Abstract
Brain tumors are heterogeneous tumors composed of differentiated tumor cells that resemble various neural cells and a small number of multipotent cancer stem cells. These tumors modify normal cells in their environment to promote tumor growth, invasion and metastases by various ways. Recent publications show that glioblastoma cells release microvesicles that contain a select subset of cellular proteins and RNAs. These microvesicles are avidly taken up by normal cells in cell culture and can change the translational profile of these cells through delivery of tumor-derived mRNAs, which are translated into functional proteins. In addition to mRNA and proteins, microvesicles have been shown to contain microRNAs, non-coding RNAs and DNA. This commentary explores the recent advances in this novel intercellular communication route and discusses the potential physiological role of microvesicles in brain tumorigenesis.
Keywords: Microvesicles, Glioblastoma, Biomarkers, Gene transfer, Tumor microenvironment, Non-coding RNAs, Retrotransposons, MicroRNAs
Introduction
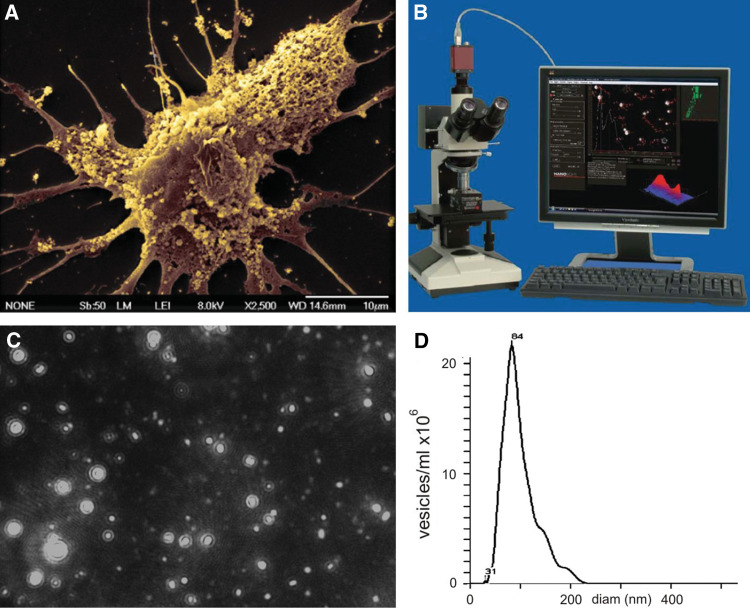
Brain tumors are believed to derive from neural stem cells (Galli et al. 2004; Germano et al. 2010), with a subset retaining their multipotent properties (cancer stem cells) and others expressing differentiated properties of various neural cell types in the nervous system. Tumors are dangerous expansions of abnormal brain cells and represent a grotesque caricature of normal brain cells, with exaggerated features that may help us understand properties of normal neural cells which may have been overlooked. A dramatic example is the finding of abundant microvesicles (MVs) on the surface of primary human glioblastoma (GBM) cells in culture (Skog et al. 2008, Fig. 1a). These MVs, which are “invisible” by standard microscopy, can be visualized by nanoparticle tracking analysis (NTA) which reveals particles with sizes in the range of 10 nm–1 μm in diameter (Fig. 1b–d), with primary GBM cells in culture releasing about 10,000 MVs per cell over 48 h (Balaj et al. 2011). These MVs contain a plethora of protein and nucleic acid information, including cytoplasmic and membrane proteins, mRNAs, microRNAs (miRNAs), non-coding RNAs (ncRNAs), and both genomic DNA and cDNAs (ibid.), which can be transferred to other cells (Fig. 2). The presence of these tumor-derived MVs in the circulation of GBM patients and their loss upon removal of the tumor (Skog et al. 2008) confirms that they are produced in vivo. More importantly, they are thought to have a critical role in manipulating normal cells in the tumor microenvironment in favor of tumor growth by increasing invasion and angiogenesis, and decreasing immune responses (Théry et al. 2009; Iero et al. 2008). These MVs are also released by neural stem cells during development (Marzesco et al. 2005) and represent a novel means of intercellular communication involving genetic information and non-secreted proteins (Simons and Raposo 2009).
Fig. 1.
Monitoring MVs. a Scanning transmission electron micrograph of primary human GBM cell (bar 10 μm) (Skog et al. 2008), b Nanosight microscope (Nanosight Ltd.), c Serum MVs diluted 1/5000 and visualized using the Nanosight Tracking Analysis, d Histogram showing distribution of MV diameter in serum sample
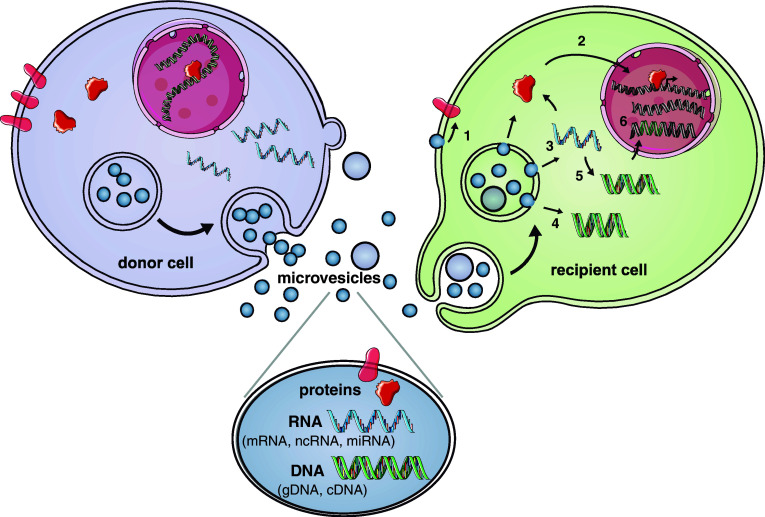
Fig. 2.
MV-mediated intercellular communication. Components of donor cells are incorporated into MVs which contain proteins (e.g., signaling proteins, transcriptional regulators, RT, and transmembrane proteins), RNAs (i.e., mRNAs, miRNAs, and ncRNAs), and DNA (i.e., cDNA and genomic DNA). MVs can be taken up by recipient cells through endocytosis and release their contents after fusing with the endosomal membrane, or fusion at the plasma membrane. 1 Transmembrane proteins can be transferred to the plasma membrane and trigger signaling. 2 Transcriptional regulators can potentially be transferred into the nucleus and regulate promoter activity. 3 mRNAs/miRNAs can be transferred and influence the translational profile. 4 Donor cell-derived cDNAs, e.g., c-Myc can be delivered directly within MVs. 5 or generated from reverse-transcribed mRNAs in the cell of origin, within MVs or possibly in the recipient cell. 6 Retrotransposon and other DNA elements from MVs may integrate into the recipient cell genome. In one scenario, the donor cell is a tumor cell and the recipient cells are normal cells in the microenvironment. These MV delivery events have the potential to change the phenotype of normal cells to make them more supportive of tumor growth (figure modified from Dr. Charles Lai; produced using Servier Medical Art, http://www.servier.com)
The definition of MVs is currently a subject of debate, so for purposes of this commentary the term will comprise membrane-bound vesicles, including exosomes, shedding MVs, microparticles, retroviral-like particles, and apoptotic bodies. These can be produced by release of vesicles contained within endosome-derived multivesicular bodies (MVBs) or by protrusion and budding from the plasma membrane (Cocucci et al. 2009). Upon release, MVs can influence many biological processes, for example, by binding to and being taken up by target cells via ligand-receptor interactions. In some cases, MVs are thought to be taken up by cells through endocytosis and to release their intravesicular content after fusing their membrane with the endosomal membrane. Alternatively, MVs may fuse directly with the cellular plasma membrane. Although the precise mechanisms of uptake are poorly understood, it is evident that release of signaling proteins and RNA from the lumen of MVs can induce activation of specific signal transduction cascades and influence the physiologic state of recipient cells. Microvesicles have an additional advantage over naturally secreted signaling molecules in that they can present multiple epitopes to the recipient cell, allowing co-stimulatory pathways to be activated. Like the two faces of Janus, MVs can serve as information packets to guide the phenotype of surrounding cells, or be used to rid a cell of unwanted components. Release of MVs is exaggerated in tumor cells, but also occurs in most normal cells. Neurons (Fauré et al. 2006), astrocytes (Taylor et al. 2007; Bianco et al. 2009), oligodendrocytes (Krämer-Albers et al. 2007; Trajkovic et al. 2008), and microglia (Potolicchio et al. 2005; Bianco et al. 2009), as well as embryonic neural stem cells (Marzesco et al. 2005; Yuan et al. 2009) have all been described to release MVs, which are thought to play an important role in development and function of the nervous system and other tissues by providing a gradient of morphogen polarity (Lakkaraju and Rodriguez-Boulan 2008). Again, in a parallel with neural stem and progenitor cells (Singer et al. 2010), as well as embryonic cells (Macia et al. 2011), brain tumor cells also have a relatively hypomethylated genome, as compared to differentiated cells, with increased expression of retrotransposon elements associated with plasticity of the genome (Cordaux and Batzer 2009). This raises the question of whether normal neural stem/progenitor cells also release retrotransposon elements within MVs to orchestrate developmental patterning and morphogenesis in the nervous system.
Microvesicle-Mediated Information Transfer and Potential Changes in the Tumor Microenvironment
Transfer of mRNAs and miRNAs by Microvesicles
In 2006 Ratajczak et al. showed that MVs derived from embryonic stem cells contained mRNA encoding pluripotent transcription factors. Furthermore, Valadi et al. (2007) showed that mRNA from mast cell-derived MVs could be translated into proteins following uptake, demonstrating the potential to alter the translational profile of the recipient cell. These surprising findings resulted in the intriguing hypothesis that, similar to viruses, MVs can transfer genetic information between cells. This type of intercellular genetic communication could play an important role in tissue development and homeostasis, as well as in tumor manipulation of its microenvironment, and might even affect distant sites by trafficking of MVs through the systemic circulation. For example, GBM cells release MVs that contain a concentrated repertoire of mRNAs associated with growth, invasion, and immune repression (Skog et al. 2008). Interestingly, although most of the RNA transcriptome of GBM cells was found in the MVs, the relative concentration of specific mRNAs was enriched in MVs, as compared to the cell of origin, suggesting that there might be a molecular process involved in selective packaging of mRNAs in MVs. These tumor-derived MVs were found to be avidly taken up by normal brain microvascular endothelial cells in culture and to promote an angiogenic phenotype (Skog et al. 2008). Both Valadi et al. (2007) and Skog et al. (2008) also provided evidence that mRNAs in MVs taken up by recipient cells can be translated within them. Microvesicle-mediated transfer of mRNA has also been demonstrated between embryonic stem cells and hematopoietic progenitors (Ratajczak et al. 2006), endothelial progenitor cells and endothelial cells (Deregibus et al. 2007) and mesenchymal stem cell and epithelial cells (Bruno et al. 2009). In addition, MVs can serve as a means of transfer of cytoplasmic proteins, e.g., GFP, as well as membrane proteins, e.g., a mutant form of epidermal growth factor receptor (EGFRvIII) to recipient cells (Al-Nedawi et al. 2008; Yuan et al. 2009).
The release of MVs containing mRNAs has been described for other tumor types including colon and gastric cancer (Hong et al. 2009), indicating that transfer of genetic information via MVs is a general phenomenon in oncogenesis. Interestingly, comparison of microvesicular RNA derived from GBM cells and colorectal cancer cells revealed a large overlap of mRNA transcripts between the two cancer cell types. Network analysis indicated that mRNAs involved in tumorigenesis-related processes, such as cell cycle regulation and metabolic processes were over represented in MVs from both cancer cell types, suggesting that they promote common oncogenic pathways.
In addition to mRNA transcripts, miRNA molecules are also present in MVs derived from tumor cells such as GBM, lung cancer, gastric cancer, and ovarian cancer (Skog et al. 2008; Rabinowits et al. 2009; Ohshima et al. 2010; Taylor and Gercel-Taylor 2008). Similar to mRNAs, the tumor-released MVs contain a miRNA signature representative of the tumor cells from which they originate, with specific miRNAs enriched in MVs compared to their cells of origin. For example, MVs isolated from serum of ovarian cancer patients showed enhanced levels of 8 miRNAs previously found to be overexpressed in ovarian cancer (Taylor and Gercel-Taylor 2008). These small regulatory miRNA molecules have been found to have a critical role in the progression of various cancers. By negatively regulating their mRNA targets through degradation or translational repression, they can act either as tumor suppressors or oncogenes depending on their targets (Croce 2009). It has been estimated that a single miRNA might interact with up to 200 different mRNAs and that relatively minor changes in levels of specific miRNAs can have a large impact on the physiologic state of cells. This indicates that transfer of miRNAs from tumor cells (or normal cells) to surrounding tissue cells through MVs could potentially play an important role in modulating normal cells in the microenvironment.
Although miRNAs have been detected in MVs from multiple cancer types, their potential to regulate translation in recipient cells is an ongoing investigation. Several recent publications demonstrate that functional miRNAs can be transferred between cells by MVs. For example, monocyte-derived MVs transfer functional miR-150 to endothelial cells, resulting in repression of c-Myb, thereby decreasing cell proliferation (Zhang et al. 2010), and MVs released by B cells infected with Epstein Barr virus transfer miRNAs which repress immunoregulatory genes in dendritic cells (Pegtel et al. 2010). Insight into MV-mediated modification of the transcriptome of normal and tumor cells in the tumor environment can provide a new venue for development of cancer therapeutics. For instance, regulatory mechanisms mediated by upregulated miRNAs may be normalized using specific antagomirs to those miRNAs and translation of mRNAs can be curtailed using siRNAs electroporated into MVs (Alvarez-Erviti et al. 2011).
The “Dark Matter of the Transcriptome”
A fascinating component of tumor MVs is their high content of non-coding (nc) RNAs (Balaj et al. 2011; Skog, unpublished data). The human genome only contains about 20,000 protein coding genes, representing less than 2% of the genome. Until recently, most mapping projects have focused on the protein-coding sequences. The authors now know that transcription at different levels occurs from most parts of the genome and that these ncRNAs have many important functions in the cell. When looking at the genomes of different organisms, it is fascinating that the ratio of non-coding to protein coding DNA sequences increases as a function of developmental complexity (Mattick 2004). Prokaryotes have less than 25% ncDNA, simple eukaryotes have between 25 and 50%, more complex multicellular organisms like plants and animals have more than 50% ncDNA, and humans have about 98.5% ncDNA (Mattick 2004) (Fig. 3). This suggests that the high content of ncRNAs in MVs may be a critical feature of intercellular communication in higher organisms.
Fig. 3.
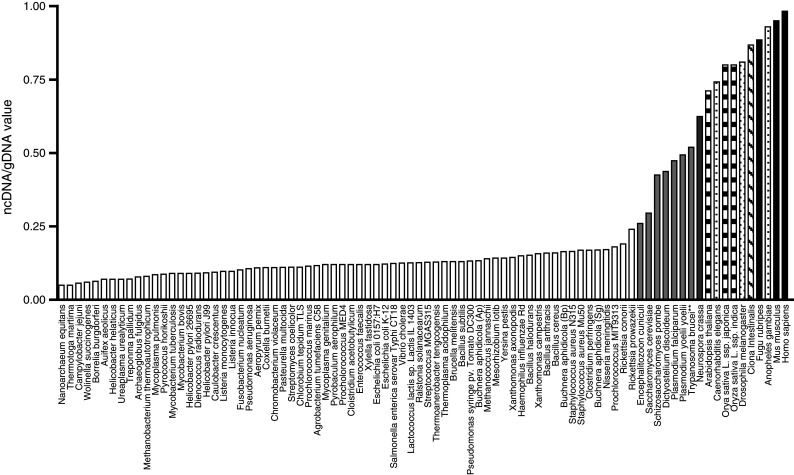
Multicellular organisms have high levels of non-coding DNA sequences. The ratio of ncDNA to total genomic DNA (ncDNA/tgDNA) increases with the biological complexity of organisms.  prokaryotes,
prokaryotes,  unicellular eukaryotes,
unicellular eukaryotes,  the multicellular fungus Neurospora crassa,
the multicellular fungus Neurospora crassa,  plants,
plants,  non-chordate invertebrates (nematodes, insects),
non-chordate invertebrates (nematodes, insects),  Ciona intestinalis (urochordate),
Ciona intestinalis (urochordate),  vertebrates. (Reproduced with permission from Taft and Mattick 2003)
vertebrates. (Reproduced with permission from Taft and Mattick 2003)
ncRNAs have been implicated in many important processes in the cell, including functioning enzymes (ribozymes), binding to specific proteins (aptamers), and regulating gene activity at both the transcriptional and post-transcriptional levels. The function of most ncRNAs has not yet been determined. Examples of ncRNA classes, their functions and the presence in MVs are shown in Table 1. Interestingly, many of the ncRNA species have multiple, seemingly non-related functions. For example, Ribonuclease P (RNase P) is a ribozyme which is involved in maturation of tRNA by cleaving the precursor tRNA, but nuclear RNaseP can also act as a transcription factor (Jarrous and Reiner 2007). In addition, bifunctional RNAs have also been described that function both as mRNA and nc regulatory RNAs (Dinger et al. 2008), or that have two different ncRNA functions (Ender et al. 2008). One example of the many long ncRNAs is the X-inactive specific transcript (Xist) expressed by the inactive X-chromosome, which is used to silence the extra X-chromosome in females (Ng et al. 2007). This RNA transcript binds to and inactivates the same X chromosome from which it is produced. Another example is the HOX antisense intergenic RNA (HOTAIR) (Rinn et al. 2007). This RNA is expressed from chromosome 12, but controls gene expression on chromosome 2, affecting the skin phenotype on the different parts of the body surface (Rinn et al. 2007), as well as being involved in cancer metastasis (Gupta et al. 2010).
Table 1.
Examples of non-coding RNAs in nature and presence in microvesicles
| Non-coding RNA | Abbreviation | Example of function | References | MVs* |
|---|---|---|---|---|
| Messenger RNA | mRNA | Coding information for translation | (Aitken et al. 2010) | Yes (see Transfer of mRNAs and miRNAs by microvesicles) |
| Transfer RNA | tRNA | Translation | (Aitken et al. 2010) | No |
| Ribosomal RNA | rRNA | Translation | (Aitken et al. 2010) | Yes (Hong et al. 2009) |
| Signal recognition particle RNA | 7SL RNA or SRP RNA | Translocation of proteins across the endoplasmatic reticulum | (Gribaldo and Brochier-Armanet 2006) | No |
| Small nuclear RNA | snRNA | Splicing | (Valadkhan 2010) | Yes (Valadi et al. 2007) |
| Small nucleolar RNA | snoRNA | Guides chemical modifications of other RNAs (like methylation and pseudouridylation) | (Kiss 2002) | Yes (Hunter et al. 2008) |
| Short interspersed repetitive elements | SINE | The most common SINE is the Alu element (~10% of the genome). Alu is upregulated in response to stress and binds RNA polymerase II to suppress transcription | (Mariner et al. 2008) | Yes (see Retrotransposons) |
| microRNA | miRNA | Post-transcriptional gene silencing | (Bartel 2009) | Yes (see Transfer of mRNAs and miRNAs by microvesicles) |
| Small interfering RNA | siRNA | Post-transcriptional gene silencing | (Elbashir et al. 2001) | Yes (Kosaka et al. 2010) |
| Piwi-interacting RNA | piRNA | Transcriptional gene silencing, defense against retrotransposons | (Taft et al. 2010) | No |
| Ribonuclease P | RNase P | Ribozyme involved in tRNA maturation | (Guerrier-Takada et al. 1983) | No |
| Ribonuclease MRP | RNase MRP | Ribozyme involved in rRNA maturation as well as mitochondrial DNA replication | (Li et al. 2002) | No |
| Y RNA | Y RNA | RNA processing, DNA replication | (Lerner et al. 1981) | No |
| Telomerase RNA | Telomere synthesis | (Feng et al. 1995) | No | |
| Antisense RNA | aRNA | Transcriptional attenuation/mRNA degradation/mRNA stabilization/translation block | (Katayama et al. 2005) | No |
| Long ncRNA, large intervening ncRNA (>200 nt) | Long ncRNA, lincRNA | Regulation of gene transcription, post-transcriptional regulation, epigenetic regulation | (Kapranov et al. 2007) | Yes (Nilsson et al. 2009) |
* Documented presence in MVs
It is thought-provoking to speculate about the functional aspect of ncRNA (as well as other RNA and DNA) in MVs as they appear to be important modulators of cellular responses. Of the different ncRNA classes, many have been detected in MVs including ribosomal RNA, small nuclear RNA, short interspersed RNA, miRNA, and siRNA. Although studies have begun to unravel the functional consequences of MV-mediated transfer of mRNAs and miRNAs, the contributions of other ncRNAs awaits further research.
Retrotransposons
Retrotransposon elements make up a major component of repetitive sequences comprising approximately 45% of the human genome and have played an important role in driving evolution and shaping the genome through altering gene content and expression (Cordaux and Batzer 2009). Only a small percentage (<0.05%) of these elements are capable of active genomic retrotranslocation (Mills et al. 2007), but their overall expression is increased both in embryonic cells and tumor cells, as compared to differentiated cells, through hypomethylation of the genome (Cordaux and Batzer 2009). In fact, retrotransposon reinsertion events in the human genome are thought to occur in about 0.1% of births (Cordaux and Batzer 2009) and can be visualized with marker proteins in neural progenitor cells (Singer et al. 2010). These new insertional events can engender positive aspects of genomic plasticity, as well as potentially negative mutagenic events. These retroposon sequences are highly upregulated in MVs from tumor cells.
Retrotransposons move by a copy and paste mechanism which involves reverse transcriptase (RT), endonuclease cuts in the DNA, and integrase activity. At the end of these steps, a new copy of the retrotransposon element is created and inserted into the genome. Most of these elements are normally silent, but under pathological condition, such as cancer, they may become transcriptionally active. Repetitive microsatellite sequences are highly upregulated in cancer, as noted for example in high-level expression of LINE-1 retrotransposons upon microsatellite deregulation (Ting et al. 2011). The most recently integrated retrotransposons, such as human endogenous retroviral sequences, HERV-H, HERV-W, and HERV-K are regulated primarily by DNA methylation (Szpakowski et al. 2009). An inverse correlation, for example, has been observed between HERV-K transcriptional activity and DNA hypomethylation levels (Lavie et al. 2005). The most recent retrotransposon entries into the genome also have the most conserved coding sequences, thus increasing their potential for mobility within the genome.
High levels of RT activity have been detected under some normal conditions, such as in preimplantation embryos (Poznanski and Calarco 1991) and placenta (Mwenda 1993), as well as under pathological conditions, such as cancer (Ruprecht et al. 2008). In contrast, RT activity is scarcely detectable in somatic tissue suggesting its correlation to cell differentiation status. Retrotransposition depends on RT activity and remarkably, treatment of cancer cells with nevirapine, an inhibitor of RT, led to a decrease in cancer cell proliferation (Mangiacasale et al. 2003), thus, suggesting a potential driving role for active retrotransposon elements in oncogenesis.
The group has recently reported that retrotransposon RNAs, especially LINE-1, HERV-K, and Alu, are not only abundant in tumor cells and even more so in MVs derived from them, as compared to normal fibroblasts and their MVs (Balaj et al. 2011). Exposure of normal human umbilical vein endothelial cells to MVs from human medulloblastoma tumor cells, which have high levels of HERV-K sequences, increased the content of HERV-K sequences up to 60-fold in the endothelial cells (ibid.). These findings open a new window into the possibility of an active role of MVs in transferring retrotransposons sequences into normal surrounding cells, thereby potentially shaping their genomes, increasing the plasticity of their phenotype and making them more cancer permissive.
DNA
Microvesicles have also been found to contain DNA. The presence of this genetic material has allowed much speculation about its transfer to, and possible integration into neighboring cell genomes. Horizontal transfer of genes has been shown in lower organisms and is important, for example, in the generation of resistance to drugs, such as antibiotics (Jain et al. 2002). Microvesicles can include small apoptotic bodies which have been shown to transfer chromosomal fragments as well as oncogenes, such as H-ras and c-Myc to neighboring phagocytic cells, possibly aiding tumor progression (Holmgren et al. 1999; Bergsmedh et al. 2001). In fact, p53-negative mouse embryonic cells lost contact inhibition in vitro and became tumorigenic in vivo when exposed to apoptotic vesicles derived from rat embryonic fibroblast transfected with the oncogene c-Myc and mutant H-ras (Bergsmedh et al. 2001). It has also been reported that MVs derived from GBM cells and astrocytes contain mitochondrial DNA which can be transferred to recipient cells (Guescini et al. 2010).
The group has recently shown that MVs from brain tumor cells contain single stranded DNA (ssDNA), including both cDNA, and genomic DNA (Balaj et al. 2011). This DNA included elevated levels of sequences from the c-Myc oncogene in MVs from medulloblastoma tumor cells that were amplified for this oncogene, as well as genomic sequences for the flanking POU5F1B locus, which is located about 319 kb from the c-Myc gene and co-amplifies with it (Storlazzi et al. 2006). The cDNA in MVs presumably results from elevated RT activity found in these tumor cells and MVs derived from them. L1 and HERV RNA elements, which encode RTs, are transcriptionally upregulated in cancer cells and enriched in MVs, as is RT activity (Balaj et al. 2011).
The source of genomic DNA in MVs is less clear, but may derive from Okazaki fragments or amplified genomic DNA sequences that enter the cytoplasm during mitosis following breakdown of the nuclear envelope and are then incorporated into MVs. It has been previously reported that ssDNA accumulates in the cytoplasm of TREX1-negative cells (the major 3′ DNA exonuclease; Yang et al. 2007) and interestingly most of these accumulated sequences were retrotransposon elements which may end up in MVs. Treatment of a tumor cell line with l-mimosine, an inhibitor of DNA replication, reduced the amount of ssDNA in MVs in a dose-dependent manner thus supporting DNA replication in the origin of genomic DNA in MVs (Balaj et al. 2011). The presence of oncogene and retrotransposon sequences in the tumor-derived MVs population increases the possibility of horizontal transfer of genetic information to normal cells in the tumor microenvironment in support of cancer cell growth and invasion.
Microvesicles as Biomarkers for Cancer
Genotyping of mutations in individual tumors has become more and more important with the expanding knowledge that each tumor contains a different constellation of mutations that are linked to tumor phenotype and response to treatment (Harris and McCormick 2010). This genotypic information will form the basis for personalized, targeted cancer therapy as reliable and accessible tumor genotyping methods become available. Typically mutations are analyzed in DNA from biopsies of the tumor themselves, which is limited by inaccessibility of some tumors, and confounded by genetic heterogeneity within and among tumors in the same individual. Further, taking repeated biopsies of a tumor during treatment, especially in the brain, is not practical, and tracking treatment response is usually limited to MRI imaging of the tumor, which can be ambiguous (Nelson and Cha 2003).
The representation of the tumor cell transcriptome in MVs and their release into the circulation (Skog et al. 2008) provides a window into the genotype and indirectly the phenotype of tumors in individual cancer patients without the need for a biopsy. Microvesicles are shed into many body fluids, including cerebrospinal fluid, blood or urine, making it possible to do repeated longitudinal samplings to assess the tumor genotype over time. Thus, sampling tumor MVs in body fluids is promising as a companion diagnostic to monitor response and tumor dynamics during treatment.
Somatic mutations/splice variants of coding genes, as well as levels of tumor-related mRNAs, miRNAs, and ncRNAs can be measured in RNA from MVs in serum samples (Skog et al. 2008). This information is critical as drug responses are linked to certain mutations. As examples, activating mutations in KRAS in colorectal cancer patients correlate with a poor response to EGFR inhibitors like Cetuximab/Erbitux (Qiu et al. 2010); the drug PLX4032 is only effective against melanomas bearing the V600E BRAF mutation (Smalley 2010); and certain EGFR tyrosine kinase inhibitors (like gefitinib/Iressa) used for treatment of lung cancer work best when EGFR is activated by a mutation in the tyrosine kinase domain (Kobayashi et al. 2005). These studies have been important in understanding the response of tumors to drug therapy depending on the mutational state of the tumor, rather than just the tissue origin of the tumor.
The genotype of tumors is dynamic and changes over time during progression and in response to treatment. For example, lung cancer patients treated with EGFR tyrosine kinase inhibitors often relapse with a tumor subtype harboring a resistance mutation (e.g., EGFRT790 M; Kobayashi et al. 2005). This finding supports the longitudinal profiling of the tumor transcriptome to guide the selection of second-line treatments.
Recently, ncRNAs have also entered the field as cancer biomarkers, with the ncRNA, PCA3 proving useful as a biomarker for prostate cancer (Day et al. 2011). Although the function of PCA3 has yet to be determined, its primarily nuclear localization suggests a gene regulatory role. PCA3 can be readily measured in the RNA from urine MVs (Nilsson et al. 2009), which can be extracted using a rapid filtration concentrator method (Miranda et al. 2010). Another ncRNA transcript overexpressed in prostate cancer is PCGEM1 (Srikantan et al. 2000), which seems to be involved in regulation of cell proliferation and inhibition of apoptosis (Fu et al. 2006). The ncRNA NEAT2/MALAT1 has been found to be upregulated during metastasis of non-small cell lung cancer, and was correlated with poor patient survival (Ji et al. 2003). This same transcript has also been found associated with trophoblast cell invasion in vitro (Tseng et al. 2009).
Summary and Implications in Neuronal Function
Microvesicles contain an abundance of genetic information which can report on the genome/transcriptome of the cell of origin and modulate the genotypic/phenotypic fate of recipient cells. These membrane bound satchels of information expand the number of ways that cells can communicate with each other to include transfer of genetic information and membrane-bound, and non-secreted proteins. Proteins transferred in MVs include oncogenic, mutant EGFRs (Al-Nedawi et al. 2008), the chemokine receptor, CCR5 (thereby expanding range of HIV infection in brain; Mack et al. 2000), transcription factors (Di Vizio et al. 2009) and sequestered cytokines, such as interleukin-1beta (IL-1beta; Bianco et al. 2009). In addition to providing a means to orchestrate environmental signals, MVs may prove clinically valuable as peripheral markers of disease states and potentially as a means to deliver therapeutic agents in vivo, e.g., siRNA across the blood–brain barrier (Alvarez-Erviti et al. 2011).
Recent studies have found MVs to participate in positive neural functions in many ways. For example, mature neurons release MVs in the vicinity of synaptic connections and their release is stimulated by exposure to a GABAA receptor antagonist, consistent with a role in neurotransmitter regulation and modulation (Lachenal et al. 2011; Smalheiser 2007). Oligodendrocytes appear to release MVs as means of suppressing myelination until appropriate signals are received from neurons during development (Bakhti et al. 2011). Schwann cells can deliver mRNA and ribosomes to injured axons via MVs to stimulate protein synthesis and regeneration (Court et al. 2008). On the other side of the coin, the disposal function of MVs may serve to spread degenerative proteins throughout the nervous system. Microvesicles have been implicated in release of alpha-synuclein and beta-amyloid from neural cells, the elevation of which are associated with neuronal degeneration in Parkinson’s disease (Emmanouilidou et al. 2010) and Alzheimer’s disease (AD; Aguzzi and Rajendran 2009), respectively, as well as of prion proteins causing transmissible spongiform encephalopathy (Alais et al. 2008). For example, proteins of exosomal origin have been found to accumulate in AD patient’s brains and Aβ peptides have been shown to associate with MVs released from cells, implicating MVs in AD pathogenesis (Rajendran et al. 2006). During brain injury astrocytes release ATP which activates release of MVs from microglia containing IL-1beta, which can trigger an inflammatory response which can be damaging to neural tissues (Bianco et al. 2009). Thus, within the nervous system MVs can modulate developmental signaling, neurotransmission and regeneration, as well as spreading neurotoxic proteins and inflammatory signals.
Brain tumors appear to have co-opted a very critical mode of communication mediated by MVs in the nervous system and other tissues. The exaggerated expression of this phenotype in tumors provides a window into deeper understanding of the various types of MVs and their array of functions. Microvesicles also offers a view into dynamic changes in gene expression associated with changes in state, as in oncogenesis, disease, and normal development/communication.
Acknowledgments
This commentary is written in deep appreciation of the inspiring mentorship of Dr. Marshall Nirenberg. The authors have tried to include exciting new scientific ideas, which he always relished. The authors would like to thank Ms. Suzanne McDavitt for skilled editorial assistance and Drs. Charles Lai and David Corey for contributing figures. The Breakefield laboratory is funded by NINDS and NCI. Dr. Skog has a grant from the American Brain Tumor Association (ABTA).
References
- Aguzzi A, Rajendran L (2009) The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 64:783–790 [DOI] [PubMed] [Google Scholar]
- Aitken CE, Petrov A, Puglisi JD (2010) Single ribosome dynamics and the mechanism of translation. Annu Rev Biophys 39:491–513 [DOI] [PubMed] [Google Scholar]
- Alais S, Simoes S, Baas D, Lehmann S, Raposo G, Darlix JL, Leblanc P (2008) Mouse neuroblastoma cells release prion infectivity associated with exosomal vesicles. Biol Cell 100:603–615 [DOI] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10:619–624 [DOI] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. doi:10.1038/nbt.1807 (Epub ahead of print) [DOI] [PubMed]
- Bakhti M, Winter C, Simons M (2011) Inhibition of myelin membrane sheath formation by oligodendrocyte-derived exosome-like vesicles. J Biol Chem 286:787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho Y-J, Pomeroy SL, Breakefield XO, Skog J (2011) Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2:180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, Holmgren L (2001) Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci USA 98:6407–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, Menna E, Saglietti L, Schuchman EH, Furlan R, Clementi E, Matteoli M, Verderio C (2009) Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J 28:1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno S, Grange C, Deregibus MC, Calogero RA, Saviozzi S, Collino F, Morando L, Busca A, Falda M, Bussolati B, Tetta C, Camussi G (2009) Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol 20:1053–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G, Meldolesi J (2009) Shedding microvesicles: artefacts no more. Trends Cell Biol 19:43–51 [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10:691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court FA, Hendriks WT, MacGillavry HD, Alvarez J, van Minnen J (2008) Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J Neurosci 28:11024–11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10:704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JR, Jost M, Reynolds MA, Groskopf J, Rittenhouse H (2011) PCA3: from basic molecular science to the clinical lab. Cancer Lett 301:1–6 [DOI] [PubMed] [Google Scholar]
- Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B, Camussi G (2007) Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 110:2440–2448 [DOI] [PubMed] [Google Scholar]
- Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, True LD, Rubin MA, Adam RM, Beroukhim R, Demichelis F, Freeman MR (2009) Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res 69:5601–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4:e1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15:188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K (2010) Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci 30:6838–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G (2008) A human snoRNA with microRNA-like functions. Mol Cell 32:519–528 [DOI] [PubMed] [Google Scholar]
- Fauré J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B, Grange J, Schoehn G, Goldberg Y, Boyer V, Kirchhoff F, Raposo G, Garin J, Sadoul R (2006) Exosomes are released by cultured cortical neurones. Mol Cell Neurosci 31:642–648 [DOI] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J (1995) The RNA component of human telomerase. Science 269:1236–1241 [DOI] [PubMed] [Google Scholar]
- Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S (2006) Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol 25:135–141 [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64:7011–7021 [DOI] [PubMed] [Google Scholar]
- Germano I, Swiss V, Casaccia P (2010) Primary brain tumors, neural stem cell, and brain tumor cancer cells: where is the link? Neuropharmacology 58:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Brochier-Armanet C (2006) The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci 361:1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–857 [DOI] [PubMed] [Google Scholar]
- Guescini M, Genedani S, Stocchi V, Agnati LF (2010) Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm 117:1–4 [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, McCormick F (2010) The molecular pathology of cancer. Nat Rev Clin Oncol 7:251–265 [DOI] [PubMed] [Google Scholar]
- Holmgren L, Szeles A, Rajnavölgyi E, Folkman J, Klein G, Ernberg I, Falk KI (1999) Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood 93:3956–3963 [PubMed] [Google Scholar]
- Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, Kim JH, Choi DS, Kim YK, Hwang D, Gho YS (2009) Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 10:556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB (2008) Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 3:e3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, Rivoltini L (2008) Tumour-released exosomes and their implications in cancer immunity. Cell Death Diff 15:80–88 [DOI] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Moore JE, Lake JA (2002) Horizontal gene transfer in microbial genome evolution. Theor Popul Biol 61:489–495 [DOI] [PubMed] [Google Scholar]
- Jarrous N, Reiner R (2007) Human RNase P: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res 35:3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22:8031–8041 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488 [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C (2005) Antisense transcription in the mammalian transcriptome. Science 309:1564–1566 [DOI] [PubMed] [Google Scholar]
- Kiss T (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145–148 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B (2005) EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352:786–792 [DOI] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T (2010) Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285:17442–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer-Albers EM, Bretz N, Tenzer S, Winterstein C, Möbius W, Berger H, Nave KA, Schild H, Trotter J (2007) Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Prot Clin 1:1446–1461 [DOI] [PubMed] [Google Scholar]
- Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, Blot B, Haase G, Goldberg Y, Sadoul R (2011) Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci 46:409–418 [DOI] [PubMed] [Google Scholar]
- Lakkaraju A, Rodriguez-Boulan E (2008) Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol 18:199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie L, Kitova M, Maldener E, Meese E, Mayer J (2005) CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J Virol 79:876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Hardin JA, Steitz JA (1981) Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 211:400 [DOI] [PubMed] [Google Scholar]
- Li X, Frank DN, Pace N, Zengel JM, Lindahl L (2002) Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA 8:740–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A, Muñoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL (2011) Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol 31:300–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack M, Kleinschmidt A, Bruhl H, Klier C, Nelson PJ, Cihak J, Plachy J, Stangassinger M, Erfle V, Schlondorff D (2000) Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med 6:769–775 [DOI] [PubMed] [Google Scholar]
- Mangiacasale R, Pittoggi C, Sciamanna I, Careddu A, Mattei E, Lorenzini R, Travaglini L, Landriscina M, Barone C, Nervi C, Lavia P, Spadafora C (2003) Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene 22:2750–2751 [DOI] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA (2008) Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29:499–509 [DOI] [PubMed] [Google Scholar]
- Marzesco AM, Janich P, Wilsch-Bräuninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB (2005) Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci 118:2849–2858 [DOI] [PubMed] [Google Scholar]
- Mattick JS (2004) RNA regulation: a new genetics? Nat Rev Genet 5:316–323 [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE (2007) Which transposable elements are active in the human genome? Trends Genet 23:183–191 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Bond DT, McKee M, Skog J, Păunescu TG, Da Silva N, Brown D, Russo LM (2010) Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 78:191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwenda JM (1993) Biochemical characterization of a reverse transcriptase activity associated with retroviral-like particles isolated from human placental villous tissue. Cell Mol Biol Noisy-le-grand 39:317–328 [PubMed] [Google Scholar]
- Nelson SJ, Cha S (2003) Imaging glioblastoma multiforme. Cancer J 9:134–145 [DOI] [PubMed] [Google Scholar]
- Ng K, Pullirsch D, Leeb M, Wutz A (2007) Xist and the order of silencing. EMBO Rep 8:34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A (2009) Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Brit J Cancer 100:1603–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T (2010) Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One 5:e13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans E, Lindengey JL, de Gruijl TD, Wurdinger T, Middeldorp JM (2010) Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 107:6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potolicchio I, Carven GJ, Xu X, Stipp C, Riese RJ, Stern LJ, Santambrogio L (2005) Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol 175:2237–2243 [DOI] [PubMed] [Google Scholar]
- Poznanski AA, Calarco PG (1991) The expression of intracisternal A particle genes in the preimplantation mouse embryo. Dev Biol 143:271–281 [DOI] [PubMed] [Google Scholar]
- Qiu LX, Mao C, Zhang J, Zhu XD, Liao RY, Xue K, Li J, Chen Q (2010) Predictive and prognostic value of KRAS mutations in metastatic colorectal cancer patients treated with cetuximab: a meta-analysis of 22 studies. Eur J Cancer 46:2781–2787 [DOI] [PubMed] [Google Scholar]
- Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 10:42–46 [DOI] [PubMed] [Google Scholar]
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K (2006) Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA 103:11172–11177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ (2006) Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20:847–856 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N (2008) Endogenous retroviruses and cancer. Cell Mol Life Sci 65:3366–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Raposo G (2009) Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol 21:575–581 [DOI] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH (2010) LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci 33:345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Würdinger T, van Rijn S, Meijer D, Gainche L, Curry WTJ, Carter BS, Krichevsky AM, Breakefield XO (2008) Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat Cell Biol 10:1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR (2007) Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct 2:35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley KS (2010) PLX-4032, a small-molecule B-Raf inhibitor for the potential treatment of malignant melanoma. Cur Opin Investig Drugs 11:699–706 [PubMed] [Google Scholar]
- Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino K, Buzard GS, Mostofi FK, McLeod DG, Moul JW, Srivastava S (2000) PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA 97:12216–12221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi CT, Fioretos T, Surace C, Lonoce A, Mastrorilli A, Strömbeck B, D’Addabbo P, Iacovelli F, Minervini C, Aventin A, Dastugue N, Fonatsch C, Hagemeijer A, Jotterand M, Mühlematter D, Lafage-Pochitaloff M, Nguyen-Khac F, Schoch C, Slovak ML, Smith A, Solè F, Van Roy N, Johansson B, Rocchi M (2006) MYC-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of MYC as the target gene. Hum Mol Genet 15:933–942 [DOI] [PubMed] [Google Scholar]
- Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, Sasaki C, Costa J, Lizardi PM (2009) Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene 448:151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Mattick JS (2003) Increasing biological complexity is positively correlated with relative genome-wide expansion of non-protein coding DNA sequences. Genome Biol. http://www.genomebiology.com/2003/5/1/P1. Accessed 1 Dec 2003
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS (2010) Non-coding RNAs: regulators of disease. J Pathol 220:126–139 [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110:13–21 [DOI] [PubMed] [Google Scholar]
- Taylor AR, Robinson MB, Gifondorwa DJ, Tytell M, Milligan CE (2007) Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. Dev Neurobiol 67:1815–1829 [DOI] [PubMed] [Google Scholar]
- Théry C, Ostrowski M, Segura E (2009) Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9:581–593 [DOI] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, Rivera MN, Bardeesy N, Maheswaran S, Haber DA (2011) Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331:593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247 [DOI] [PubMed] [Google Scholar]
- Tseng JJ, Hsieh YT, Hsu SL, Chou MM (2009) Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod 15:725–731 [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659 [DOI] [PubMed] [Google Scholar]
- Valadkhan S (2010) Role of the snRNAs in spliceosomal active site. RNA Biol 7:345–353 [DOI] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE (2007) Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 1313:873–886 [DOI] [PubMed] [Google Scholar]
- Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, Farber DB (2009) Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One 4:e4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY (2010) Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 39:133–144 [DOI] [PubMed] [Google Scholar]