Abstract
Summary: Because an enormous amount of sequence data is being collected, a method to effectively display sequence variation information is urgently needed. tasuke is a web application that visualizes large-scale resequencing data generated by next-generation sequencing technologies and is suitable for rapid data release to the public on the web. The variation and read depths of multiple genomes, as well as annotations, can be shown simultaneously at various scales. We demonstrate the use of TASUKE by applying it to 50 rice and 100 human genome resequencing datasets.
Availability and implementation: The tasuke program package and user manual are available from http://tasuke.dna.affrc.go.jp/.
Contact: taitoh@affrc.go.jp
1 INTRODUCTION
Recent advances in next-generation sequencing (NGS) technologies have allowed the rapid production of a tremendous amount of genomic sequence data at a low cost. This has naturally led to the resequencing of hundreds or thousands of genomes, such as the 1000 human genomes (http://www.1000genomes.org/) and the 1001 genomes project in Arabidopsis (http://www.1001genomes.org/). A method for comparing dozens of genomes in an effective manner is, therefore, urgently needed. Although a few stand-alone programs for comparative genome visualization have been developed (Fiume et al., 2010; Preston et al., 2012; Thorvaldsdóttir et al., 2013), to our knowledge, there is no web-based application that can handle dozens or more resequencing data of large genomes from higher eukaryotes.
The basic requirements of a visualization program for genome-wide resequencing data are as follows. First, a large amount of data obtained from tens or hundreds of samples from a species with genomes of >100 Mb need to be displayed in a smooth manner. An overview of NGS read mapping results needs to be shown so that users can grasp read coverage of a genome at the hundred- to million-base scale at a glance. Second, the use of storage and memory resources for the data browser should be minimal and small enough to be handled by the average computer server. It is not realistic to load an enormous amount of mapped read data from individual samples one by one with a stand-alone program on PC. Therefore, a client-server system, in which an efficient program runs on the server, is preferred. Third, it should be possible to share the data with collaborators or the public. In general, data from resequencing studies that are published as figures and tables in an article are not sufficient to reproduce a study’s results, whereas raw short reads registered in sequence read archive databases and resultant single-nucleotide polymorphism (SNP) data are informative. For experimental researchers who seek polymorphisms at the genome-wide level, a browser that can effectively address hundreds of genomes and display a large number of polymorphic sites is needed.
Here, we present tasuke, a web application for the visualization of large-scale resequencing data obtained from at least 100 genomes. This application allows users to rapidly release their own data on the web. Variant frequencies, read coverage and gene annotation information are shown simultaneously at various scales. tasuke uses a window analysis so that users can get a bird’s-eye view of the SNP density.
2 FUNCTIONS AND APPLICATION
For the sake of ease of use, tasuke was designed as web application implemented in HTML5. In this way, researchers can easily share data via general web browsers using a graphical interface. The input files required are as follows: a reference genome in FASTA format, Variant Call Format (VCF) files (Danecek et al., 2011) and depth files created by the ‘depth’ command of SAMtools (Li et al., 2009). Annotation files in General Feature Format (GFF, http://www.sanger.ac.uk/resources/software/gff/) are optional. A MySQL database is also required for the website’s backend data management. tasuke helps bioinformatics researchers of genome-wide resequencing projects to visualize a large amount of polymorphisms on multiple genomes and to release the data to the public.
On the upper pane, the reference genome and annotation information are displayed (Fig. 1b and c). Users can choose a specific position by clicking on the selected chromosome or moving a slider in the upper right region. Alternatively, the top menu bar provides users with a search function to find identifiers or genomic positions (Fig. 1a). Nucleotide variations (SNPs and length polymorphisms) and depth of mapped reads are presented in the lower main pane, which can be dragged to the left or right (Fig. 1e). The depth information is important to distinguish whether the region has no SNPs or no mapped reads, which are generally omitted in VCF format. The frequency of variation occurrence and/or average depth are shown in a block that corresponds to a region scalable from 1 bp to 100 kb with colored gradations: blue for SNPs, red for insertions/deletions and gray or yellow for depth (Fig. 1f). The maximum number of blocks displayed in a window is 200, so that up to 20 Mb can be viewed. At the most precise level, individual nucleotides and translated amino acids can be shown (Fig. 1B). By clicking on a block, a window of detailed information about nucleotide variations and depth pops up (Fig. 1l). To find mutations that possibly affect phenotypes, the effect information of each variant, such as non-synonymous changes and frame shifts, which can be added to a VCF file by snpEff (Cingolani et al., 2012), is shown by selecting ‘snpEFF’ in the menu bar (Fig. 1l and m). If a sample name is clicked, the reference genome is reset to the selected sample and variant frequencies are recalculated for all genomes. This reference switch function is useful to look over variations derived from different origins. From the ‘Tools’ menu (Fig. 1a), users can export a list of variant information, which is described in a tab-delimited file of a specified region of up to 200 kb. An image file of the displayed area is also downloadable.
Fig. 1.
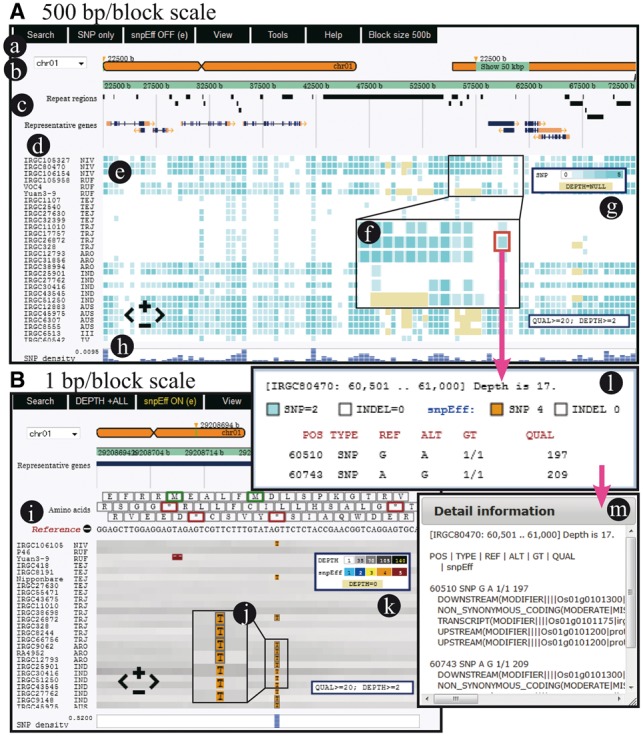
Screenshots of tasuke. (A) A view showing variants of 500 bp/block scale. (a) Menu bar for various functions. (b) Chromosomal positions. (c) Annotation tracks. (d) Sample names and related information. (e) Main panel for variant frequencies of block regions. (f) Magnified view of blocks. Blocks without reads are yellow. (g) Indicator for variant frequency and depth. (h) Overall SNP density. (B) A view showing variants and depth of 1 bp/block scale. (i) Amino acids and nucleotides on a reference genome. (j) Variants and their effects. (k) Indicator of levels of variant effects. (l) Variants and average depth information are shown by clicking on a block. (m) Variant effects are shown by clicking on the sub-window of (l)
As a demonstration, we applied tasuke to resequencing data from rice and human samples so that users can experience the functions of tasuke. First, we used resequencing data from 50 rice genomes at ∼15× coverage (Xu et al., 2011), which was downloaded from the DDBJ Sequence Read Archive (Kodama et al., 2010). The short-reads of rice were mapped to the reference genome (Sakai et al., 2013) by BWA (Li and Durbin, 2009). Second, alignments of human genome resequencing data generated by the 1000 Genomes Project were downloaded (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/), and 100 individuals who represent subpopulations were arbitrarily selected. Variant and read depth information were obtained from both datasets by using SAMtools. The annotations from the Rice Annotation Project Database (Sakai et al., 2013) and Ensembl (Flicek et al., 2013) were also stored in MySQL databases. These datasets are accessible through tasuke at http://tasuke.dna.affrc.go.jp/.
3 CONCLUSION
tasuke is designed for the visualization and rapid release of large-scale resequencing data on the web. This application allows users to see variant frequencies, read depth and annotation information in a scalable and smooth manner. We demonstrated its functionality through application to resequencing data from the rice and human genomes. This application is useful for the analysis of other genome-wide NGS data obtained from large samples. In future, to cope with growing resequencing data as well as RNA-seq and other NGS data, we will further improve tasuke.
ACKNOWLEDGEMENT
The authors thank J. Yonemaru, H. Numa, T. Tanaka, H. Sakai and Y. Kawahara for valuable comments and suggestions.
Funding: This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, GIR1001; Development of Genome Information Database System for Innovation of Crop and Livestock Production).
Conflict of Interest: none declared.
REFERENCES
- Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume M, et al. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–1944. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, et al. Biological databases at DNA data bank of Japan in the era of next-generation sequencing technologies. Adv. Exp. Med. Biol. 2010;680:125–135. doi: 10.1007/978-1-4419-5913-3_15. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MD, et al. VarB: a variation browsing and analysis tool for variants derived from next-generation sequencing data. Bioinformatics. 2012;28:2983–2985. doi: 10.1093/bioinformatics/bts557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, et al. Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:1–11. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, et al. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2011;30:1–10. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]


