Abstract
Summary: Systematic studies of drug repositioning require the integration of multi-level drug data, including basic chemical information (such as SMILES), drug targets, target-related signaling pathways, clinical trial information and Food and Drug Administration (FDA)-approval information, to predict new potential indications of existing drugs. Currently available databases, however, lack query support for multi-level drug information and thus are not designed to support drug repositioning studies. DrugMap Central (DMC), an online tool, is developed to help fill the gap. DMC enables the users to integrate, query, visualize, interrogate, and download multi-level data of known drugs or compounds quickly for drug repositioning studies all within one system.
Availability: DMC is accessible at http://r2d2drug.org/DMC.aspx.
Contact: STWong@tmhs.org
1 INTRODUCTION
Drug repositioning has emerged as a cost-effective and time-efficient strategy to find new indications for old drugs (Ashburn and Thor, 2004; Iorio et al., 2010; Jin et al., 2012; Sirota et al., 2011). Because development of a new drug is time consuming (∼12–15 years), expensive (>$1 billion), and has a low success rate (between 1/10 000 and 1/5000), researchers are applying more systematic approaches to address new treatment opportunities by using the available multi-level data and information of known drugs, including their chemical structures, targets, target-related signaling pathways, clinical trial information and FDA-approval information. Several available databases have been developed for the aim of supporting drug repositioning research, including PharmDB (http://pharmdb.org), PROMISCUOUS (von Eichborn et al., 2011) and DrugPredict (Simon et al., 2012).
However, clinical trial and FDA-approval information as well as signaling pathway information are often missing from these available drug-repositioning databases. It is inconvenient and time-consuming for researchers without any pharmacological background to interrogate and assimilate these multi-level drug data distributed across different data sources for drug repositioning studies. Moreover, these data often do not share consistent entry names. When a specific biological question is asked, e.g. which drugs that are not in clinical trial phase II or later phases target on the HER2 pathway, the researcher has to go to different databases to check the data of clinical trials, targets and pathways. In this article, we present a united data source, DrugMap Central (DMC), with online visualization interfaces to query and search the integrated drug data quickly. Furthermore, DMC integrates the standardized disease information from MeSH (http://www.nlm.nih.gov/mesh/) into clinical trial and FDA-approval information to facilitate checking known disease information of drugs or compounds for drug repositioning studies. The unique multi-level drug data with visualization interfaces are available at http://r2d2drug.org/DMC.aspx with testing demos.
2 METHODS
We collected the multi-level drug data from a variety of public databases by using Python based codes. The basic drug information, such as chemical structures and targets, was collected from three data sources—Drugbank (Knox et al., 2011), PharmGKB (Hewett et al., 2002) and TTD (Chen et al., 2002), and uses the chemical IDs curated from PubChem. The signaling pathway data were derived from three databases—KEGG (Kanehisa and Goto, 2000) (only data prior to 2011 available), PID (Schaefer et al., 2009) and BioCarta (http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways). Clinical trial information was curated from ClinicalTrial.gov, and FDA approval information was collected from Drugs@FDA (http://www.accessdata.fda.gov/scripts/cder/drugsatfda/) and DailyMed (http://dailymed.nlm.nih.gov/).
The multi-level drug data were integrated together using the drug terms in MeSH (http://www.nlm.nih.gov/mesh/), in which the drug target information was linked to signaling pathway data. We further used the MeSH disease terms to annotate the disease information in the clinical trials and FDA-approval data.
Visualization interfaces were designed to visualize the integrated multi-level drug data using GraphViz software (http://www.graphviz.org). The network structure is constructed through a recursive method that starts from the initial drug to further levels, e.g. targets, pathways, clinical trials and FDA, until no more contiguous interactions are found in the database. The visualization method translates all curated database entities to the GraphViz language and feeds them into the GraphViz program. The GraphViz dot program applies an automatic layout algorithm to generate the visualization interface. An example graph is shown in Figure 1.
Fig. 1.
The search and query methods of the DMC database. (A) The multi-field search. (B) The visualization interface for the multi-level drug data
DMC database search engine is built on Microsoft IIS web server 7 and SQL server 2005. Full-text indexing is enabled for optimal information retrieval.
3 RESULTS AND DISCUSSIONS
The DMC provides two major methods to search and query the integrated multi-level drug data, as shown in Figure 1. The first method supports multiple-field search to retrieve drug data of interest. For instance, users can input ‘ERBB2’ in the target field and ‘breast cancer’ in the clinical trial options—disease field. The multi-filed search then returns all of the drugs with targets of ‘ERBB2’ testing for ‘breast cancer’ in clinical trials. The next interface is the visualization of the multi-level drug data (see Methods and Fig. 1B). The Graphviz-based interface enables users to query or download the fields-of-interest quickly, including the information for targets, clinical trials and FDA-approval applications. This visualization interface is also helpful for users to access the original data sources easily.
DMC provides two types of visualization interfaces, namely, All-Data-Integration-Builder (DMC-All, as shown in Fig. 1B) and Drug-Target-Pathway-Builder (DMC-DTP, as shown in Fig. 2), to integrate, query and download multi-dimensional data and information of known drugs or compounds. The DMC-All interface enables interactive visualization of six types of drug information: drug (chemical structure), target, clinical trial, FDA, targeted signaling pathways and targeted diseases under clinical trials or approved for use, whereas the DMC-DTP specifically represents the information of drug targets with their targeted signaling pathways. In the reported DMC database, we provide the DMC-All interface for interactively querying and visualizing information of about 2000 known drugs, with available clinical trial information and FDA-approval information, and the DMC-DTP interface for querying and visualizing available targets and signaling pathway information of >10 000 known drugs.
Fig. 2.
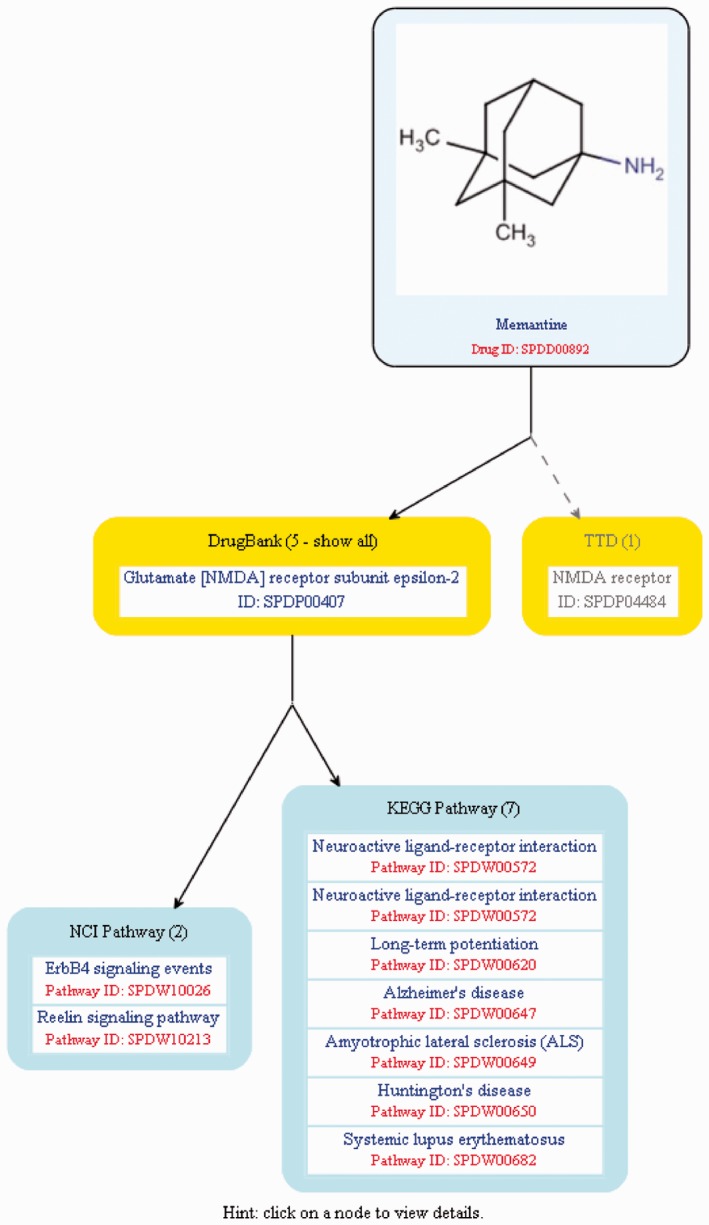
The visualization interface for drug-target-pathway information
The integrated multi-level drug data with the convenient visualization-based search and query tools will support researchers and scientists to interrogate available drug information quickly. The availability of DMC will benefit greatly both computational scientists and experimental researchers to conduct drug repositioning studies.
ACKNOWLEDGEMENTS
We are grateful to many SMAB members, especially Hong Zhao, who provided advice and helped test the visualization interface of DMC.
Funding: NIH [U54CA149196]; TT & WF Chao Foundation (to S.T.C.W.).
Conflict of Interest: none declared.
REFERENCES
- Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Chen X, et al. TTD: therapeutic target database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett M, et al. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl Acad. Sci. USA. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, et al. A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res. 2012;72:33–44. doi: 10.1158/0008-5472.CAN-11-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, et al. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon Z, et al. Drug effect prediction by polypharmacology-based interaction profiling. J. Chem. Inf. Model. 2012;52:134–145. doi: 10.1021/ci2002022. [DOI] [PubMed] [Google Scholar]
- Sirota M, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eichborn J, et al. PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res. 2011;39:D1060–D1066. doi: 10.1093/nar/gkq1037. [DOI] [PMC free article] [PubMed] [Google Scholar]



