Abstract
Summary: The human complement system is increasingly perceived as an intricate protein network of effectors, inhibitors and regulators that drives critical processes in health and disease and extensively communicates with associated physiological pathways ranging from immunity and inflammation to homeostasis and development. A steady stream of experimental data reveals new fascinating connections at a rapid pace; although opening unique opportunities for research discoveries, the comprehensiveness and large diversity of experimental methods, nomenclatures and publication sources renders it highly challenging to keep up with the essential findings. With the Complement Map Database (CMAP), we have created a novel and easily accessible research tool to assist the complement community and scientists from related disciplines in exploring the complement network and discovering new connections.
Availability: http://www.complement.us/cmap.
Contact: lambris@upenn.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The complement system is an intricate protein network comprising more than 50 plasma and membrane-bound proteins that interact in a cascade manner (Supplementary Fig. S1). Activation of the system can be achieved by three major initiation pathways (classical, alternative and lectin) and culminate in the generation of biologically active molecules that play important roles in many physiological and pathological processes (Ricklin et al., 2010). Apart from its direct role in the maintenance of body homeostasis, clearance of pathogens, and induction of inflammation, the complement system also cross-talks with and modulates the functions of other biological systems, such as the coagulation cascade or toll-like receptor (TLR)-induced signaling pathways (Ricklin et al., 2010).
The proper operation of such an intricate system relies on tight control of complement activation by regulatory proteins. Although excessive complement activation can lead to severe damage to the host tissue with consequent induction of inflammatory and autoimmune diseases (Ricklin and Lambris, 2013), lack of complement activation, as observed in complement deficiency states, are also correlated with pathologies, such as hereditary angioedema, systemic lupus erythematosus, kidney diseases, recurrent infections and others (Reis et al., 2006; Skattum et al., 2011).
The extensive numbers of reported molecular interactions that build the base of the complement cascade, as well as the increasing connections observed between proteins from the complement and other biological systems, render it progressively challenging to keep up with current information, even for researchers in the field. To offer a comprehensive view of the entire complement-related network and facilitate systems research, we constructed the Complement Map Database (CMAP, Supplementary Fig. S2), a unique repository focused on documented molecular interactions described within the complement cascade and between complement and other biological systems.
2 DATA MODEL
Data in CMAP are organized into three levels: entities, relations and pathways. Entities are defined as proteins, small molecules and complexes (protein–protein, protein-small molecule). Relations are interactions between entities such as complex associations, covalent modifications, substrate conversion (catalytic) and activation and inhibition processes. Pathways in CMAP are composed of a main map (Supplementary Fig. S1), which is presented as a network model, and several focus maps that visualize networks involved in cross-talk, immune evasion or disease states (Table 1 and Supplementary Figs S3–S6). Each connection between complement components and their ligands shown in the fully searchable maps is hyperlinked and contains information about the publication sources (e.g. link to PubMed) and the analytical methods used for describing the interaction (Supplementary Table S1). Information contained in CMAP is entirely based on published experimental data and is fully revised by experts in the field. Additionally, links to external databases (e.g. NCBI, UniProt, Geo, HGNC and others) provided for each component further ensure rapid and focused gathering of information for a particular connection. The web interface, which was inspired by popular online search and cartography services, allows users to quickly pan, zoom and search the map. Further, specific CMAP interactions can be exported in XML and JSON format. CMAP is based on established technologies to ensure high compatibility: data are stored in a MySQL database, and the user interface is written in PHP and JavaScript (see Supplementary Methods for additional technical details).
Table 1.
CMAP pathway description
| Database ID | Path name | Path type | Molecules | Relations | Citations | Export |
|---|---|---|---|---|---|---|
| CMAP0001 | Complement | Main | 193 | 244 | 365 | XML, JSON |
| CMAP0002 | Coagulation | Cross-talk | 30 | 21 | 21 | XML, JSON |
| CMAP0003 | TLR | Cross-talk | 14 | 5 | 16 | XML, JSON |
| CMAP0004 | S.aureus | Cross-talk | 28 | 32 | 38 | XML, JSON |
| CMAP0005 | Cancer | Cross-talk | 16 | 4 | 7 | Image |
Note: The XMLSchema is available at http://www.complement.us/cmap/XML/CMAPSchema.xsd.
3 APPLICATION
3.1 Case study: C5a
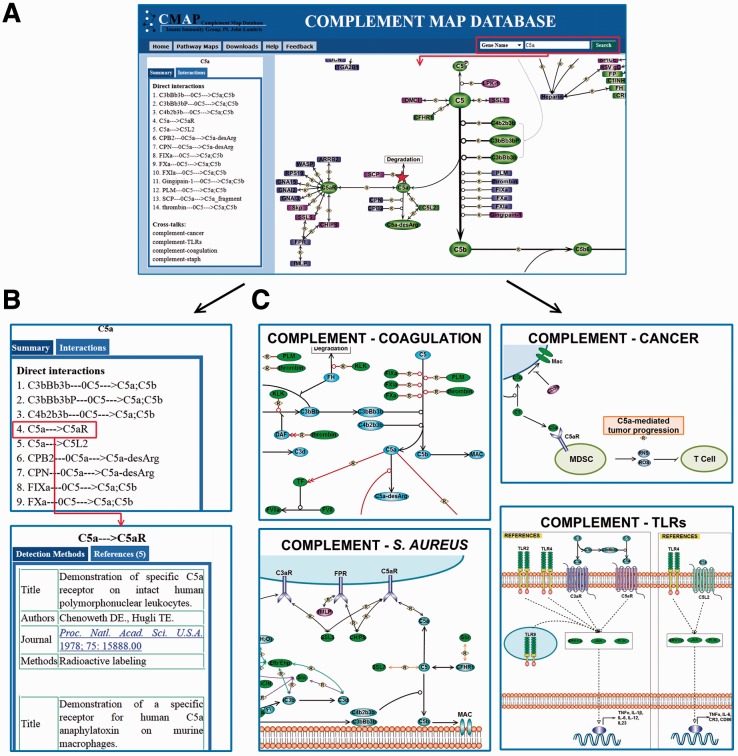
CMAP can be used to learn biological functions of complement components, assist in experimental design and provide information that supports experimental observations. The potent pro-inflammatory mediator C5a may serve as an important real-life example of CMAP’s usability. By searching for the keyword ‘C5a’, users get a table containing all the direct interactions between C5a and other proteins, such as the anaphylatoxin receptors C5aR and C5L2 (Fig. 1A and B). The same table also reveals that C5a is a fragment of complement component C5 and gets generated not only by the endogenous C5 convertases (e.g. C3bBb3b) but also by coagulation factors and the microbial proteins gingipain-1 and streptococcal peptidase. Another section of the information panel indicates that C5a is involved in the modulation of the coagulation system and TLR-induced responses, in microbial immune evasion (Staphylococcus aureus), and in cancer and tumor development (Fig. 1C). Users can find additional information on C5a-mediated functions and interactions by exploring the cross-talk maps and clicking on the references symbol next to individual functions or interaction links (Fig. 1C). Finally, buttons on each connection arrow reveal the experimental methods and publications on which the observations are founded. With the help of CMAP, researchers can learn the role of complement proteins in different processes quickly, intuitively and comprehensively.
Fig. 1.
An example of CMAP usage to study C5a. (A) Search result of C5a. Molecular information, interaction table and the general pathway map are given in the search result. (B) The interaction table and reference table. (C) Studying the role of C5a in different physiological and pathological processes via CMAP cross-talk pathways
4 CONCLUSION
CMAP is a novel and easily accessible research tool to assist the complement community and scientists from related disciplines in exploring the complement network and discovering new connections. In contrast to many other pathway maps, CMAP is tailored to the complement system and its cross-talk with endogenous pathways and exogenous modulators, logically organized according to the hierarchy of the cascade, and entirely based on experimental interaction data that have been reported in scientific journals and reviewed by expert scientists before addition to the database. The user-friendly interface allows easy accessibility and exploration of the site. Although offering a highly comprehensive and well-founded overview in its current form, we expect CMAP to improve even further with the help of and input from the complement community, thereby truly putting complement on the map for many research disciplines.
Funding: National Institutes of Health [AI030040, AI068730, AI072106, EY020633 and GM097747 to J.D.L., AI097805 to D.R., GM078005 to S.S.].
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Reis S, et al. Clinical aspects and molecular basis of primary deficiencies of complement component C3 and its regulatory proteins factor I and factor H. Scand. J. Immunol. 2006;63:155–168. doi: 10.1111/j.1365-3083.2006.01729.x. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J. Immunol. 2013;190:3831–3838. doi: 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, et al. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skattum L, et al. Complement deficiency states and associated infections. Mol. Immunol. 2011;48:1643–1655. doi: 10.1016/j.molimm.2011.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.