse (or forward) primer,2μL bisulfite treated DNA, and the PCR buffer mentioned above. DNA was amplified at proper PCR conditions for each assay. For the second step, we used 100 nmol/L forward (or reverse) primer, 100 nmol/L biotinylated universal primer, 0.5 μL 1st step PCR product, and PCR buffer and ran at proper conditions. Primer sequences are shown in Supplementary Table 1. The final biotin-labeled PCR product was captured by Streptavidin Sepharose HP (Amersham Biosciences, Uppsala, Sweden). PCR product bound on the bead was purified and made single stranded in a Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Inc., Westborough, MA). The sequencing primer (0.3 μmol/L; Supplementary Table 1) was annealed to the single-stranded PCR product, and pyrosequencing was done using the PSQ HS 96 Pyrosequencing System (Pyrosequencing). Quantification of cytosine methylation was done using the provided software (PSQ HS96A 1.2).
Response definition
The modified International working group (IWG) criteria for response in MDS7 were used to assess responses. A CR required normalization of the bone marrow and peripheral counts with 5% or less marrow blasts, a granulocyte count of 1 × 109/L or more, and a platelet count of 100 × 109/L or more, lasting for at least four weeks. A PR was similar to CR except for persistent marrow blasts above 5%, but which were reduced by 50% or more from baseline.8, 9 A marrow CR referred to reduction of marrow blasts to 5% or less without normalization of peripheral counts. Hematologic improvements (HIs) were coded as the modified IWG criteria: HI-E referred to a hemoglobin increase by at least 15 g/L (1.5 g/dL) or transfusion independence; HI-P referred to an absolute increase of platelet counts from less than 20 to more than 20 × 109/L and by at least 100%, or if more than 20 × 109/L, by an absolute increase of at least 30 × 109/L;HI-N referred to a granulocyte increase by at least 100% and by an absolute increase of at least 0.5 × 109/L. HIs were required to last for at least two months
Statistical consideration
The primary trial objective was to assess efficacy of decitabine in MDS post azacitidine failure. Since the expected response rate in this population is unknown, a response rate (CR + PR +HI) of 20% or more was considered of interest. Descriptive statistics were used for patient characteristics and responses. Survival was estimated by Kaplan-Meier method. Times to best response were measured from study entry. Durations of response were measured from initial response to relapse.
Methylation studies: The global methylation data generated was expressed as a percentage and showed a normal distribution. The data was summarized (mean, median, standard error of the mean) using the Excel software (Microsoft, Redmond, WA). For evaluation of the methylation levels of several genes in the same sample, Z-score analysis was used to normalize the data and allow the derivation of a mean methylation score. Z score of methylation for each gene was calculated as follows: Z-score=(methylation density of each sample-mean value of methylation density)/ SD of methylation density. For analysis of multiple genes we used the average of Z scores for each gene.
Mann-Whitney U test was used to compare methylation at different time points as appropriate. A P value of .05 was considered significant.
RESULTS
Patient characteristics
Fourteen patients with MDS and prior azacitidine therapy were enrolled at the time of this interim analysis. Azacitidine was discontinued in one patient because of grade 3 skin toxicity, in two because of loss of response, in four due to progression of disease, and in seven for lack of response. Apart from azacitidine ± growth factors, three patients received additional treatments for MDS: one with bevacizumab, one with lenalidomide and one with thalidomide and tipifamib. The median number of cycles of prior azacitidine among all patients was four (range, 1-10). The patients who discontinued azacitidine due to lack of response also received the same median number of prior azacitidine. Twelve patients had received ≥ three courses of azacitidine. The patient who received one course had grade 3 skin rash with azacitidine. All except one patient were over the age of 60 years. Most had IPSS risk category intermediate-2 or high. The patient characteristics are summarized in Table 1.
Table 1.
Patient Characteristics
| N=14 | ||
|---|---|---|
| Parameter | No (Percent) | Median (Range) |
| Diagnosis | ||
| MDS | 13 (93) | |
| CMML | 1(7) | |
| Age (years) | 74.5 (58-85) | |
| No. Azacitidine courses | 4 (1-10) | |
| IPSS Risk Score | 2 (0.5-3) | |
| Bone marrow blasts ≥10% | 12 (86) | |
| 2 or more cytopenias | 10 (72) | |
| Cytogenetics: IPSS Risk category | ||
| Low | 8 (57) | |
| Intermediate | 2 (14) | |
| High | 4 (29) | |
| Reason off Azacitidine | ||
| No response | 7 (59) | |
| Progression | 4 (29) | |
| Relapse | 2 (14) | |
| Toxicity | 1 (7) | |
Abbreviations: MDS= myelodysplastic syndrome, CMML= chronic myelomonocytic leukemia, IPSS= international prognostic scoring system
Responses
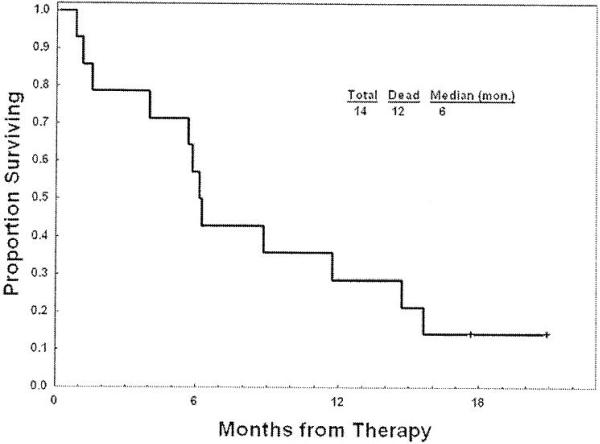
According to study a 20% response at interim analysis was to be considered of sufficient interest. Four patients (28%) responded according to the modified IWG criteria: Three (21%) achieved CR and one had a marrow CR with hematological improvement in platelet (7%) (HI-P) (Table 2). Of the responders, 1 stopped prior 5-azacitidine due to disease progression, 2 for no response and 1 for severe skin toxicity. The median survival in all patients was six months (range, 1 to 14.8 months)(Fig.1) and the median time to progression was four months (range 1 to 11.3 months).
Table 2.
Response Summary
| No (percent) | Median (range) | |
|---|---|---|
| Responses | ||
| CR | 3 (21) | |
| Marrow CR with HI | 1 (7) | |
| Stable disease | 5 (36) | |
| Progressive disease / death | 4/1 (29/7) | |
| No of DAC courses to response | 3 (1-5) | |
| Median survival (months) | 6 (1 -14.8) |
Abbreviations: CR= complete remission, HI= hematological improvement, DAC= decitabine
Fig. 1.
Overall survival of all14 patients
In the four patients responding to decitabine, the median number of courses of decitabine to response was three (range, 1-5). Among these four patients, one patient, who had discontinued azacitidine because of severe skin toxicity, achieved CR after 1 cycle of decitabine. Two patients were treated with 4 and 8 cycles of prior azacitidine, and achieved CR after 5 and 3 cycles of decitabine respectively. Two of these 3 patients eventually had progressive disease after 9.7 and 10.2 months; the third was taken off study after 11 .3 months of CR because of commercial availability of decitabine. The fourth patient had a marrow CR with improvement in platelet counts after 3 cycles of decitabine and proceeded for a stem cell transplant. Table 3 summarizes the details of the responders.
Table 3.
Characteristics of Responders
| No. prior Aza courses | Best response Aza | Reason off Aza/Weeks off Aza | Weeks from prior Aza before DAC | Best response DAC/Courses to response | Response Duration (months) | Percent marrow Blasts Pre/at response | Platelets Pre/at response | ANC Pre/at response | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 8 | Marrow CR | PD | 3 | CR/3 | 9.7 | 15/1 | 24/336 | 1.1/3.2 |
| 2 | 4 | SD | NR | 11 | Marrow CR/3 | 8.2 | 8/4 | 65/95 | 1.8/5.1 |
| 3 | 4 | SD | NR | 9 | CR/5 | 11.3+ | 12/3 | 80/234 | 0.6-1.4 |
| 4 | 1 | N/A | Toxicity | 5 | CR/1 | 10.2 | 13/4 | 24/110 | 0.38/2.8 |
Abbreviations: CR= complete remission, Aza= azacitidine, DAC= decitabine, SD=stable disease, PD= progressive disease, NR= no response, ANC= absolute neutrophil count
Two of the three patients who achieved CR. had abnormal cytogenetics at enrollment. These were trisomy 8 in one and complex cytogenetics with deletions of chromosomes 7,4 and 12 in the other. At CR both patients had normal cytogenetics.
Toxicities
Four patients experienced Grade 3/4 non-hematological toxicities. These included two episodes of fatigue, one syncope, one elevated liver enzymes and one hypokalemia. Febrile neutropenias were encountered in five (33%) patients and resolved with antimicrobial therapy. There were no treatment-related deaths. The only death that occurred while on study happened under unclear circumstances during Hurricane Katrina. Four patients needed dose reduction of decitabine due to cytopenias. Twelve patients needed recombinant erythrpoetin and/or filgrastim for treatment of cytopenias.
Methylation studies
Global methylation (LINE 1) and methylation of six genes (Npm2, PGRA , PGRB, Clorfl02, OLIG2 and CDH13) were studied by bisulfite pyrosequencing, before each decitabine cycle on all available samples.
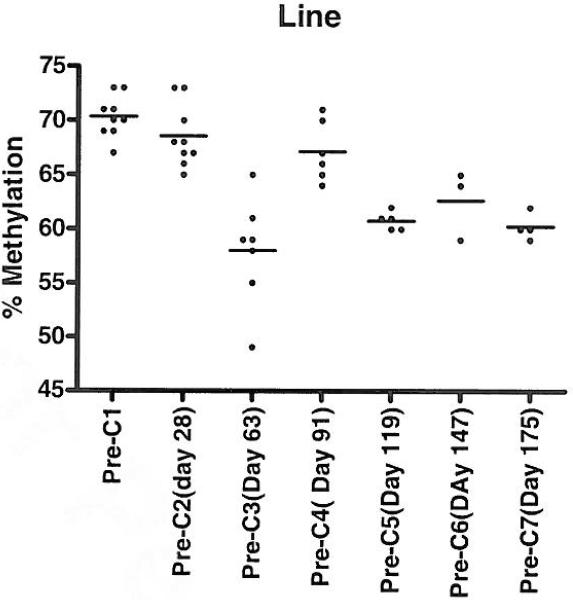
Pre-treatment LINE methylation for all samples (n=l4) was 71.43 +/-0.71 (mean +/- standard error of the mean [SEM]). There was no difference in pre-treatment LINE methylation between responders (70.83 +/- 2.83) and non responders ( 71.87 +/- 0.71 ) (p=0.6). LINE methylation pre-Cycle 2 showed a decrease to a mean +/- (SEM) 68.55 +/- 1.41; it decreased further pre-Cycle 3 to a mean +/- (SEM) 58 +/- 0.71(p <0.01 compared to pretreatment). LINE methylation between responders and non-responders at the end of cycle 2 and beyond was not significantly different (p= 0.8). Mean values for methylation prior to cycle 2 and subsequent cycles remained consistently lower than pretreatment, with some variation (Fig. 2).
Fig. 2. Serial LINE methylation studies reflecting changes in global methylation.
This scatter plot analysis shows LINE methylation changes as treatment cycles went on. The highest decrease in methylation occurred after 2 cycles. It is also important to note that at any given cycle LINE methylation was lower than baseline.
Gene specific methylation was measured at baseline, using a cut-off of 10% (15% for PGRB) methylation measurement to be positive. Methylation was noted in 3/14 samples for Npm2, 8/14 for PGRA, 4/14 for PGRB, 0/14 for Clorfl02 ,11/14 for OLIG2 and 11/14 for CDH13. There was no association between methylation of any gene at baseline and response.
Methylation of all six genes was averaged by z-score correction to derive a composite methylation score. There was no correlation of this score with response (p=0.1579). With regards to changes in gene-specific methylation after treatment, generally, too few genes were informative for this analysis. Using strict criteria to define a change (>20% methylation density at baseline, decrease in absolute methylation by at least 10%), only one gene (PGRB) changed significantly after treatment in 2/3 informative patients but these two patients did not respond to the therapy. When analyzing the three patients with CR separately, they generally had low levels of methylation at baseline. One patient showed decreased methylation of OLIG2 from 17% to 10% after 2 cycles and CDH13 from 17% to 12% after 2 cycles. The second patient had persistent methylation of PGRB, OLIG2 and CDH13 after 2 cycles, despite achieving a cytogenetic CR. The third patient had no methylation of any of the genes analyzed. Thus, overall, in this limited analysis, we were unable to find a consistent relation between gene-specific hypomethylation induction and response.
DISCUSSION
In phase III studies,10, 11 azacitidine and decitabine were associated with better overall reponses and CRs and lower rates of transformation to acute leukemia compared with supportive care. With optimized dosing schedules, the use of both agents result in comparable responses.12, 13 Efficacy of one agent in the setting of failure of another has not been reported. In this study, decitabine therapy resulted in a response rate of 28% in patients with MDS after failure/lack of response to or intolerance of azacitidine. LINE methylation was reduced after treatment with decitabine but lack of data regarding methylation studies during prior therapy with azacitidine limits our ability to comment on lack cross-resistance between these two hypomethylating agents. There was no difference in LINE or individual gene methylation among responders and non-responders.
Data with azacitidine showed that the median number of cycles to first response was three with 75% of responders achieving a response by cycle 4.13 Similarly, the median number of courses of decitabine to achieve CR in MDS was three.10, 12 Three out of four responders in our report received 4 or more cycles of prior azacitidine. However, based on the knowledge that 25% of patients on azacitidine can respond after 4 cycles of therapy, we can not completely rule out the possibility that continuation of azacitidine therapy could have produced response among the 7 patients included in this study who had discontinued azacitidine due to lack of response. Repetitive sequences called long interspersed nucleotide elements (LINE) and Alu are normally methylated, and can be quantified by bisulfite-pyrosequencing or other methods.14 Changes in LINE methylation can be used as a surrogate marker of global methylation changes.14 We have examined global and gene-specific DNA methylation changes in patients with leukemi a and MDS treated with decitabine.15, 18
In these studies,16 decitabine induced significant LINE hypomethylation as early as day 5, and the degree of hypomethylation was most prominent in patients receiving 20 mg/m2 IV daily X 5, a dosing schedule which also produced the best clinical response rate. The degree of LINE hypomethylation was similar in patients with or without clinical response, suggesting that LINE hypomethylation is a good pharmacodynamic surrogate of decitabine's hypomethylating activity, but is not necessarily a biological surrogate of its clinical activity. A likely explanation for this is that events downstream of hypomethylation are key to achieving responses.
Our global methylation data are incomplete in that no samples were available immediately post treatment to establish a pharmacodynamic correlation. Nevertheless, global methylation was clearly reduced after decitabine (most significantly after 2 cycles), indicating that none of the patients had pharmacodynamic resistance to the drug.
In summary, our data show that clinical response as well as reduction in global methylation with decitabine therapy can be seen in patients with MDS lacking response to or progressing on azacitidine. The study continues to accrue patients but we believe that only studies with randomized cross-over design will eventually define the clinical efficacy of one hypomethylating agent in the setting of failure of another. We present the results of our interim analysis in this report to generic interest towards such a randomized study.
Supplementary Material
Condensed Abstract.
Fourteen patients with myelodysplastic syndrome were treated with decitabine post failure with azacitidine. Overall 3 patients achieved a complete remission and 1 patient had hematologic improvement with minimal grade 3-4 toxicity, for an overall response rate of 28%.
References list
- 1.Christman JK, Mendelsohn N, Herzog D, Schneiderman N. Effect of 5-azacytidine on differentiation and DNA methylation in human promyelocytic leukemia cells (HL-60). Cancer Res. 1983;43(2):763–9. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=6184156. [PubMed] [Google Scholar]
- 2.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Bioi Chem. 1982;257(4):2041–8. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=6173384. [PubMed] [Google Scholar]
- 3.Leone G, Teofili L, Voso MT, Lubbert M. DNA methylation and demethylating drugs in myelodysplastic syndromes and secondary leukemias. Haematologica. 2002;87(12):1324–41. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12495905. [PubMed] [Google Scholar]
- 4.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–88. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9058730. [PubMed] [Google Scholar]
- 5.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22(15):2990–7. doi: 10.1093/nar/22.15.2990. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8065911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35(1):146–50. doi: 10.2144/03351md01. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12866414. [DOI] [PubMed] [Google Scholar]
- 7.Cheson BD, Greenberg PL, Bennett JM, Lowenberg B, Wijermans PW, Nimer SD, et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood. 2006;108(2):419–25. doi: 10.1182/blood-2005-10-4149. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=l6609072. [DOI] [PubMed] [Google Scholar]
- 8.Cheson BD, Bennett JM, Kantarjian H, Pinto A, Schiffer CA, Nimer SD, et al. Report of an international working group to standardize response criteria for myelodysplastic syndromes. Blood. 2000;96(12):3671–4. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11090046. [PubMed] [Google Scholar]
- 9.Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–9. doi: 10.1200/JCO.2003.04.036. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=l4673054. [DOI] [PubMed] [Google Scholar]
- 10.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106(8):1794–803. doi: 10.1002/cncr.21792. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16532500. [DOI] [PubMed] [Google Scholar]
- 11.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20(10):2429–40. doi: 10.1200/JCO.2002.04.117. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=l2011120. [DOI] [PubMed] [Google Scholar]
- 12.Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O'Brien S, Cortes J, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–7. doi: 10.1182/blood-2006-05-021162. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=l6882708. [DOI] [PubMed] [Google Scholar]
- 13.Silverman LR, McKenzie DR, Peterson BL, Holland JF, Backstrom JT, Beach CL, et al. Further analysis of trials with azacitidine in patients with myelodysplastic syndrome: studies 8421, 8921, and 9221 by the Cancer and Leukemia Group B. J Clin Oncol. 2006;24(24):3895–903. doi: 10.1200/JCO.2005.05.4346. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16921040. [DOI] [PubMed] [Google Scholar]
- 14.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14973332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66(10):5495–503. doi: 10.1158/0008-5472.CAN-05-2385. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16707479. [DOI] [PubMed] [Google Scholar]
- 16.Oki Y, Kantarjian H, Davis J, Ravandi F, Verstovsek S, Cortes J. Hypomethylation induction in MDS after treatment with decitabine at three different doses. J Clin Oncol. 2005;16S [abstr 6546] [Google Scholar]
- 17.Kantarjian HM, O'Brien S, Cortes J, Giles FJ, Faderl S, Issa JP, et al. Results of decitabine (5-aza-2′deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer. 2003;98(3):522–8. doi: 10.1002/cncr.11543. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12879469. [DOI] [PubMed] [Google Scholar]
- 18.Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103(5):1635–40. doi: 10.1182/blood-2003-03-0687. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=l4604977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.