Abstract
A large amount of medication information resides in the unstructured text found in electronic medical records, which requires advanced techniques to be properly mined. In clinical notes, medication information follows certain semantic patterns (eg, medication, dosage, frequency, and mode). Some medication descriptions contain additional word(s) between medication attributes. Therefore, it is essential to understand the semantic patterns as well as the patterns of the context interspersed among them (ie, context patterns) to effectively extract comprehensive medication information. In this paper we examined both semantic and context patterns, and compared those found in Mayo Clinic and i2b2 challenge data. We found that some variations exist between the institutions but the dominant patterns are common.
Keywords: medication extraction, electronic medical record, natural language processing
Introduction
Electronic medical records (EMRs) have grown rapidly and as a result a large amount of clinical data is stored in free-text format. Natural language processing (NLP) techniques, which can convert unstructured text to a structured format, have been successfully applied in various clinical applications, including patient medical status extraction,1,2 sentiment analysis,3 decision support,4,5 genome-wide association studies,6,7 and diagnosis code assignment.8,9
Over the past decade, multiple clinical NLP systems have been developed and applied in the clinical domain, including cTAKES,10 YTEX,11 MTERMS,12 HiTEXT,13 and MedLEE.14 Although clinical NLP systems have proven to be successful in many information extraction tasks, their performance often varies across institutions and sources of data.15,16 As some researchers have demonstrated, this may be due to the fact that clinical language is not homogeneous but instead consists of heterogeneous sublanguage characteristics.17–21
Medication information is one of the major data components in EMRs. Patient medication history is of great concern for future medical treatments and plays a vital role in enabling the secondary use of EMRs for clinical and translational research. Although some medication information can be extracted from structured data, a substantial amount resides in an EMR’s unstructured text and requires advanced techniques—such as NLP—to extract. Since medication information is described in various ways in clinical notes, understanding the semantic patterns—ie, description patterns of medication name and attributes, which we call a semantic pattern in that we view attributes as subclasses of medication information19,22—and context patterns—ie, context variations between the semantic patterns—is essential to effectively extract comprehensive medication information and develop a general and customizable medication extraction tool.
In this paper, we investigated both the semantic and context patterns of medication descriptions in clinical notes from two different sources: Mayo Clinic and the 2009 i2b2 medication extraction challenge.23 We provide various pattern statistics and compare them at both the section level and the institution level to better understand the usage of semantic and context variations.
Background
Early research on medication extraction focused on extracting the medication name itself24,25 and mapping it to a standardized nomenclature.26 The 2009 i2b2 medication extraction challenge focused on extracting comprehensive medication information (ie, medication name, as well as attributes such as dosage, mode, frequency, duration, and reason) from clinical discharge summaries.23 In this challenge, most top-performing teams used rule-based systems developed based on the examination of medication description patterns.23 Because many of these systems performed well on most attributes, we may assume that medication information in clinical notes follows some patterns.
Some studies show that sublanguage-based clinical text processing systems such as MedLEE14 perform as accurately as medical experts on clinical concept extractions.13 A sublanguage theory hypothesizes that the information content and structure from specific domains can be defined using a sublanguage grammar specifying both domain-specific syntactic and semantic information.19,27 Aligned with sublanguage grammars, Xu et al28 performed a preliminary study on parsing medication sentences by assigning the probability to semantic-based grammar rules for medication detection. Their study showed promising results to disambiguate the complex sublanguage grammars of medication sentences.
Patterson and Hurdle29 experimentally demonstrated that different clinical domains use their own sublanguages in clinical narratives. They conducted clinical document clustering in 17 different clinical domains and showed that note types in a broad clinical scope form the same cluster, whereas note types in a narrow clinical extent form different clusters. This study suggests that the performance of a clinical NLP system may depend on the source of the clinical notes and that the semantic and context information of target notes should be seriously considered in the tool development process.
Previous studies support the idea that medication descriptions also exhibit sublanguage characteristics, and therefore an understanding of those characteristics in clinical notes within and across institutions is fundamental to properly extract medication information.
Materials and Methods
This study used annotated medication corpora from two sources: Mayo Clinic and the 2009 i2b2 medication extraction challenge data. The Mayo medication data consisted of 159 clinical notes randomly selected from clinical notes. There were 659 manually annotated medication mentions with attributes. For the i2b2 medication data, 253 discharge summaries from Partners Healthcare were manually annotated by challenge teams and released to the community for research.30 In the combined corpora, there were 9,003 medication mentions along with attributes. The semantic types of medication mentions and their abbreviations for both data sets are described in Table 1 (i2b2 annotations are excerpted from the Uzuner study).30
Table 1.
Semantic type annotation of medication information in mayo and i2b2.
| i2b2 | Mayo | ||
|---|---|---|---|
|
|
|
||
| Semantic type | Definition | Semantic type | Definition |
| medication (m) | medication name | medication (m) | medication name |
| dosage (do) | the amount of a single medication used in each administration (e.g., “one tab” “4 units” “30 mg”) | dosage (do) | how many of each medication the patient is taking (e.g., “2” in “2 daily”, “1” in “1 tablet”) |
| strength number (sn) | e.g., “30” in “30 mg” | ||
| strength unit (su) | e.g., “mg” in “30 mg” | ||
| frequency (f) | how often each dose of the medication should be taken (e.g., “daily” “once a month” “3 times a day”) | freq number (fn) | e.g., “twice” in “twice a day” |
| freq unit (fu) | e.g., “day” in “twice a day” | ||
| mode (mo) | the route for administering the medication (e.g., “oral” “intravenous” “topical”) | route (mo) | same as the i2b2 |
| duration (du) | how long the medication is to be administered (e.g., “for a month” “during spring break”) | duration (du) | same as the i2b2 but not include preposition (e.g., “a month” in “for a month) |
| form† (fm) | the physical appearance of the medication (e.g., aerosol, capsule, cream, tablet) | ||
Notes: Parenthesized strings in the semantic type column are abbreviations.
In i2b2, there is no form annotation but it may be part of the medication or dosage (eg, m: “Aspirin tablet”, do: “1 tab”).
We investigated both semantic and context patterns of medication descriptions in various aspects. The Mayo clinical notes consist of multiple sections explicitly separated by section tags. The i2b2 medication annotations include format information where the medication information is described—ie, list versus narrative. We included pattern variations within an individual section, as well as a list versus narrative format.
Semantic pattern parsing
The medication description in clinical notes consists of a medication name and its attributes, such as dosage, frequency, mode, etc. We refer to a sequence of such semantics tags (ignoring other text between them) as the medication semantic pattern. For example,

We investigated these patterns in each data set and also compared them across the data sets (Mayo versus i2b2). Since the two corpora have different annotation guidelines, we mapped semantically equivalent Mayo annotations to i2b2 annotations in the following way (Mayo → i2b2): mo → mo; sn, su, do → do; fn, fu →f; fm → N/A. In order to simplify the expression of semantic patterns we assigned a short symbol to dominant patterns. Table 2 contains those pairs and actual text examples.
Table 2.
Examples of medication semantic patterns and symbols.
| Symbol | Semantic pattern | Example |
|---|---|---|
| A | m | <m>Aspirin</m> |
| B | m|do|mo|f | <m>Zocor</m> <do>5 mg</do> <mo>p.o.</mo> <f>q.h.s.</f> |
| C | m|do|f | <m>Glipizide</m> <do>5 mg</do> <f>b.i.d</f>. |
| D | m|mo | <m>Lasix</m> <mo>drip</mo> |
| E | m|do | <m>Coumadin</m> <do>alternating doses of 4 mg</do> |
| F | mo|m | <mo>IV</mo> <m>ACE inhibitors</m> |
| G | do|m | <do>one unit</do> of <m> packed red cells</m> |
| H | m|du | <m>Dilantin</m> <du>for less than a year</du> |
| I | m|f | <m>Pepcid AC</m> <f>QHS</f> |
| J | m|do|mo|f|du | <m>KEFLEX (CEPHALEXIN)</m> <do>250 MG</do> <mo>PO</mo> |
| <f>QID</f> <du>X 12 doses</du> | ||
| K | m|do|mo | <m>acetylsalicylic acid</m> <do>325 mg</do> <mo>p.o.</m> |
| L | m|do|f|du | <m>Lasix</m> <do>40 mg</do> <f>QD</f> <du>x3 doses</du> |
| M | du|m | <du>two days</du> of <m>Indocin</m> |
| N | f|m | stable on <f>b.i.d.</f> <m>torsemide</m> |
| O | m|mo|f | <m>nitroglycerin</m> <mo>sublingual</m> <f>p.r.n.</f> |
| P | m|mo|do|f | <m>Lovenox</m> <mo>subcutaneously</mo> <do>90 mg</do> <f>daily</f> |
| Q | do|mo|m | <do>10 mg</do> of <mo>IV</mo> <m>Lopressor</m> |
| R | mo|m|du | <mo>IV</mo> <m>cefotaxime</m> <du>for the 7-day period</du> |
| S | do|m|f | <do>low dose</do> <m>dilaudid</m> <m>as needed</m> |
| T | m|do|du | <m>heparin</m> <do>500 units</do> <du>for 48 hours</du> |
Note: In example, semantic types are annotated as XML-style tags.
Context pattern discovery
We use the phrase medication context pattern to refer to the semantic pattern plus the text that occurs between the semantic tags. Any text that occurs before the first semantic tag or after the last semantic tag is excluded. For example, the phrase “The patient said that Aspirin was reduced to 1 tablet po twice daily then stopped it” was patterned as follows: <m> was reduced to <do><mo><f> (i2b2) and <m> was reduced to <do><fm><mo><fn><fu> (Mayo).
Large corpus analysis (Mayo)
Mayo 159 clinical notes have a lower number of medication descriptions (659 in Mayo versus 9,003 in i2b2) and it might therefore cause sampling bias. To alleviate this issue we also examined 10,000 Mayo Clinic notes randomly selected from the Mayo patient cohort in the eMERGE study.31 They were processed by the drug NER annotator in cTAKES10 and their semantic patterns were parsed for comparison with i2b2. Drug NER uses a rule-based pattern match implemented by finite state machine to parse medications and their attributes. These semantic patterns were not manually reviewed.
Results
Medication semantic patterns
Mayo data
In the Mayo corpus, there are a total of 89 unique semantic patterns, but 85% of the medication mentions occur in the 20 most frequent patterns. Seventy percent of the medication mentions occurr in the 5 most frequent patterns: m (322 mentions); m|sn|su|fu (85); m|fu (19); m|sn|su|fn (17); and m|sn|su|fn|fu (16). Medication alone was the most dominant pattern, comprising almost half of all the medication mentions.
The Mayo clinical notes consist of multiple sections, and each section contains specific content. To further examine the content-based medication description, we analyzed each section separately and obtained section-specific semantic patterns. These results appear in Table 3.
Table 3.
Section-level statistics for mayo medication semantic patterns.
| Section | # medications | # patterns | Patterns† (≥5) |
|---|---|---|---|
| Impression/report/plan | 227 | 53 | m (97), m|sn|su|fu (42), m|sn|su (8), m|fu (5), |
| Current medications | 183 | 38 | m (52), m|sn|su|fu (28), m|fu (11), m|sn|su|mo|fu (9), m|sn|su|do|fu (9), m|sn|su|do|fn (8), m|sn|su|fn (6), m|sn|su (6), m|sn|su|fn|fu (5) |
| History of present illness | 139 | 26 | m (82), m|sn|su|fu (14), m|sn|su|fn (8), mo|m (5) |
| Allergies | 39 | 1 | m (39) |
| Past medical/surgical history | 21 | 4 | m (15) |
| Problem oriented hosp. course | 19 | 8 | m (10) |
| Social history | 10 | 3 | m (8) |
| Chief complaint/reason for visit | 7 | 3 | m (5) |
| The other sections | 14 | 1 | m (14) |
Note:
Number within ( ) denotes # medications of a given pattern.
There are 14 sections that contain medications. Almost 90% of the medications appear in the four most dominant sections: Impression/Report/Plan; Current Medications; History of Present Illness; and Allergies. If we consider only patterns that occur five or more times throughout all sections, the Current Medication section is the most diverse, with nine patterns. With the exception of the three most dominant sections, the other sections contain only one semantic pattern—m (medication)—that occurs five or more times. This pattern comprises all 39 mentions in the Allergies section.
i2b2 data
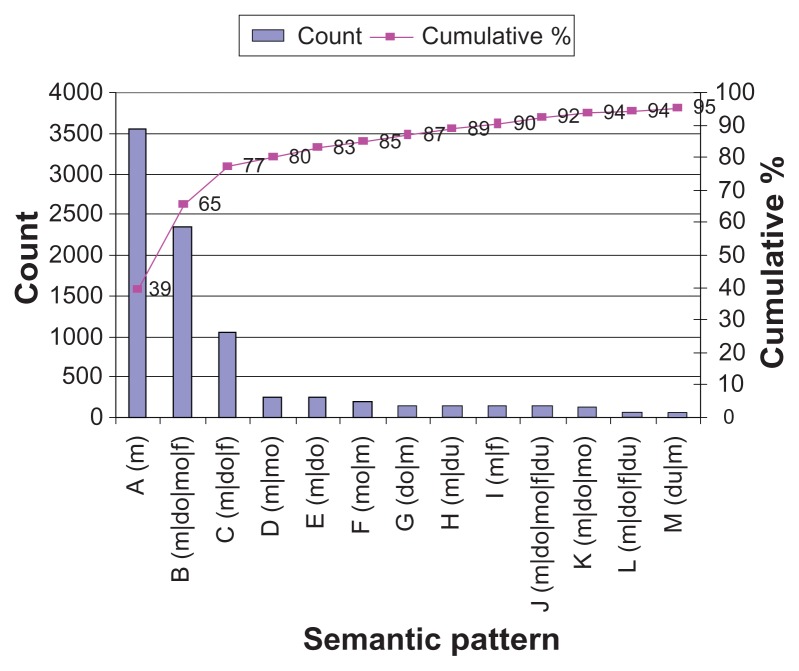
A similar dominance of a few patterns exists in the i2b2 corpus. Figure 1 shows the medication semantic patterns and their counts that cover 95% of the medication mentions in the 253 discharge summaries. A cumulative percentage denotes what portion of the counts from the total medication mentions appears in the given series of patterns up until that point. The semantic pattern in the figure is described as a short symbol with the actual semantic pattern in parentheses. There are a total of 69 unique semantic patterns, but 95% of the medication mentions occur in the 13 most frequent patterns.
Figure 1.
i2b2 medication semantic patterns (top 95%).
Mayo vs. i2b2
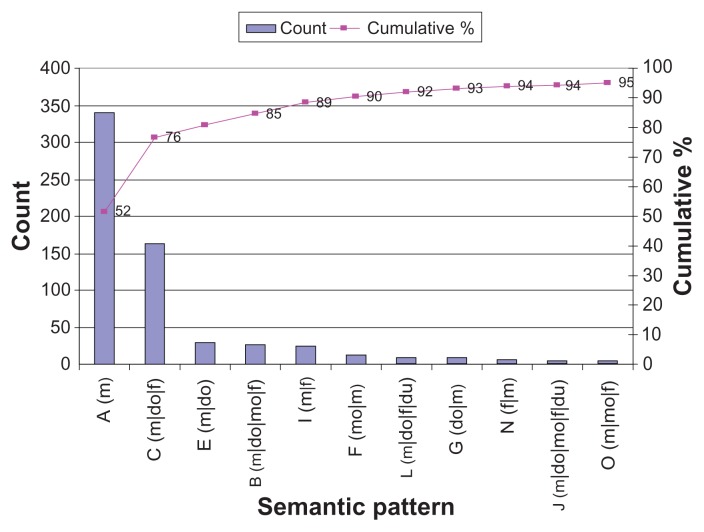
In order to properly compare Mayo semantic patterns with i2b2 semantic patterns, the Mayo medication semantic tags were mapped to i2b2 tags. The distribution of these semantic patterns is presented in Figure 2. After mapping, 85% of the medication mentions occur in four dominant patterns (m, m|do|f, m|do, m|do|mo|f), and 95% of them occur in 11 patterns.
Figure 2.
Mayo medication semantic patterns mapped to i2b2 (top 95%).
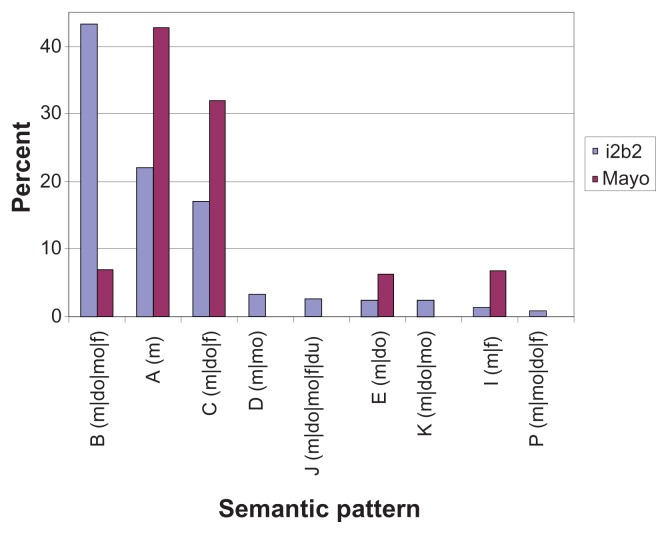
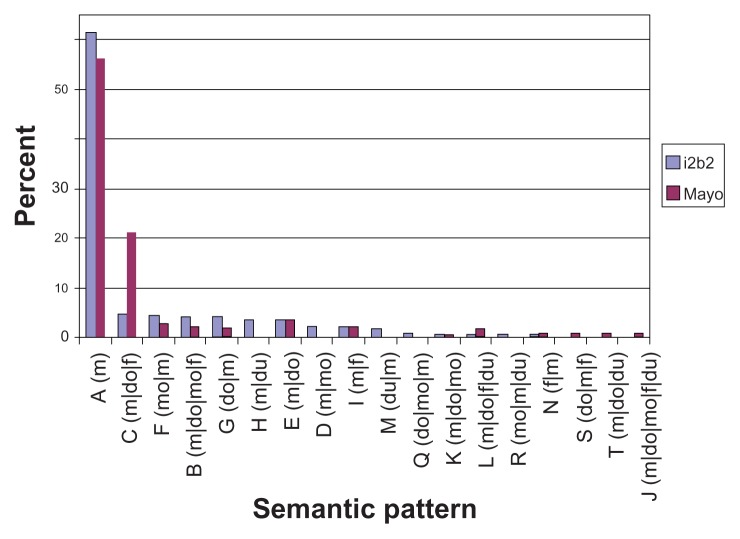
Each i2b2 annotation also contains format information, indicating whether the mention occurred in a list or narrative context. Our observations of Mayo data show that the Current Medication and Allergies sections have a list format for the medication description, whereas the other sections have a narrative format. To compare the pattern variations in different formats, we characterized Mayo’s Current Medication and Allergies sections as list and the others as narrative. As before, Mayo’s semantic tag set was mapped to that of i2b2. The comparisons for list and narrative formats are shown in Figures 3 and 4, respectively. The y-axis denotes the percentage of medication mentions out of the total medication mentions for a given pattern. We show the top 95% most frequent medication semantic patterns.
Figure 3.
Mayo vs. i2b2 medication semantic patterns in list (top 95%).
Figure 4.
Mayo vs. i2b2 medication semantic patterns in narrative (top 95%).
In list-formatted sections, Mayo data have five semantic patterns that cover 95% of the medication mentions, whereas i2b2 data have nine (Figure. 3). There are five semantic patterns that appear in both Mayo and i2b2. The most frequent pattern in the Mayo data is “m”, while “m|do|mo|f” is the most frequent in the i2b2 data.
In the narrative sections, Mayo data have 13 semantic patterns that cover 95% of the medication mentions, whereas i2b2 data have 15 (Figure. 4). There are 10 common semantic patterns that appear in both the Mayo and i2b2 data. The different pattern distributions between Mayo and i2b2 in Figure 3 and 4 show the sublanguage characteristics of the two institutions; additionally, Mayo and i2b2 have a different type of data source—ie, Mayo data are clinical notes that contain a variety of sections (see in Table 3) but i2b2 data are all from discharge summaries.
To see the distribution of both Mayo and i2b2 semantic patterns in a larger data set, we also examined a large corpus of Mayo data. Ten thousand Mayo clinical notes were processed by the cTAKES drug NER module to annotate medication information; we refer to the results as the Mayo 10K corpus. Note that these are system-generated results and were not manually reviewed by human experts.
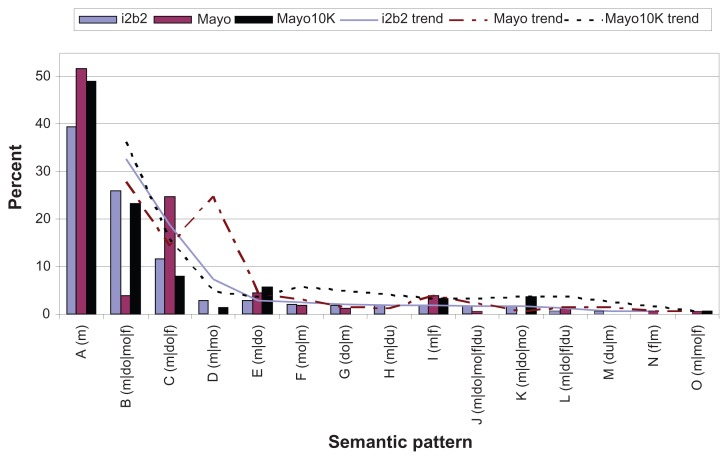
Figure 5 shows the i2b2, Mayo, and Mayo 10K medication semantic patterns and also includes trends of those patterns, the moving average with period two. It should be noted that Figure 5 includes only the top 95% of patterns—the lowest 5% do not appear even if they exist in a given corpus. The Mayo medication annotations were also mapped to the i2b2 annotations. In the top 95%, of 15 semantic patterns, nine appear in both Mayo and i2b2, four patterns appear only in i2b2 (m|mo, m|du, m|do|mo, du|m), and two patterns appear only in Mayo (f|m, m|mo|f). All data sets have the pattern of the medication name appearing alone as the most frequent pattern. However, the second most frequent pattern in Mayo is “m|do|f,”; however, in both Mayo 10K and i2b2, it is “m|do|mo|f”, showing a notable difference in trends. In Mayo 10K, there are a total of 311,004 medication mentions, and 95% of all the medication mentions appear in nine sections. After mapping the Mayo annotations to i2b2, 95% of all the medication mentions occur in eight semantic patterns. Intriguingly, the Mayo 10K’s three most dominant patterns (m, m|do|mo|f, m|do|f) exactly match the three most dominant patterns of i2b2, covering 80% and 77% of total medication mentions in Mayo 10K and i2b2 respectively. Comparing the trend to the Mayo 159 notes, this larger Mayo corpus looks more similar to the i2b2 semantic patterns.
Figure 5.
i2b2 vs. Mayo vs. Mayo10K medication semantic patterns with moving average trend (top 95%).
We also compared the semantic patterns in list versus narrative for both data sets. In the list sections, Mayo’s top three patterns (m|do|mo|f, m, and m|do|f) also exactly matched the top three from i2b2; they covered 75% and 82% of the medication mentions in Mayo and i2b2, respectively. In the narrative sections, Mayo had only five patterns that covered 95% of the medication mentions; all five patterns appeared in i2b2. Mayo’s top two patterns (m, m|do|f), which covered 84% of the medication mentions, matched i2b2’s top two patterns.
Medication context patterns
Medications in clinical notes are sometimes mentioned with additional words inserted between medication attributes. Those words may provide clues to medication semantics, so we examined this text as well.
Mayo data
The Mayo data have a total of 659 medication mentions. Out of those, only 93 cases (14%) have one or more words between medication semantic attributes. Of those 93 cases, 82 are unique. Since the individual words between the medication attributes are so varied, we did not notice a high frequency of exact context patterns. Therefore, we examined the semantic characteristics of the dominant partial context patterns. Table 4 shows statistics of dominant partial context patterns. When the medications are followed by verb forms, the verbs are often related to medication status changes—ie, change of medication attributes. For example, <m> was increased to <sn>. The other minor patterns not in Table 4 are as follows: <m> another medication <fu>|<fn>; x <du>|<do>; <m> at <do>; <m> from <sn>; <m> be given <fu>; and <m> were|used for <fm>.
Table 4.
Medication context patterns in mayo (number denotes frequency).
| Partial pattern | Status change pattern* |
|---|---|
| 22 per|a <fu> | 6 <m> INCREASE indication <sn> |
| 12 for (period of|an additional) <du> | 1 <m> DECREASE indication <sn> |
| 7 <fu> times (a) <fu> | 3 <m> START indication <sn> |
| 6 <do>|<mo> every <fn> | |
| 5 <m> dose|cycle (of|at) <sn> | |
| 5 <m> to <sn> | |
| 5 <su>|<du>|<fm> of <m> |
Note:
Lexical variations were normalized.
i2b2 data
Since i2b2 medication annotations are more coarse-grained than Mayo, we were able to find a high frequency of exact context patterns. In i2b2, there are a total of 9,003 medication mentions, and only 390 of those (4%) contain word(s) between medication attributes; of these 390 cases, 231 are unique context patterns. Table 5 shows the major exact and partial context patterns. Approximately one third of the total exact patterns belong to the exact patterns in Table 5. Similar to the Mayo data, when the medications are followed by verb forms, many are related to medication status changes. For example, <m> was started…,du., <m> was increased to <do>, and <m> was reduced to <do>. Another medication mention also appears between medication semantics, for example, <m> and Lipitor <du>. The other minor patterns include <m> on <do>, and <mo> with <m>.
Table 5.
Medication context patterns in i2b2 (number denotes frequency).
| Exact pattern | Partial pattern | Status change pattern* |
|---|---|---|
| 71 <do> of <m> | 141 <do>|<du> of <m> | 16 <m> INCREASE indication <do> |
| 28 <du> of <m> | 21 <m> at <do> | 2 <m> INCREASE indication <f> |
| 9 <do> of <m> <f> | 8 <m> (dose) from <do> | 1 <m><do> INCREASE indication <f> |
| 8 <du> of <mo> <m> | 5 <m> from <do> | 8 <m> DECREASE indication <do> |
| 6 <m> at <do> | 6 <m> to <do> | 1 <m> DECREASE indication <du> |
| 6 <do> of <mo> <m> | 4 <m> with <do> | 1 <m> DECREASE indication <f> |
| 5 <m> at <do> <mo> <f> | 8 <m> START indication <do> | |
| 5 <m> at <do> <f> | 3 <m> START indication <du> | |
| 5 <do> of <m> <du> | 1 <du> START indication <m> |
Note:
Lexical variations were normalized.
Discussion
Medication information was expressed in numerous ways in the clinical notes. We have investigated the medication description patterns in manually annotated data from 159 Mayo clinical notes and 253 i2b2 discharge summaries. Additionally, we have further compared these with a large medication corpus compiled by cTAKES drug NER from Mayo’s 10K clinical notes.
Notes from Mayo and i2b2 share most of the common semantic patterns, but differences exist. Overall, the medication name alone is the most dominant pattern in both data sets. In Mayo, about 90% of the medication mentions occur in four sections, and all but the Allergies section contains a variety of semantic patterns. The Allergies and the other sections mostly contain the medication name alone.
In lists, i2b2 has “m|do|mo|f” as the most frequent pattern, but in the Mayo data it is the medication name. This might be because Mayo’s samples of 159 notes contain a relatively lower portion of Current Medication sections, which in fact contain most of the medication information, as well as the most common list-format section. After mapping the Mayo annotations to the i2b2 annotations, the most frequent pattern in the Current Medication section is “m|do|f.” Based on these observations, we could conclude that the list-format medication descriptions tend to provide at least the minimum information necessary to guide the proper medication intake (ie, medication with dosage, frequency, and possibly mode information).
In narratives, the pattern of the medication name alone is the most frequent in both data sets. Generally, narratives in clinical notes describe the physician’s impression, report, and plan, as well as the patient’s medical history and diagnosis. In content of this nature, we may assume that physicians often simply mention medication names without further detailed attributes in clinical narratives as they do in describing other conditions. The second dominant pattern in both data sets is “m|do|f”—21% and 5% of the total medication mentions in Mayo and i2b2 respectively. In both data sets, the rest of the patterns cover a lower number of the medication mentions.
After mapping the Mayo annotations to i2b2, it can be seen that 12% (N = 11) of the semantic patterns cover 95% of the medication mentions. In i2b2, 19% (N = 13) of the semantic patterns cover 95% of the medication mentions. The i2b2 medication mentions are slightly more diverse than Mayo’s. Covering 95% of the entire medication mentions from both data sets are a total of 15 semantic patterns, nine of which are common in both.
In both Mayo’s 159 notes and i2b2, the medication descriptions do not contain many words between the medication attributes—14% and 4% of medication mentions contain some words between them in Mayo and i2b2, respectively. One reason for the higher context pattern for the Mayo data is due to different annotations. For example, if we look at duration, Mayo does not contain prepositions (eg, for a week → duration is “a week”) but i2b2 does (duration is “for a week”). Some prepositions such as “of” are commonly used in combination with medication and dosage/duration (eg, dosage or duration of medication). When a medication is followed by a verb, it can be related to a medication status change in many cases. Such medication status change descriptions often follow a pattern, eg, medication + status change verb + dosage or frequency.
When we examined a large Mayo corpus of 10,000 clinical notes, those semantic patterns were even closer to i2b2. Although there is more similarity between the two, Mayo 10K clinical notes have not been reviewed manually and therefore this might make the results slightly less reliable. The trends of i2b2 and Mayo 10K in Figure 5 seem to be very similar; the top three semantic patterns, which cover the majority of the medication mentions, are exactly matched with i2b2’s. The portion of the list of medication descriptions is also closer to i2b2 than Mayo 159 notes. In Mayo 10K, 49% of the medication mentions belong to the list format (34% in Mayo 159 note); in i2b2, 56% belong to the list format. These observations imply that a medication extraction tool based on the semantic patterns in Mayo data may work reasonably well for i2b2 and vice versa.
In our semantic pattern analysis, we considered only one medication mention at a time. For example, “Aspirin or Tylenol 1 tablet prn” generates two instances of the semantic pattern “m|do|f” instead of “m|m|do|f”. In the future, we will further compile and analyze these kinds of patterns.
As shown in this study, the sublanguage of medication description in general can be well characterized through semantic patterns. The statistics of medication patterns obtained in this study can be used as a confidence measure to determine whether a given medication and nearby attributes are truly associated or not. If its description pattern belongs to one of the majority patterns, we have high confidence in considering it a correct association.
Conclusion
The semantic patterns that are used to describe medications vary in clinical notes as a whole, and some variations also exist between institutions. However, dominant patterns do exist that account for most of the medication descriptions, and most of those patterns are common between Mayo and i2b2. Extra context between the medication attributes is not common in the medication descriptions, but it does occur in some particular patterns primarily associated with medication, including the dosage or duration “of” medication and a medication followed by a status change verb. Those semantic and context patterns could be utilized to properly parse medication and its attributes, as well as to correctly associate them.
Footnotes
Author Contributions
SS, HL conceived and designed the experiments. SS, HL, SM acquired data. All authors analysed and interpreted the data and results. SS wrote the first draft of the manuscript and all authors contributed to the writing of the manuscript. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests. This manuscript is intended for the special issue of Biomedical Informatics Insights—on Computational Semantics in Clinical Text.
Funding
This manuscript was supported by Strategic Health IT Advanced Research Projects (SHARP) Program (90TR002), National Science Foundation ABI:0845523, National Institute of Health 5R01LM009959, and NLM 1K99LM011389.
References
- 1.Sohn S, Kocher JPA, Chute CG, Savova GK. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc. 2011;18(Suppl 1):144–9. doi: 10.1136/amiajnl-2011-000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sohn S, Savova GK. Mayo Clinic Smoking Status Classification System: Extensions and Improvements. Paper presented at: AMIA Annual Symposium; 2009; San Francisco, CA. [PMC free article] [PubMed] [Google Scholar]
- 3.Sohn S, Torii M, Li D, Wagholikar K, Wu S, Liu H. A Hybrid Approach to Sentiment Sentence Classification in Suicide Notes. Biomed Inform Insights. 2012;(Suppl 1):43–50. doi: 10.4137/BII.S8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demner-Fushman D, Chapman W, McDonald C. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42(5):760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronsky D, Fiszman M, Chapman WW, Haug PJ. Combining decision support methodologies to diagnose pneumonia. Proc AMIA Symp. 2001:12–6. [PMC free article] [PubMed] [Google Scholar]
- 6.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17(5):568–74. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5(9):e13011. doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pakhomov SVS, Buntrock JD, Chute CG. Automating the assignment of diagnosis codes to patient encounters using example-based and machine learning techniques. J Am Med Inform Assoc. 2006;13(5):516–25. doi: 10.1197/jamia.M2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savova G, Masanz J, Ogren P, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.YTEX. Yale cTAKES Extension. http://code.google.com/p/ytex/
- 12.Zhou L, Plasek JM, Mahoney LM, et al. Using Medical Text Extraction, Reasoning and Mapping System (MTERMS) to Process Medication Information in Outpatient Clinical Notes. AMIA Annu Symp Proc. 2011;2011:1639–48. [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng Q, Goryachev S, Weiss S, Sordo M, Murphy S, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6(1):30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan J, Prasad R, Yabut RM, et al. Part-of-speech tagging for clinical text: wall or bridge between institutions? AMIA Annu Symp Proc. 2011;2011:382–91. [PMC free article] [PubMed] [Google Scholar]
- 16.Wagholikar K, Torii M, Jonnalagadda S, Liu H. Feasibility of pooling annotated corpora for clinical concept extraction. AMIA Summits Transl Sci Proc. 2012;2012:38. [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman C. A broad coverage natural language processing system. Proc AMIA Symp. 2000:270–4. [PMC free article] [PubMed] [Google Scholar]
- 18.Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage. Proc AMIA Symp. 2002:742–6. [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform. 2002;35(4):222–35. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 20.Harris ZS. A Grammar of English on Mathematical Principles. New York: Wiley; 1982. [Google Scholar]
- 21.Harris ZS. A theory of Language and Information: A Mathematical Approach. Oxford: Clarendon Press; 1991. [Google Scholar]
- 22.Chun H, Fuller S, Friedman C, Hersh W, editors. Knowledge Management and Data Mining in Biomedicine. New York: Springer; 2005. Semantic Text Parsing for Patient Records. [Google Scholar]
- 23.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17(5):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chhieng D, Day T, Gordon G, Hicks J. Use of natural language programming to extract medication from unstructured electronic medical records. AMIA Annu Symp Proc. 2007 Oct 11;:908. [PubMed] [Google Scholar]
- 25.Sirohi E, Peissig P. Study of effect of drug lexicons on medication extraction from electronic medical records. Pac Symp Biocomput. 2005:308–18. doi: 10.1142/9789812702456_0029. [DOI] [PubMed] [Google Scholar]
- 26.Levin MA, Krol M, Doshi AM, Reich DL. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA Annu Symp Proc. 2007:438–42. [PMC free article] [PubMed] [Google Scholar]
- 27.Harris ZS. The structure of science information. J Biomed Inform. 2002;35(4):215–21. doi: 10.1016/s1532-0464(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, AbdelRahman S, Lu Y, Denny JC, Doan S. Applying semantic-based probabilistic context-free grammar to medical language processing— a preliminary study on parsing medication sentences. J Biomed Inform. 2011;44(6):1068–75. doi: 10.1016/j.jbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson O, Hurdle JF. Document clustering of clinical narratives: a systematic study of clinical sublanguages. AMIA Annu Symp Proc. 2011;2011:1099–107. [PMC free article] [PubMed] [Google Scholar]
- 30.Uzuner Ö, Solti I, Xia F, Cadag E. Community annotation experiment for ground truth generation for the i2b2 medication challenge. J Am Med Inform Assoc. 2010;17(5):519–23. doi: 10.1136/jamia.2010.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarty C, Chisholm R, Chute C, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4(1):13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]