Abstract
Because of privacy concerns and the expense involved in creating an annotated corpus, the existing small-annotated corpora might not have sufficient examples for learning to statistically extract all the named-entities precisely. In this work, we evaluate what value may lie in automatically generated features based on distributional semantics when using machine-learning named entity recognition (NER). The features we generated and experimented with include n-nearest words, support vector machine (SVM)-regions, and term clustering, all of which are considered distributional semantic features. The addition of the n-nearest words feature resulted in a greater increase in F-score than by using a manually constructed lexicon to a baseline system. Although the need for relatively small-annotated corpora for retraining is not obviated, lexicons empirically derived from unannotated text can not only supplement manually created lexicons, but also replace them. This phenomenon is observed in extracting concepts from both biomedical literature and clinical notes.
Keywords: natural language processing, distributional semantics, concept extraction, named entity recognition, empirical lexical resources
Background
One of the most time-consuming tasks faced by a Natural Language Processing (NLP) researcher or practitioner trying to adapt a machine–learning based NER system to a different domain is the creation, compilation, and customization of the needed lexicons. Lexical resources, such as lexicons of concept classes, are considered necessary to improve the performance of NER. It is typical for medical informatics researchers to implement modularized systems that cannot be generalized.1 As the work of constructing or customizing lexical resources needed for these highly specific systems is human-intensive, automatic generation is a desirable alternative. It might be possible that empirically created lexical resources would incorporate domain knowledge into a machine-learning NER engine and increase its accuracy.
Although many machine–learning based NER techniques require annotated data, semi-supervised and unsupervised techniques for NER have long been explored due to their value in domain robustness and minimizing labor costs. Some attempts at automatic knowledgebase construction included automatic thesaurus discovery efforts,2 which sought to build lists of similar words without human intervention to aid in query expansion or automatic lexicon construction.3 More recently, the use of empirically derived semantics for NER is used by Finkel and Manning,4 Turian et al,5 and Jonnalagadda et al6 Finkel’s NER tool uses clusters of terms built apriori from the British National corpus7 and English gigaword corpus8 for extracting concepts from newswire text and PubMed abstracts for extracting gene mentions from biomedical literature. Turian et al5 also showed that statistically created word clusters9,10 could be used to improve named entity recognition. However, only a single feature (cluster membership) can be derived from the clusters. Semantic vector representations of terms had not been used for NER or sequential tagging classification tasks before.5 Although Jonnalagadda et al6 use empirically derived vector representation for extracting concepts defined in the GENIA11 ontology from biomedical literature using rule-based methods, it was not clear whether such methods could be ported to extract other concepts or incrementally improve the performance of an existing system. This work not only demonstrates how such vector representation could improve state-of-the-art NER, but also that they are more useful than statistical clustering in this context.
Methods
We designed NER systems to identify treatment, tests, and medical problem entities in clinical notes and proteins in biomedical literature. Our systems are trained using (1) sentence-level features using training corpus; (2) a small lexicon created, compiled, and curated by humans for each domain; and (3) distributional semantics features derived from a large unannotated corpus of domain-relevant text. Different models are generated through different combinations of these features. After training for each concept class, a Conditional Random Fields (CRF) machine-learning model12 is created to process input sentences using the same set of NLP features. The output is the set of sentences with the concepts tagged. We evaluated the performance of the different models in order to assess the degree to which human-curated lexicons can be substituted by the automatically created list of concepts.
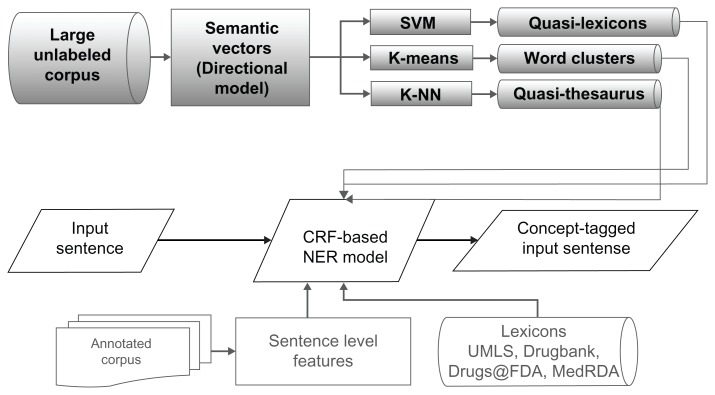
The architecture of the system is shown in Figure 1 and the different components and settings are detailed in Table 1. We first used a state-of-the-art NER algorithm, CRF, as implemented by MALLET,13 that extracts concepts from both clinical notes and biomedical literature using several sentence-level orthographic and linguistic features derived from respective training corpora. Then, we studied the impact on the performance of the baseline after incorporating manual lexical resources and empirically generated lexical resources. The CRF algorithm classifies words according to IOB or IO-like notations (I = inside, O = outside, B = beginning) to determine whether they are part of a description of an entity of interest, such as a treatment or protein. We used four labels for clinical NER: “Iproblem,” “Itest,” and “Itreatment,” for tokens that were inside a problem, test, or treatment respectively, and “O” if they were outside any clinical concept. For protein tagging, we used the IOB notation, ie, the three labels “Iprotein,” “Bprotein,” and “O.”
Figure 1.
Overall Architecture of the System.
Notes: The design of the system to identify concepts using machine learning and distributional semantics. The top three components are related to distributional semantics.
Table 1.
Description of different components and settings of the system.
| Name | Description |
|---|---|
| Clinical NER | |
| Conditional Random Fields (CRF)12 | CRF is a sequential deterministic machine learning algorithm that is considered state of the art for concept extraction in general English, biomedical literature and clinical narratives. We use the MALLET13 toolkit’s implementation of our CRF paper. |
| Sentence-level orthographic and linguistic features | These machine learning features used by all the settings are generated through NLP tasks such as tokenization, part-of-speech tagging, chunking and parsing. We used Apache OpenNLP14 library for implementing these sentence-level tasks. |
| MED_noDict | MED_noDict is the CRF-based clinical NER system with all the sentence-level orthographic and syntactic features generated from OpenNLP. |
| Lexicons for clinical concept extraction | Compiled from UMLS Metathesaurus15—built from the electronic versions of various thesauri, classifications, code sets, and lists of controlled terms; MedDRA16—medical terminology for medical products used by humans; DrugBank17—combines detailed drug (ie, chemical, pharmacological and pharmaceutical) data with comprehensive drug target; Drugs@FDA18—FDA-approved brand name and generic prescription and over-the-counter human drugs. |
| MED_Dict | The clinical NER system with several sentence-level orthographic and syntactic features, along with features from the above four lexicons. |
| Semantic vectors26 | Semantic Vectors creates semantic vector spaces of individual tokens and documents from free natural language text. This package is extended in this paper to empirically construct three different types of lexical resources for this project: Quasi-lexicons using SVM, Word clusters using K-means, Quasi-thesaurus using K-nearest neighbor. |
| MED_Dict+SVM | The quasi-lexicons from Semantic Vectors are used in addition to the features in MED_Dict. |
| MED_Dict+NN | The quasi-thesaurus from Semantic Vectors are used in addition to the features in MED_Dict. |
| MED_Dict+CL | The word clusters from Semantic Vectors are used in addition to the features in MED_Dict. |
| MED_Dict+NN+SVM | The quasi-lexicons and quasi-thesaurus from Semantic Vectors are used in addition to the features in MED_Dict. |
| MED_noDict+NN+SVM | The quasi-lexicons and quasi-thesaurus from Semantic Vectors are used in addition to the features in MED_noDict. |
| Protein NER | |
| BANNER21 | One of the best CRF-based protein-tagging systems.22 |
| BioCreative II gene normalization training set23 | The source for the 344,000 single-word lexicon used by BANNER by default (called BANNER_Dict in this paper). |
| BANNER_Dict+DistSem | The system that uses both manual and empirical lexical resources. |
| BANNER_noDict | The system that uses neither manual nor empirical lexical resources. |
| BANNER_noDict+DistSem | The system that uses only empirical lexical resources. |
Several sentence-level orthographic and linguistic features such as lower-case tokens, lemmas, prefixes, suffixes, n-grams, patterns such as “beginning with a capital letter” and parts of speech were adapted from the OpenNLP14 package to build the NER model and tag the entities in input sentences. This configuration is referred to as MED_noDict for clinical NER and BANNER_noDict for protein tagging.
The UMLS Metathesaurus,15 MedDRA,16 DrugBank,17 and Drugs@FDA18 are used to create dictionaries for medical problems, treatments, and tests. The guidelines of the i2b2/VA NLP entity extraction task19 are followed to identify the corresponding UMLS semantic types for each of the three concepts. The other three resources are used to add more terms to our manual lexicon. In an exhaustive evaluation on the nature of the resources by Gurulingappa et al,20 UMLS and MedDRA were found to be the best resources for extracting information about medical problems among several other resources. For protein tagging, BANNER,21 one of the best protein-tagging systems,22 uses the 344,000 single-word lexicon constructed using the BioCreative II gene normalization training set.23 This configuration is referred to as MED_Dict for clinical NER and as BANNER_Dict for protein tagging.
Distributional Semantic Feature Generation
Here, we implemented automatically generated distributional semantic features based on a semantic vector space model trained from unannotated corpora. This model, referred to as the directional model, uses a sliding window that is moved through the text corpus to generate a reduced-dimensional approximation of a token-token matrix, such that two terms that occur in the context of similar sets of surrounding terms will have similar vector representations after training. As the name suggests, the directional model takes into account the direction in which a word occurs with respect to another by generating a reduced-dimensional approximation of a matrix with two columns for each word, with one column representing the number of occurrences to the left and the other column representing the number of occurrences to the right. The directional model is therefore a form of sliding-window based Random Indexing,24 and is related to the Hyperspace Analog to Language.25 Sliding-window Random Indexing models achieve dimension reduction by assigning a reduced-dimensional index vector to each term in a corpus. Index vectors are high dimensional (eg, dimensionality on the order of 1,000), and are generated by randomly distributing a small number (eg, on the order of 10) of +1’s and −1’s across this dimensionality. As the rest of the elements of the index vectors are 0, there is a high probability of index vectors being orthogonal, or close-to-orthogonal to one another. These index vectors are combined to generate context vectors representing the terms within a sliding window that is moved through the corpus. The semantic vector for a token is obtained by adding the contextual vectors gained at each occurrence of the token, which are derived from the index vectors for the other terms it occurs with within the sliding window. The model was built using the open source Semantic Vectors package.26 Random indexing is more suitable than Latent Semantic Analysis (LSA) or topic models (LDA, etc.) when applied to a huge unannotated corpus, such as tens of thousands of clinical narratives or clinical abstracts.28
The performance of distributional models depends on the availability of an appropriate corpus of domain-relevant text. For clinical NER, 447,000 Medline abstracts that are indexed as pertaining to clinical trials are used as the unlabeled corpus. In addition, we have also used clinical notes from the Mayo Clinic and the University of Texas Health Science Center to understand the impact of the source of unlabeled corpus. For protein NER, 8,955,530 Medline citations in the 2008 baseline release that include an abstract27 are used as the large unlabeled corpus. Previous experiments28 revealed that using a directional model with 2000-dimensional vectors, five seeds (number of +1’s and –1’s in the vector), and a window radius of six is better suited for the task of NER. While a stop-word list is not employed, we have rejected tokens that appear only once in the unlabeled corpus or have more than three nonalphabetical characters.
SVM: quasi-lexicons of concept classes using SVM
A support vector machine (SVM)29 is designed to draw hyper-planes separating two class regions such that they have a maximum margin of separation. Creating the quasi-lexicons (automatically generated word lists) is equivalent to obtaining samples of regions in the distributional hyperspace that contain tokens from the desired (problem, treatment, test, and none) semantic types. In clinical NER, each token in a training set can belong to either one or more of the classes: problem, treatment, test, or none of these. Each token is labeled as “Iproblem,” “Itest,” “Itreatment,” or “Inone.” To remove ambiguity, tokens that belong to more than one category are discarded. For example, based on the information that “thoracic cancer” is a problem, “CT of the thoracic cavity” is a test and “thoracic surgery” is a treatment, “thoracic” is discarded, “cancer” is labeled as problem, “CT” is labeled as test, and “surgery” is labeled as treatment. Each token has a representation in the distributional hyperspace of 2,000 dimensions. Six (C[4, 2] = 4!/[2!*2!]) binary SVM classifiers are generated for predicting the class of any token among the four possible categories. During the execution of the training and testing phase of the CRF machine-learning algorithm, the class predicted by the SVM classifiers for each token is used as a feature for that token.
CL: clusters of distributionally similar words over K-means
The K-means clustering algorithm30 is used to group the tokens in the training corpus into 200 clusters using distributional semantic vectors. As an illustration, cluster number 33 contains the tokens: Sept, August, January, December, October, March, April, November, June, July, Nov, February, and September. Cluster number 46 contains the tokens: staphylococcus, faecium, enterococci, staphylococci, hemophilus, streptococcus, pneumoniae, klebsiella, bacteroides, coli, enterobacter, mycoplasma, aureus, anitratus, influenzae, calcoaceticus, serratia, aeruginosa, diphtheroids, proteus, methicillin, enterococcus, cloacae, oxacillin, mucoid, escherichia, mirabilis, fragilis, citrobacter, staph, acinetobacter, faecalis, pseudomonas, legionella, coagulase, and viridans. The cluster identifier assigned to the target token is used as a feature for the CRF-based system for NER. This feature is similar to the Clark’s automatically created clusters,10 used by Finkel and Manning,31 where the same number of clusters are used. We focused on using features generated from semantic vectors as they allow us to also create the other two types of features.
NN: quasi-thesaurus of distributionally similar words using nearest neighbors
Cosine similarity of vectors is used to find the 20 nearest tokens for each token. These nearest tokens are used as features for the respective target token. Figure 2 shows the top few tokens closest in the word space to “haloperidol” to demonstrate how well the semantic vectors are computed. Each of these nearest tokens is used as an additional feature whenever the target token is encountered. Barring evidence from other features, the word “haloperidol” would be classified as belonging to the “medical treatment,” “drug,” or “psychiatric drug” semantic class based on other words belonging to that class sharing nearest neighbors with it.
Figure 2.
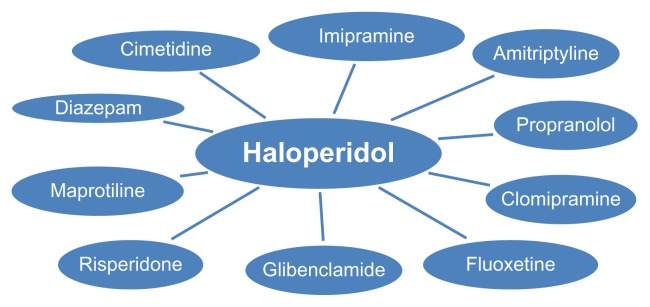
Nearest Tokens to Haloperidol.
Notes: The closest tokens to haloperidol in the word space are psychiatric drugs. Using the nearest tokens to haloperidol as features, when haloperidol is not a manually compiled lexicon or when the context is unclear, would help to still infer (statistically) that haloperidol is a drug (medical treatment).
Evaluation strategy
The previous sub-sections detail how the manually created lexicons are compiled and how the empirical lexical resources are generated from semantic vectors (2000 dimensions). In the machine-learning system for extracting concepts from literature and clinical notes, each manually created lexicon (three for the clinical notes task) contributes one binary feature whose value depends on whether a term surrounding the word is present in the lexicon. Each quasi-lexicon will also contribute one binary feature whose value depends on the output of the SVM classifier discussed before. Together, the distributional semantic clusters contribute a feature whole value that is the id of the cluster which the word belongs to. The quasi-thesaurus contributes 20 features which are the 20 words distributionally similar to the word for which features are being generated.
As a gold standard for clinical NER, the fourth i2b2/VA NLP shared-task corpus19 for extracting concepts of the classes—problems, treatments, and tests—was used. The corpus contains 349 clinical notes as training data and 477 clinical notes as testing data. For protein tagging, the BioCreative II Gene Mention Task32 corpus is used. The corpus contains 15,000 training set sentences and 5,000 testing set sentences.
Results
Comparison of different types of lexical resources on extracting clinical concepts
Table 2 shows that the F-score of the clinical NER system for exact match increases by 0.3% after adding quasi-lexicons, whereas it increases by 1.4% after adding the quasi-thesaurus. The F-score increases slightly more with the use of both these features. The F-score for an inexact match follows a similar pattern. Table 2 also shows that the F-score for an exact match increases by 0.5% after adding clustering-based features, whereas it increases by 1.6% after adding quasi-thesaurus and quasi-lexicons. The F-score decreases slightly with the use of both the features. The F-score for an inexact match follows a similar pattern.
Table 2.
Clinical NER: comparison of SVM-based features and clustering-based features with N-nearest neighbors– based features.
| Setting | Exact F | Inexact F | Exact increase | Inexact increase |
|---|---|---|---|---|
| MED_Dict | 80.3 | 89.7 | ||
| MED_Dict+SVM | 80.6 | 90 | 0.3 | 0.3 |
| MED_Dict+NN | 81.7 | 90.9 | 1.4 | 1.2 |
| MED_Dict+NN+SVM | 81.9 | 91 | 1.6 | 1.3 |
| MED_Dict+CL | 80.8 | 90.1 | 0.5 | 0.4 |
| MED_Dict+NN+SVM+CL | 81.7 | 90.9 | 1.4 | 1.2 |
Notes: MED_Dict is the baseline, which is a machine-learning clinical NER system with several sentence-level orthographic and syntactic features, along with features from lexicons such as UMLS, Drugs@FDA, and MedDRA. In MED_Dict+SVM, the quasi-lexicons are also used. In MED_Dict+NN, the quasi-thesaurus is used. In MED_Dict+CL, the clusters automatically generated are used in addition to other features in MED_Dict. Exact F is the F-score for exact match as calculated by the shared task software. Inexact F is the F-score for inexact match or matching only a part of the other. Exact Increase is the increase in Exact F from previous row. Inexact Increase is the increase in Inexact F from previous row.
Overall impact on extracting clinical concepts
Table 3 shows how the F-score increased over the baseline (MED_noDict, which uses various sentence-level orthographic and syntactic features). After manually constructed lexicon features are added (MED_Dict), it increased by 0.9%. On the other hand, if only distributional semantic features (quasi-thesaurus and quasi-lexicons) were added without using manually constructed lexicon features (MED_noDict+NN+SVM), it increased by 2.0% (P < 0.001 using Bootstrap Resampling33 with 1,000 repetitions). It increases only by 0.5% more if the manually constructed lexicon features were used along with distributional semantic features (MED_Dict+NN+SVM). The F-score for an inexact match follows a similar pattern.
Table 3.
Clinical NER: impact of distributional semantic features.
| Setting | Exact F | Inexact F | Exact increase | Inexact increase |
|---|---|---|---|---|
| MED_noDict | 79.4 | 89.2 | ||
| MED_Dict | 80.3 | 89.7 | 0.9 | 0.5 |
| MED_noDict+NN+SVM | 81.4 | 90.8 | 2.0 | 1.6 |
| MED_Dict+NN+SVM | 81.9 | 91.0 | 2.5 | 1.8 |
Notes: MED_noDict is the machine-learning clinical NER system with all the sentence-level orthographic and syntactic features, but no features from lexicons such as UMLS, Drugs@FDA, and MedDRA. MED_noDict+NN+SVM also has the features generated using SVM and the nearest neighbors algorithm.
Moreover, the improvement was consistent even across different concept classes, namely medical problems, tests, and treatments. Each time the distributional semantic features are added, the number of TPs increases, and the number of FPs and FNs decreases.
Impact of the source of the unlabeled data
We utilized three sources for creating the distributional semantics models for NER from i2b2/VA clinical notes corpus. The first source is the set of Medline abstracts indexed as pertaining to clinical trials (447,000 in the 2010 baseline). The second source is the set of 0.8 million clinical notes (half of the total available) from the clinical data warehouse at the School of Biomedical Informatics, University of Texas Health Sciences Center, Houston, Texas ( http://www.uthouston.edu/uth-big/clinical-data-warehouse.htm). The third source is the set of 0.8 million randomly chosen clinical notes written by clinicians at Mayo Clinic in Rochester. Table 4 shows the performance of the systems that use each of these sources for creating the distributional semantics features. Each of these systems has a significantly higher F-score than the system that does not use any distributional semantic feature (P < 0.001 using Bootstrap Resampling33 with 1,000 repetitions and a difference in F-score of 2.0%). The F-scores of these systems are almost the same (differing by <0 .5%).
Table 4.
Clinical NER: impact of the source of unlabeled corpus.
| Unlabeled corpus | Exact F | Inexact F |
|---|---|---|
| None | 80.3 | 89.7 |
| Medline | 81.9 | 91.0 |
| UT Houston | 82.3 | 91.3 |
| Mayo | 82.0 | 91.3 |
Notes: None = The machine-learning clinical NER system that does not use any distributional semantic features. Medline = The machine-learning clinical NER system that uses distributional semantic features derived from the Medline abstracts indexed as pertaining to clinical trials. UT Houston = The machine-learning clinical NER system that uses distributional semantic features derived from the notes in the clinical data warehouse at University of Texas Health Sciences Center. Mayo = The machine-learning clinical NER system that uses distributional semantic features derived from the clinical notes of Mayo Clinic, Rochester, MN.
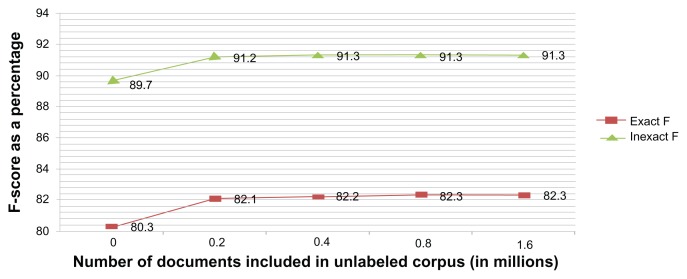
Impact of the size of the unlabeled data
Using a set of 1.6 million clinical notes from the clinical data warehouse at the University of Texas Health Sciences Center (after adding 0.8 million clinical notes to those in the previous experiment) as the baseline, we studied the relationship between the size of the unlabeled corpus used and the accuracy achieved. We randomly created subsets of size one-half, one-fourth, and one-eighth the original corpus and measured the respective F-scores. Figure 3 depicts the F-score for exact match and inexact match, suggesting a monotonic relationship with the number of documents used for creating the distributional semantic measures. While there is a leap from not using any unlabeled corpus to using 0.2 million clinical notes, the F-score is relatively constant from there. We might infer that by incrementally adding more documents to the unlabeled corpus, one would be able to determine what size of corpus is sufficient.
Figure 3.
Impact of the size of the unlabeled corpus.
Notes: On the X-axis, N represents the system created using distributional semantic features from N-unlabeled documents. N = 0 refers to the system that does not use any distributional semantic feature.
Impact on extracting protein mentions
In Table 5, the performance of BANNER with distributional semantic features (row 3) and without distributional semantic features (row 9) is compared with the top ranking systems in the most recent gene-mention task of the BioCreative shared tasks. Each system has an F-score that has a statistically significant comparison (P < 0.05) with the teams indicated in the Significance column. The significance is estimated using Table 1 in the BioCreative II gene mention task.32 The performance of BANNER with distributional semantic features and no manually constructed lexicon features is better than BANNER with manually constructed lexicon features and no distributional semantic features. This demonstrates again that distributional semantic features (that are generated automatically) are more useful than manually constructed lexicon features (that are usually compiled and cleaned manually) as a means to enhance supervised machine learning for NER.
Table 5.
Protein tagging: impact of distributional semantic features on BANNER.
| Rank | Setting | Precision | Recall | F-score | Significance |
|---|---|---|---|---|---|
| 1 | Rank 1 system | 88.48 | 85.97 | 87.21 | 6–11 |
| 2 | Rank 2 system | 89.30 | 84.49 | 86.83 | 8–11 |
| 3 | BANNER_Dict+DistSem | 88.25 | 85.12 | 86.66 | 8–11 |
| 4 | Rank 3 system | 84.93 | 88.28 | 86.57 | 8–11 |
| 5 | BANNER_noDict+DistSem | 87.95 | 85.06 | 86.48 | 10–11 |
| 6 | Rank 4 system | 87.27 | 85.41 | 86.33 | 10–11 |
| 7 | Rank 5 system | 85.77 | 86.80 | 86.28 | 10–11 |
| 8 | Rank 6 system | 82.71 | 89.32 | 85.89 | 10–11 |
| 9 | BANNER_Dict | 86.41 | 84.55 | 85.47 | – |
| 10 | Rank 7 system | 86.97 | 82.55 | 84.70 | – |
| 11 | BANNER_noDict | 85.63 | 83.10 | 84.35 | – |
Notes: The significance column indicates which systems are significantly less accurate than the system in the corresponding row. These values are based on the Bootstrap re-sampling calculations performed as part of the evaluation in the BioCreative II shared task (the latest gene or protein tagging task). BANNER_Dict+DistSem is the system that uses both manual and empirical lexical resources. BANNER_noDict+DistSem is the system that uses only empirical lexical resources. BANNER_Dict is the system that uses only manual lexical resources. This is the system available prior to this research, and the baseline for this study. BANNER_noDict is the system that uses neither manual nor empirical lexical resources. BANNER_Dict+DistSem is the system that is significantly more accurate than the baseline. It is equally important to the improvement that the accuracy of BANNER_noDict+DistSem is better than BANNER_noDict. The most significant contribution in terms of research is that an equivalent accuracy (BANNER_noDict+DistSem and BANNER_Dict) could be achieved even without using any manually compiled lexical resources apart from the annotated corpora.
Discussion
The evaluations for clinical NER reveal that the distributional semantic features are better than manually constructed lexicon features. Some examples of the differences in the output are shown in Table 6. The accuracy further increases when both manually created dictionaries and distributional semantic feature types are used, but the increase is not very significant (P = 0.15 using Bootstrap Resampling33 with 1,000 repetitions). This shows that distributional semantic features could supplement manually built lexicons, but the development of the lexicon, if it does not exist, might not be as critical as previously believed. We speculate that the improvement is because the empirically constructed lexical resources provide additional semantic information about the concept (bradycardia in example 2, cannula in example 3) and enhance the confidence of the machine learning system about an existing lexicon entry (mensa in example 1). Further, the n-nearest neighbor (quasi-thesaurus) features are better than SVM-based (quasi-lexicons) and clustering-based (quasi-clusters) features for improving the accuracy of clinical NER (P < 0.001 using Bootstrap Resampling33 with 1,000 repetitions). For the protein extraction task, the improvement after adding the distributional semantic features to BANNER is also significant (P < 0.001 using Bootstrap Resampling33 with 1,000 repetitions). The absolute ranking of BANNER with respect to other systems in the BioCreative II task improves from 8 to 3. The F-score of the best system is not significantly better than that of BANNER with distributional semantic features. We again notice that distributional semantic features are more useful than manually constructed lexicon features alone. The purpose of using protein mention extraction in addition to NER from clinical notes is to verify that the methods are generalizable. Hence, we only used the nearest neighbor or quasi-thesaurus features (as the other features contributed little) for protein mention extraction and have not studied the impact of the source or size of the unlabeled data separately. The advantages of our features are that they are independent of the machine-learning system used and can be used to further improve the performance of forthcoming algorithms.
Table 6.
Example outputs of the additional true positives found in the clinical NER system that uses distributional semantic features over the one that does not.
| Annotation | Sentence | Quasi-thesaurus |
|---|---|---|
| Concept = mesna; type = treatment | She also received Cisplatin 35 per meter squared on 06/19 and Ifex and Mesna on 06/18 |
Mesna Etoposide DTIC Cisplatinum Cisplatin CDDP 5-fu Hydroxyurea Gemcitabine Ceftriaxone Mitoxantrone VP-16 Ifo Irinotecan Ifosfamide Carboplatin Idarubicin Epirubicin Dexamethasone Prednisolone |
| Concept = mild bradycardia; type = problem | May start beta-blocker at a low dose given mild bradycardia at atenolol 50 mg p.o. q day |
Bradycardia Hypotension Dysphagia Hemorrhages Edema Bleeding Dyspnea Agitation Hypoxemia Fever Diarrhea Hyponatremia Nephrotoxicity Atelectasis Sedation Cough Pruritus Neurologic Proteinuria Ar |
| Concept = 2 liters nasal cannula oxygen; type = treatment | She needs home oxygen and is currently at 2 liters nasal cannula oxygen |
Cannula Syringe Prosthesis Plate Flap Electrode Stimulus Reservoir Filter Bar Catheter Sensor Probe Tube Preparation Endoscope Device Port Apparatus Dressing |
Notes: These examples are from the annotated corpus that belongs to Partners Healthcare. We were allowed to share them publicly after removing the protected health information.
The improvement in F-scores after adding manually compiled dictionaries (without distributional semantic features) is only around 1%. However, many NER tools, both in the genomic domain21,34 and in the clinical domain35,36 use dictionaries. This is partly because systems trained using supervised machine-learning algorithms are often sensitive to the distribution of data, and a model trained on one corpus may perform poorly on those trained from another. For example, Wagholikar37 recently showed that a machine-learning model for NER trained on the i2b2/VA corpus achieved a significantly lower F-score when tested on the Mayo Clinic corpus. Other researchers recently reported this phenomenon for part of speech tagging in clinical domain.38 A similar observation was made for the protein-named entity extraction using the GENIA, GENETAG, and AIMED corpora,39,40 as well as for protein-protein interaction extraction using the GENIA and AIMED corpora.41,42 The domain knowledge gathered through these semantic features might make the system less sensitive. This work showed that empirically gained semantics are at least as useful for NER as the manually compiled dictionaries. It would be interesting to see if such a drastic decline in performance across different corpora could be countered using distributional semantic features.
Currently, very little difference is observed between using distributional semantic features derived from Medline and unlabeled clinical notes for the task of clinical NER. Future research would study the impact of using clinical notes related to a specific specialty of medicine. We hypothesize that the distributional semantic features from clinical notes of a subspecialty might be more useful than the corresponding literature. Our current results lack qualitative evaluation. As we repeat the experiments in a subspecialty such as cardiology, we would be able to involve the domain experts in the qualitative analysis of the distributional semantic features and their role in the NER.
Conclusion
Our evaluations using clinical notes and biomedical literature validate that distributional semantic features are useful to obtain domain information automatically, irrespective of the domain, and can reduce the need to create, compile, and clean dictionaries, thereby facilitating the efficient adaptation of NER systems to new application domains. We showed this through analyzing results for NER of four different classes (genes, medical problems, tests, and treatments) of concepts in two domains (biomedical literature and clinical notes). Though the combination of manually constructed lexicon features and distributional semantic features provides a slightly better performance, suggesting that a manually constructed lexicon should be used if available, the de-novo creation of a lexicon for purpose of NER is not needed.
The distributional semantics model for Medline and the quasi-thesaurus prepared from the i2b2/VA corpus and the clinical NER system’s code is available at (http://diego.asu.edu/downloads/AZCCE/) and the updates to the BANNER system are incorporated at http://banner.sourceforge.net/.
Acknowledgements
We thank the developers of BANNER (http://banner.sourceforge.net/), MALLET (http://mallet.cs.umass.edu/) and Semantic Vectors (http://code.google.com/p/semanticvectors/) for the software packages and the organizers of the i2b2/VA 2010 NLP challenge for sharing the corpus.
Footnotes
Author Contributions
Conceived and designed the experiments: SJ. Analyzed the data: SJ, TC, GG. Wrote the first draft of the manuscript: SJ. Contributed to the writing of the manuscript: SJ, TC, SW, HL, GG. Agree with manuscript results and conclusions: SJ, TC, SW, HL, GG. Jointly developed the structure and arguments for the paper: SJ, GG. Made critical revisions and approved final version: SJ, TC, SW, HL, GG. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests. The submission is intended for the special issue of Biomedical Informatics Insights—on Computational Semantics in Clinical Text. It is a revised and extended version of the long paper accepted at the Computational Semantics in Clinical Text workshop (editors: Wu and Shah).
Funding
This work was possible because of funding from possible sources: NLM HHSN276201000031C (PI: Gonzalez), NCRR 3UL1RR024148, NCRR 1RC1RR028254, NSF 0964613 and the Brown Foundation (PI: Bernstam), NSF ABI:0845523, NLM R01LM009959A1 (PI: Liu) and NLM 1K99LM011389 (PI: Jonnalagadda).
References
- 1.Stanfill MH, Williams M, Fenton SH, Jenders RA, Hersh WR. A systematic literature review of automated clinical coding and classification systems. J Am Med Inform Assoc. 2010;17(6):646–51. doi: 10.1136/jamia.2009.001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grefenstette G. Explorations in Automatic Thesaurus Discovery. Norwell, MA, USA: Kluwer Academic Publishers; 1994. [Google Scholar]
- 3.Riloff E. An empirical study of automated dictionary construction for information extraction in three domains. Artif Intell. 1996;85(1–2):101–34. [Google Scholar]
- 4.Finkel JR, Manning CD. Joint parsing and named entity recognition. Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics on ZZZ; Association for Computational Linguistics; 2009. pp. 326–34. [Google Scholar]
- 5.Turian J, Opérationnelle R, Ratinov L, Bengio Y. Word representations: A simple and general method for semi-supervised learning. ACL. 2010;51:61801. [Google Scholar]
- 6.Jonnalagadda S, Leaman R, Cohen T, Gonzalez G. A distributional semantics approach to simultaneous recognition of multiple classes of named entities. Computational Linguistics and Intelligent Text Processing (CICLing) 2010;6008/2010 Lecture Notes in Computer Science. [Google Scholar]
- 7.Aston G, Burnard L. The BNC handbook: Exploring the British National Corpus with SARA. Edinburgh Univ Pr; 1998. [Google Scholar]
- 8.Graff D, Kong J, Chen K, Maeda K. Linguistic Data Consortium. Philadelphia: 2003. English gigaword. [Google Scholar]
- 9.Brown PF, Desouza PV, Mercer RL, Pietra VJ, Lai JC. Class-based n-gram models of natural language. Computational linguistics. 1992;18(4):467–79. [Google Scholar]
- 10.Clark A. Inducing syntactic categories by context distribution clustering. Proceedings of the 2nd Workshop on Learning language in Logic and the 4th conference on Computational Natural Language learning-Volume 7; NJ, USA: Association for Computational Linguistics Morristown; 2000. pp. 91–4. [Google Scholar]
- 11.Kim JD, Ohta T, Tsujii J. Corpus annotation for mining biomedical events from literature. BMC Bioinformatics. 2008;9:10. doi: 10.1186/1471-2105-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutton C, McCallum A. Introduction to Statistical Relational Learning. Cambridge, Massachusetts, USA: MIT Press; 2007. An introduction to conditional random fields for relational learning. [Google Scholar]
- 13.McCallum A. MALLET: A Machine Learning for Language Toolkit. 2002. [Accessed May 9, 2010]. Available at: http://mallet.cs.umass.edu.
- 14.Apache. OpenNLP. The Apache OpenNLP library. Available at: http://opennlp.apache.org/
- 15.Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Libr Assoc. 1993;81(2):170–7. [PMC free article] [PubMed] [Google Scholar]
- 16.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA) Drug Saf. 1999;20(2):109–17. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 17.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Food US. Drug Administration. Drugs@FDA. FDA Approved Drug Products. 2009. Available at: http://www.accessdata.fda.gov/scripts/cder/drugsatfda.
- 19.Uzuner O, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011. 2011 Sep-Oct;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurulingappa H, Klinger R, Hofmann-Apitius M, Fluck J. An empirical evaluation of resources for the identification of diseases and adverse effects in biomedical literature. 2nd Workshop on Building and Evaluating Resources for Biomedical Text Mining; 2010. p. 15. [Google Scholar]
- 21.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pacific Symposium in Bioinformatics. 2008 [PubMed] [Google Scholar]
- 22.Kabiljo R, Clegg AB, Shepherd AJ. A realistic assessment of methods for extracting gene/protein interactions from free text. BMC Bioinformatics. 2009;10:233. doi: 10.1186/1471-2105-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan A, Lu Z, Wang X, et al. Overview of BioCreative II gene normalization. Genome Biology. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanerva P, Kristoferson J, Holst A. Random indexing of text samples for latent semantic analysis. Proceedings of the 22nd annual conference of the cognitive science society; 2000. p. 1036. [Google Scholar]
- 25.Lund K, Burgess C. Hyperspace analog to language (HAL): A general model of semantic representation. Language and Cognitive Processes. 1996 [Google Scholar]
- 26.Widdows D, Cohen T. The semantic vectors package: new algorithms and public tools for distributional semantics. Fourth IEEE International Conference on Semantic Computing. 2010;1:43. [Google Scholar]
- 27.NLM. MEDLINE®/PubMed®Baseline Statistics. 2010. Available at: http://www.nlm.nih.gov/bsd/licensee/baselinestats.html.
- 28.Jonnalagadda S, Cohen T, Wu S, Gonzalez G. Enhancing clinical concept extraction with distributional semantics. J Biomed Inform. 2012;45(1):129–40. doi: 10.1016/j.jbi.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995;20:273–97. [Google Scholar]
- 30.MacQueen J. Some methods for classification and analysis of multivariate observations. Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability; 1967. [Google Scholar]
- 31.Finkel JR, Manning CD. Nested named entity recognition. Proceedings of the 2009 Conference on Empirical Methods in Natural Language Processing; 2009. [Google Scholar]
- 32.Wilbur J, Smith L, Tanabe T. BioCreative 2 Gene Mention Task. Proceedings of the Second BioCreative Challenge Workshop. 2007:7–16. [Google Scholar]
- 33.Noreen EW. Computer-intensive Methods for Testing Hypotheses: An Introduction. New York: John Wiley & Sons, Inc; 1989. [Google Scholar]
- 34.Torii M, Hu Z, Wu CH, Liu H. BioTagger-GM: A gene/protein name recognition system. J Am Med Inform Assoc. 2009;16(2):247–55. doi: 10.1197/jamia.M2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman C. Towards a comprehensive medical language processing system: methods and issues. AMIA. 1997 [PMC free article] [PubMed] [Google Scholar]
- 36.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagholikar KB, Torii M, Jonnalagadda SR, Liu H. Pooling annotated corpora for clinical concept extraction. J Biomed Semantics. 2013;4(1):3. doi: 10.1186/2041-1480-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan J, Prasad R, Yabut RM, et al. Part-of-speech tagging for clinical text: wall or bridge between institutions? AMIA Annu Symp Proc. 2011;2011:382–91. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Kim J-D, Sætre R, Pyysalo S, Tsujii J. Investigating heterogeneous protein annotations toward cross-corpora utilization. BMC Bioinformatics. 2009;10(1):403. doi: 10.1186/1471-2105-10-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohta T, Kim J-D, Pyysalo S, Wang Y, Tsujii J. Incorporating GENETAG-style annotation to GENIA corpus. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing; BioNLP ’09; Stroudsburg, PA, USA: Association for Computational Linguistics; 2009. pp. 106–7. Available at: http://portal.acm.org/citation.cfm?id=1572364.1572379. [Google Scholar]
- 41.Jonnalagadda S, Gonzalez G. BioSimplify: an open source sentence simplification engine to improve recall in automatic biomedical information extraction. AMIA Annual Symposium Proceedings. 2010 [PMC free article] [PubMed] [Google Scholar]
- 42.Jonnalagadda S, Gonzalez G. Sentence simplification aids protein-protein interaction extraction. Languages in Biology and Medicine. 2009 [Google Scholar]