Abstract
Converting information contained in natural language clinical text into computer-amenable structured representations can automate many clinical applications. As a step towards that goal, we present a method which could help in converting novel clinical phrases into new expressions in SNOMED CT, a standard clinical terminology. Since expressions in SNOMED CT are written in terms of their relations with other SNOMED CT concepts, we formulate the important task of identifying relations between clinical phrases and SNOMED CT concepts. We present a machine learning approach for this task and using the dataset of existing SNOMED CT relations we show that it performs well.
Keywords: SNOMED CT, clinical phrases, relation identification, natural language processing
Introduction
Many clinical applications, including clinical decision support, medical error detection, answering clinical queries, generating patient statistics, and biosurveillance, would be automated if the clinical information locked in natural language clinical text could be converted into computer-amenable structured representations. To enable this, a long-term goal is to convert entire natural language clinical documents into structured representations. As an important step in that direction, in this paper we focus on a task that can help in converting clinical phrases into a structured representation. Systemized Nomenclature of Medicine—Clinical Terms (SNOMED CT)1 is a standardized representation for clinical concepts whose extensiveness and expressivity makes it suitable for precisely encoding clinical phrases. A concept in SNOMED CT is defined in terms of its relations with other concepts, and SNOMED CT currently includes around 400,000 pre-defined clinical concepts. If a natural language clinical phrase represents a concept which is already present in SNOMED CT then the conversion process reduces to a matching function; some previous work2,3 as well as existing SNOMED CT browsers such as CliniClue,a can automatically perform such matching. Our focus in this paper is instead on the task of creating new SNOMED CT concepts for clinical phrases for which no SNOMED CT concept already exists.
Since new concepts in SNOMED CT can be created by identifying their relations with existing SNOMED CT concepts, we formulate the important task of identifying relations between clinical phrases and SNOMED CT concepts. That is, given a clinical phrase (for example, “acute gastric ulcer with perforation”) and a description of a SNOMED CT concept (for example, “stomach structure”), whether a particular kind of relation (for example, “finding site”) is present between them or not (in this example it is present). To the best of our knowledge, there is no other work which has attempted this type of relation identification task. Note that this task is very different from the relation extraction task.4 In that task, two entities are given in a sentence and the system determines whether the two entities are related or not mostly based on what the sentence says. In contrast, there is no sentence in this task and the presence of a relation is determined entirely based on the two entities.
Since several thousand relations already exist in SNOMED CT, we used these existing relations to form our dataset. Both training and test relation example pairs were obtained from this dataset. To identify each kind of relation, we separately trained a machine learning method. We employed the Support Vector Machine (SVM)5 machine learning method in combination with a new kernel that we specifically designed for this relation identification task. The experimental results show that the trained system obtains a good accuracy.
Such a system could be used for creating precise SNOMED CT expressions for clinical phrases. For example, “acute gastric ulcer with perforation” could be represented as an “acute gastric ulcer”, whose finding site is “stomach structure” and whose associated morphologies are “perforated ulcer” and “acute ulcer” (this is also shown in Table 1 under phrase (c)). In this example, “is a”, “finding site” and “associated morphology” are the identified relations, and “acute gastric ulcer”, “stomach structure”, “perforated ulcer” and “acute ulcer” are already present concepts in SNOMED CT. This representation would be obtained by efficiently testing the phrase for all the relations and with all the existing SNOMED CT concepts.
Table 1.
Some examples of natural language clinical phrases and their corresponding SNOMED CT expressions.
| Natural language clinical phrase | SNOMED CT expression |
|---|---|
| (a) severe pain in the stomach | 116680003 |is a| = 22253000 |pain| {363698007 |finding site| = 69695003 | stomach structure |, 272141005 |severity| = 24484000 |severe| } |
| (b) neoplasm of right lower lobe of lung | 116680003 |is a| = 126713003 |neoplasm of lung| {116676008 |associated morphology| = 108369006 |neoplasm|, 363698007 |finding site| = 266005 |structure of right lower lobe of lung|} |
| (c) acute gastric ulcer with perforation | 116680003 |is a| = 95529005 |acute gastric ulcer| {363698007 |finding site| = 69695003 | stomach structure |, 116676008 |associated morphology| = 26317001 |acute ulcer|, 116676008 |associated morphology| = 91182001 |perforated ulcer|} |
| (d) family history of aplastic anemia | 116680003 |is a| 243796009 |situation with explicit context| {246090004 |associated finding| = 306058006 |aplastic anemia| {408732007 |subject relationship context| = 303071001 |person in the family|} } |
Notes: The numbers are the SNOMED CT concept and relation identifiers and their natural language descriptions are shown for human readability. The “=” character indicates relation kind on its left side and the related concept on its right side.
Background and Related Work
Realizing the importance of unlocking the clinical information present in free-text clinical reports, researchers started working on automatically converting them into structured representations years ago. Previous systems to convert natural language clinical information into structured representations, such as the Linguistic String Project6,7 MedLEE,8,9 and Menelas,10,11 were manually built by linguistically and medically trained experts over a long course of time. The builders manually encoded how different natural language patterns should convert into the target structured representations. They also developed their own suitable structured representations12,13 which restrict their systems from being useful elsewhere where a different type of structured representation is in use. Although we are limiting ourselves to clinical phrases instead of full sentences at this stage, we use machine learning techniques to minimize the manual cost of building such a system. For the structured representation, we are using the standardized clinical terminology, SNOMED CT, which is already widely in use.
SNOMED CT1 is the most comprehensive clinical terminology in the world today and is widely used in electronic health record systems for documentation purposes and reporting.14 Its extensive content and expressivity makes it suitable for precisely encoding clinical concepts. Not only does it specify approximately 400,000 pre-defined medical concepts and relations between them, but also its compositional grammar15 can be used to build new expressions that represent new medical concepts in terms of the existing concepts. SNOMED CT has been developed in the description logic formalism16 which also makes it suitable for automated reasoning. For all these reasons, we think that it is the best structured representation into which natural language clinical phrases may be converted. There are browsers and tools available that can help users search SNOMED CT as well as interactively build new expressions, such as CliniClue. Lee et al17 presented a method for manually encoding text with SNOMED CT. There also has been recent work in automatically mapping text to SNOMED CT pre-defined concepts2,3,18 or Unified Medical Language System (UMLS) pre-defined concepts.19 However, these systems at best do an approximate match from clinical phrases to pre-defined concepts, also known as pre-coordinated expressions. In contrast, the system presented in this paper can help to automatically map natural language clinical phrases which do not match any pre-defined concepts into their semantically equivalent new SNOMED CT expressions. The new SNOMED CT expressions are also known as post-coordinated expressions. We did a preliminary analysis of the i2b2 2010 clinical text corpus20 and found that out of around 8300 unique annotated concepts (noun phrases) in it, only around 1600 were pre-defined concepts in SNOMED CT. This shows that new phrases are abundantly present in clinical text and hence the ability to convert them into new SNOMED CT expressions is important.
Identifying SNOMED CT Relations
Formulation of the task
Table 1 shows some examples of clinical phrases and their associated SNOMED CT expressions. The expressions are shown using the syntax of SNOMED CT’s compositional grammar.15 The numbers are the unique SNOMED CT concept and relation identifiers. Each concept in SNOMED CT has at least one natural language description. A description for each concept and relation is shown within vertical bars for human readability. The “=” character denotes relation kind on its left side and the related concept on its right side. The “is a” relation identifies the basic concept a clinical phrase represents and this is then further qualified using more relations which are shown in “{}” brackets. Note that there could be multiple relations of the same kind in an expression, for example, in phrase (c) the “associated morphology” relation occurs twice. Similarly, even the “is a” relation can occur more than once because SNOMED CT allows multiple inheritance of concepts. There is more than one way to write an expression in SNOMED CT, ranging from close-to-user form to normal form.1 We have shown close-to-user forms in Table 1 which are simpler and easier for humans to understand. For the record, the concepts for phrases (b) and (c) are already present in the current version of SNOMED CT but the concepts for phrases (a) and (d) are not present.
As it can be observed, relations are the basis for forming SNOMED CT expressions. Hence, in this paper, we formulate the task of identifying relations between clinical phrases and SNOMED CT concepts. A new SNOMED CT expression could then be formed for a new clinical phrase by identifying its relations with existing concepts. We present a machine learning method for training a separate relation identifier for each of the relations present in SNOMED CT (for example, “is a”, “finding site”, etc.). Since every concept in SNOMED CT has a basic type (for example, “substance”, “disorder”, “body structure”, etc.), and the basic type can also be determined for every clinical phrase (either directly from the context it is used in or by using a trained classifier),b we treat each relation with different types separately. For example, the “finding site” relation that relates “disorder” to “body structure” is treated separately from the “finding site” relation that relates “finding” to “body structure”. The first column of Table 2 shows the most frequent relations in SNOMED CT along with their types which we used in our experiments.
Table 2.
Maximum F-measures over the precision-recall curves obtained by the similarity baseline and by the trained system for the most frequent SNOMED CT relations.
| Relation | Similarity baseline (%) | Trained system (%) |
|---|---|---|
| Associated morphology (disorder, morphologic abnormality) | 66.67 | 84.95 |
| Causative agent (disorder, substance) | 82.94 | 90.98 |
| Finding site (disorder, body structure) | 66.67 | 87.13 |
| Finding site (finding, body structure) | 66.67 | 90.35 |
| Has active ingredient (product, substance) | 85.92 | 91.38 |
| Is a (body structure, body structure) | 79.82 | 90.24 |
| Is a (disorder, disorder) | 78.18 | 86.00 |
| Is a (finding, finding) | 78.98 | 88.18 |
| Is a (organism, organism) | 66.67 | 82.20 |
| Is a (procedure, procedure) | 73.49 | 87.87 |
| Is a (product, product) | 67.48 | 88.52 |
| Is a (substance, substance) | 66.67 | 74.30 |
| Part of (body structure, body structure) | 66.67 | 90.56 |
| Procedure site direct (procedure, body structure) | 66.67 | 89.26 |
Since several hundred thousand concepts and the relations between them are already present in SNOMED CT, we decided to use them as our dataset for training and testing our method. Every concept in SNOMED CT has a unique identifier and is also given a unique fully specified natural language name. In addition, it may have several natural language descriptions which are essentially a few different ways of expressing the same concept. To create our dataset, for every kind of relation, we randomly took some pairs of related concepts as positive examples and some pairs of unrelated concepts as negative examples. For each of the two concepts in a relation example, we randomly selected one description (phrase) out of all the descriptions it may have (including its fully specified name). We did so because a clinical phrase may not always be a fully specified name and the method should also be trained to work with alternate descriptions. Then the task of relation identification is: given the two descriptions of two concepts of particular types, determine whether they are related by a particular relation or not. We are not aware of any other work that has considered such a relation identification task for SNOMED CT.
Machine learning approach for the task
For every kind of relation along with its types, we built a separate relation identifier. It may be noted that sometimes a presence of a relation can be identified simply by detecting overlap between the words in the two descriptions. For example, for the phrase (a) in Table 2, the word “pain” overlaps, hence “severe pain in the stomach” is a “pain”. Similarly for the phrase (b), “neoplasm of right lower lobe of lung” is a “neoplasm of lung”. However, this is not the case for many other relations. For example, the phrase (c) does not contain “stomach structure” which is its “finding site”. Hence besides mere overlap, the relation identifier system should be able to use several other clues. In the previous example, it should know that “gastric” generally means related to “stomach structure”. As it will be a formidable task to manually encode every piece of such knowledge, we use machine learning approach so that the system would automatically learn this kind of knowledge from training examples.
Another kind of knowledge a relation identifier would need is what words in the clinical phrase indicate what relations. For example, the word “in” in a “disorder” concept would usually indicate a “finding site” relation to a “body structure” concept. The machine learning system is expected to also learn this kind of knowledge from training examples. In our experiments, we used a baseline for comparison that uses only the amount of overlap for identifying relations.
We decided to use SVM5 as our learning algorithm because it has been shown to work well with thousands of features and hence has been widely used in natural language processing tasks which often involve use of several thousand features, for example, words and their combinations. An additional advantage of SVM is that one can implicitly specify potentially infinite number of features without actually enumerating them by defining a similarity function between the examples, called a kernel. This is also known as the kernel trick. For our relation identification task, we designed a specific kernel to enable the SVM learner to learn the kinds of knowledge it needs to learn which were mentioned earlier. It also incorporates word overlap which is sometimes a good indication of a relation.
The kernel is defined as follows. Let A and B be two examples. Let c1A and c2A be the descriptions of the first and the second concepts of the example A respectively. Thus, if an example has “acute gastric ulcer with perforation” as the description of the first concept and “stomach structure” as the description of the second concept then these two respective phrases will be c1A and c2A. Similarly, let c1B and c2B be the descriptions of the first and the second concepts of the example B respectively. Then the kernel K(,) between examples A and B is defined as:
| (1) |
where sim(,) is the similarity function. In our experiments, we defined similarity as the number of common words. We also tried defining it as the number of common word subsequences,21,22 but it did not result in any gain in the performance. The cw(,) function computes the set of common words between the two phrases. Note that the above is a well-defined kernel because products and summations of kernels are also well-defined kernels.23 The kernel is normalized by dividing it by the square-root of K(A,A) *K(B,B).
We now explain this kernel and what its implicit features are. The first term of the addition is a product of the number of common words between the first concepts of the two examples and the second concepts of the two examples. This essentially counts the number of common word-pairs present in the two examples such that in each example the first word is present in the first concept and the second word is present in the second concept. For example, if both examples have “gastric” present in the first concept and “stomach” present in the second concept, then it will count “gastric, stomach” as a feature present in both the examples. Thus this kernel term implicitly captures pairs of words, one in each concept, as features. Based on these features, the learner may learn what combinations of word pairs indicate a relation.
The second term simply treats the number of words overlapping between the two concepts of an example as a feature. The product then indicates how similar the two examples are along this feature. As was indicated earlier, overlap is an important feature because often the descriptions of the related concepts have overlap of words. While this term considers the number of overlapping words between the two concepts as a feature, it ignores the actual words that overlap. Overlap of certain words could be a good indicator of a relation present as opposed to overlap of some other words. For example, if the word “pain” is common between the two concepts then it is a good indication of “is a” relation, ie, a particular pain is a type of pain. In order to allow the learning process to learn such knowledge from the training data, the third term implicitly captures words common between an example’s two concepts as features. The cw(,) function computes the set of common words for each of the two examples and then the sim(,) function counts how many of these common words are common across the two examples.
In the results we show the contribution of each of the three kernel terms through an ablation study. In general, the terms could be weighed differently, however, presently we did not experiment with different weights and we simply let SVM learn appropriate weights as part of its learning process.
Experiments
In this section, we describe our experiments on the relation identification task for SNOMED CT relations.
Methodology
As was noted earlier, we formed our dataset utilizing the existing relations present in SNOMED CT. There are hundreds of different kinds relations present in SNOMED CT, some of them are more important than others (examples of some of the unimportant relations are “duplicate concept” and “inactive concept”). We report our results on the 14 important and most frequent relations, each of which had more than 10,000 instances. The “is a (procedure, procedure)” relation had the highest number of 93,925 instances. Since we had enough examples to choose our training and test examples from, instead of doing standard cross-validation, we ran five folds and in each fold we randomly selected 5000 training and 5000 test examples. Training beyond 5000 examples would lead to memory problems, but as our learning curves showed, the learning would generally converge by 5000 training examples.
For each relation, positive examples for both training and testing were randomly selected without replacement as pairs of concepts for which the relation is known to exist. Then equal number of negative examples were randomly selected without replacement as pairs of concepts of the required types which are not related by that relation. There was no overlap between training and testing datasets. We employed SVM using the LibSVM packagec along with the user-defined kernel as defined in the previous section.
We measured precision and recall. Precision is the percentage of correctly identified relations out of all the identified relations. Recall is the percentage of correctly identified relations out of all the relations present in the test set. The evaluation was done for the combined output of all the folds. SVM can also give the confidences for its classification decisions through Platt’s method.24 We used these confidences to measure precision and recall at every confidence level and plotted precision-recall curves. We also measured the maximum F-measure across the precision-recall curves, where F-measure is the harmonic mean of precision and recall.
We compared our approach with a baseline method which only uses the amount of word overlap between the two concepts to identify a relation between them. It is not a learning-based approach. It outputs its confidence on a relation as the degree of overlap between the two concepts (ie, the number of common words after normalization for word lengths). We call this baseline the similarity baseline. Note that the similarity scores are already included as features in the kernel used in the learning approach. Since there were equal number of positive and negative examples, the accuracy of a random classifier would be 50%.
Results and discussion
Table 2 shows the maximum F-measures obtained across the precision-recall curves for the similarity baseline and for the trained system for the 14 most frequent relations in SNOMED CT. It may be first noted that the baseline does well on a few of the relations, obtaining close to 80% or more on five relations. This shows that the similarity baseline is not a trivial baseline although on some other relations it does not do well at all. Note that 66.67% F-measure can be also obtained by a random classifier by calling every relation as positive which would result in 50% precision and 100% recall. The learned approach does substantially better than the baseline on every relation. On nine of the 14 relations it exceeds the baseline’s performance by more than 10% (absolute). On the five remaining relations it exceeds by more than 5%. The performance is better than 90% on five relations and better than 80% on 13 relations. The only relation on which the performance is not high is the “is a (substance, substance)” relation. We found that this is mostly because a lot of new names are used for substances and from the names themselves it is not easy to identify that a particular substance is a type of another substance, for example, “lacto-n tetrasylceramide, type 2 chain” is a “blood group antigen precursor”.
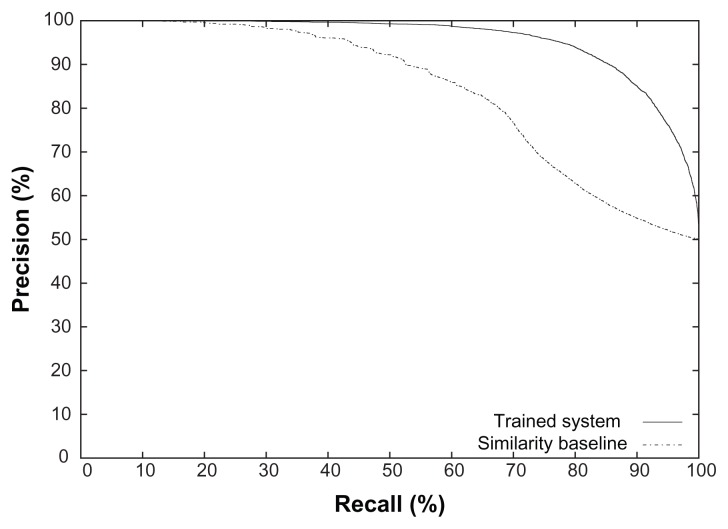
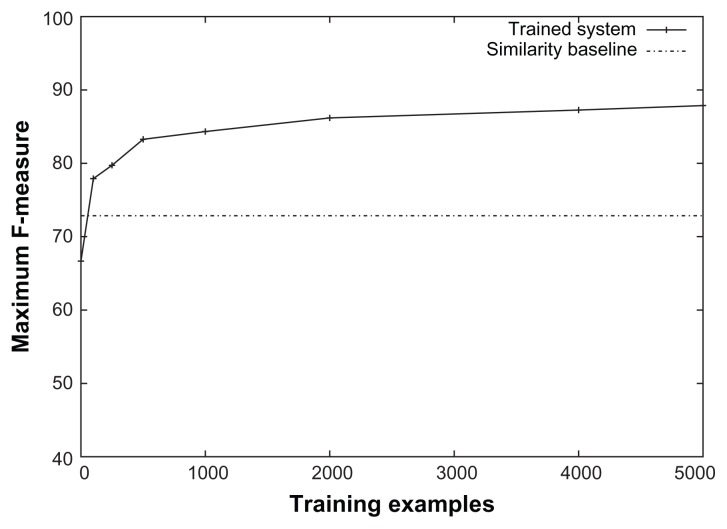
Figure 1 shows the entire precision-recall curves obtained using the trained system and the similarity baseline for the “is a (procedure, procedure)” relation. We are not showing these graphs for other relations due to space limitations, but this graph shows the typical curves obtained by the two methods. It may be noted looking at the lower part of the recall side that there are examples on which the relation can be identified with high precision even by the similarity baseline. But the learned approach continues to obtain high precision even on the high recall side when the precision of the baseline drops off. Figure 2 shows the learning curves for the same relation for the maximum F-measures on the precision-recall curves. Since the baseline method is not a learning method, its learning curve is horizontal. It can be seen that the learning method has almost converged and more training examples are unlikely to improve the performance substantially. It may also be noted that even with a few hundred training examples, the trained system already does much better than the baseline.
Figure 1.
Precision-recall curves for the “is a (procedure, procedure)” relation obtained using the similarity baseline and using the trained system.
Figure 2.
Learning curve for the “is a (procedure, procedure)” relation obtained using the trained system.
Note: The similarity baseline is shown for comparison.
In Table 3 we show the contributions of different types of implicit features captured through the different terms of the kernel in Equation 1. The numbers are the averages of the maximum F-measures across the 14 relations. Without word-pairs features, without similarity score feature and without common word features correspond to omitting first, second and third terms respectively from Equation 1. It can be seen that all three types of features contribute towards improving the performance. However, the word-pairs are the most important features without which the performance drops to only a little better than the baseline.
Table 3.
Ablation results showing contributions of the different types of implicit features corresponding to the different terms of the kernel in Equation 1.
| System | Average maximum F-measure (%) |
|---|---|
| Baseline | 72.39 |
| Without word-pairs features | 73.89 |
| Without similarity score feature | 84.27 |
| Without common words features | 85.54 |
| All features | 87.28 |
Notes: The numbers are the averages of the maximum F-measures across the 14 relations. Without word-pairs features, without similarity score feature and without common word features correspond to omitting first, second and third terms respectively from Equation 1.
Future Work
There are several avenues for future work. Currently, our method does not do any syntactic analysis of the phrases. Clearly the syntactic structure of the phrase indicates the presence of relations with other concepts. Hence it will be potentially useful information to exploit. One may do this by using syntactic tree kernels25,26 to compute similarities between descriptions. Another way to improve the performance could be by incorporating the hierarchical structure of concepts in SNOMED CT as additional features. This may help the learner generalize across concepts at similar places in the hierarchy. In future, we also want to evaluate the performance of our method on clinical phrases which are not in SNOMED CT. This will, however, require manual evaluation by experts which may be doable only on a small scale.
In future, we plan to apply the SNOMED CT relation identification method to convert clinical phrases into their SNOMED CT expressions. We have already done some preliminary experiments towards this end. In order to identify relations for a new phrase, the system needs to check every relation with every other concept. Given that there are around 400,000 concepts in SNOMED CT, doing this is computationally very intensive (testing an example in SVM requires computing kernels with all the training examples which have non-zero support vectors). However, we tested the idea on a subset of SNOMED CT with around 3000 concepts whose all relations are preserved within the subset. We obtained maximum F-measures for the relation identification task in this setting in the range of 10%–20%. But given that this test dataset contains a few thousand negative examples for every positive example (random guessing will perform less than 1%), this is in fact not a bad performance, although it needs to be improved. One way to improve will be to design a top level classifier that will filter out several obvious negative examples. Some of the SNOMED CT expressions require nested use of relations, for example, the expression for the phrase (d) in Table 1. In order to compositionally build a nested SNOMED CT expression, in future one may leverage ideas from semantic parsing,27 the task of converting natural language utterances into complete meaning representations.
Conclusions
We formulated the task of identifying SNOMED CT relations as a means for converting natural language clinical phrases into SNOMED CT expressions. We presented a machine learning approach for identifying relations and also introduced an appropriate kernel for the task. Experimental results showed that the trained system obtains a good performance on the relation identification task.
Footnotes
Alternatively, the type of a clinical phrase could also be identified by first determining the “is a” relation.
Author Contributions
Conceived and designed the experiments: RJK. Analysed the data: RJK. Wrote the first draft of the manuscript: RJK. Contributed to the writing of the manuscript: RJK. Agree with manuscript results and conclusions: RJK. Developed the structure and arguments for the paper: RJK. Made critical revisions and approved final version: RJK. Author reviewed and approved of the final manuscript.
Competing Interests
Author discloses no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the author has provided signed confirmation of compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests. This is an extended version of the paper accepted at the Computational Semantics in Clinical Text Workshop at the International Conference on Computational Semantics 2013. The submission is intended for the special issue of Biomedical Informatics Insights for the Computational Semantics in Clinical Text.
Funding
Author discloses no funding sources.
References
- 1.SNOMED Clinical Terms User Guide—July 2009 International Release. Technical Report. Copenhagen, Denmark: The International Health Terminology Standards Development Organization; 2009. [Google Scholar]
- 2.Stenzhorn H, Pacheco EJ, Nohama P, Schulz S. Automatic mapping of clinical documentation to SNOMED CT. Stud Health Technol Inform. 2009;150:228–32. [PubMed] [Google Scholar]
- 3.Barrett N, Weber-Jahnke J, Thai V. Automated clinical coding using semantic atoms and topology. Proceedings of the 25th International Symposium on Computer-Based Medical Systems (CBMS); Jun 20–22, 2012; Rome, Italy. 2012. pp. 1–6. [Google Scholar]
- 4.Zelenko D, Aone C, Richardella A. Kernel methods for relation extraction. Journal of Machine Learning Research. 2003;3:1083–106. [Google Scholar]
- 5.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and Other Kernel-based Learning Methods. Cambridge, UK: Cambridge University Press; 2000. [Google Scholar]
- 6.Sager N, Friedman C, Lyman MS. Medical Language Processing: Computer Management of Narrative Data. Addison-Wesley Publishing Company; 1987. [Google Scholar]
- 7.Sager N, Lyman M, Nhàn NT, Tick LJ. Medical language processing: applications to patient data representations and automatic encoding. Methods Inf Med. 1995;34(1–2):140–6. [PubMed] [Google Scholar]
- 8.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spyns P, Zweigenbaum P, Willems JL. Representation and extraction of information from patient discharge summaries by means of natural language processing. Proceedings of the MIC 92. 1992:309–16. Dutch. [Google Scholar]
- 11.Spyns P, Willems JL. Dutch medical language processing: discussion of a prototype. Proceedings of the Eight World Congress on Medical Informatics. 1995:37–40. [PubMed] [Google Scholar]
- 12.Friedman C, Cimino JJ, Johnson SB. A conceptual model for clinical radiology reports. Proc Annu Symp Comput Appl Med Care. 1993:829–33. [PMC free article] [PubMed] [Google Scholar]
- 13.Zweigenbaum P, Bachimont B, Bouaud J, Charlet J, Boisvieux JF. A multi-lingual architecture for building a normalised conceptual representation from medical language. Proc Annu Symp Comput Appl Med Care. 1995:357–61. [PMC free article] [PubMed] [Google Scholar]
- 14.Benson T. Principles of Health Interoperability HL7 and SNOMED (Health Informatics) London, UK: Springer-Verlag; 2010. [Google Scholar]
- 15.Compositional Grammar for SNOMED CT Expressions in HL 7 Version 3. External Draft for Trial Use Version 0.006, Dec 2008. Technical Report. Cophenhagen, Denmark: The International Health Terminology Standards Development Organization; 2008. [Google Scholar]
- 16.Baader F, Calvanese D, McGuinness D, Nardi D, Patel-Schneider P, editors. The Description Logic Handbook: Theory, Implementation, and Applications. Cambridge, UK: Cambridge University Press; 2003. [Google Scholar]
- 17.Lee DH, Lau FY, Quan H. A method for encoding clinical datasets with SNOMED CT. BMC Med Inform Decis Mak. 2010;10:53. doi: 10.1186/1472-6947-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung S, Kim S, Yoo S, Choi J. Toward the automatic generation of the entry level CDA documents. Journal of Korean Society of Medical Informatics. 2009;15(1):141–51. [Google Scholar]
- 19.Aronson AR, Lang FM. An overview of MetaMap: historical perspectives and recent advances. J Am Med Inform Assoc. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Inform Assoc. 2011;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodhi H, Saunders C, Shawe-Taylor J, Cristianini N, Watkins C. Text classification using string kernels. Journal of Machine Learning Research. 2002;2:419–44. [Google Scholar]
- 22.Bunescu RC, Mooney RJ. Subsequence kernels for relation extraction. In: Weiss Y, Scholkopf B, Platt P, editors. Advances in Neural Information Processing Systems; Proceedings of the 2005 Conference (NIPS); Cambridge, MA: MIT Press; 2006. p. 18. [Google Scholar]
- 23.Haussler D. Technical Report UCSC-CRL-99-10. Santa Cruz, CA: University of California, Santa Cruz; 1999. Convolution Kernels on Discrete Structures. [Google Scholar]
- 24.Platt JC. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In: Smola AJ, Bartlett P, Schölkopf B, Schuurmans D, editors. Advances in Large Margin Classifiers. Cambridge, MA: MIT Press; 1999. pp. 185–208. [Google Scholar]
- 25.Collins M, Duffy N. Advances in Neural Information Processing Systems (NIPS’2001) Cambridge MA: MIT Press; 2001. Convolution kernels for natural language; pp. 625–32. [Google Scholar]
- 26.Moschitti A. Making tree kernels practical for natural language learning. Proceedings of the 11th Conference of the European Chapter of the Association for Computational Linguistics (EACL-06); Apr 3–7, 2006; Trento, Italy. 2006. pp. 113–20. [Google Scholar]
- 27.Mooney RJ. In: Gelbukh A, editor. Learning for semantic parsing; Computational Linguistics and Intelligent Text Processing: Proceedings of the 8th International Conference (CICLing 2007); Mexico City. Feb 18–24, 2007; Berlin: Springer Verlag; 2007. pp. 311–24. [Google Scholar]