Abstract
Cellular responses to DNA damage induced by intrinsic and extrinsic genotoxic stresses are highly regulated by complex signaling pathways, such as activation of the phosphoinositide-3-kinase-like protein kinase family and their downstream genes. Disruption of these signaling pathways leads to genome instability and cell death, and thus may provide potential novel strategies for cancer therapy. Here, we find that the expression of a human microRNA (miRNA), hsa-miR-185, is downregulated in response to ionizing radiation. Elevation of miR-185 sensitizes renal cell carcinoma cells to X-rays both in vitro and in vivo. Bioinformatic analysis shows that the ATM- and Rad3-related (ATR) kinase, a master conductor of cellular responses to DNA damage and DNA replication stresses, is a target of miR-185. This prediction was validated by luciferase reporter and mutation assays. We also demonstrated that miR-185 negatively regulates ATR expression at post-transcriptional level. miR-185 enhances radiation-induced apoptosis and inhibition of proliferation by repressing ATR pathway. In conclusion, our findings indicate a previously unreported regulatory mechanism for ATR expression mediated by miR-185 and shed light on the potential application of miRNAs both as direct cancer therapeutics and as tools to sensitize tumor cells to radiotherapy.
Keywords: ATR pathway, miR-185, apoptosis, proliferation
DNA damage is a direct consequence of genotoxic stresses, such as ultraviolet B (UVB), ionizing radiation and DNA replication interference. The cellular responses to DNA damage are orchestrated by complex signaling pathways, also known as DNA damage checkpoints. The phosphoinositide-3-kinase-like protein kinases such as ataxia telangiectasia mutated (ATM), ATM- and Rad3-related (ATR) and DNA-dependent protein kinase (DNA-PK) are major members of these signaling pathways.1 It has been demonstrated that ATM phosphorylates its effector kinase checkpoint kinase 2 (Chk2) and ATR phosphorylates its effector kinase Chk1, which in turn activate other potential substrates linked to DNA damage responses.2, 3, 4 Compared with ATM, which is primarily activated by DNA double-strand breaks, ATR responds to a broader spectrum of DNA damage and replication interference, including single-stranded DNA (ssDNA), double-stranded DNA (dsDNA) adjacent to ssDNA, adducts, cross-links and inhibition of DNA polymerase.5, 6 The signaling pathways have a central roles in detecting DNA damage, regulating DNA repair and coordinating with other cellular processes.7, 8, 9 Disruption of these signaling pathways leads to genome instability and cell death, and thus may provide a potential therapeutic strategy for cancer treatment.
MicroRNAs (miRNAs), a class of small non-coding RNA species, function in diverse biological processes including development, cellular proliferation, differentiation, apoptosis, tumorigenesis and cancer progression by post-transcriptional regulation of their target genes. It has been demonstrated that miRNAs regulate the expression of many vital factors. For instance, miRNAs are involved in the p53 tumor-suppressor network,10, 11 and miR-21 regulates the expression of PTEN tumor-suppressor gene in human hepatocellular cancer.12 Moreover, miRNA expression can be changed by radiation.13, 14, 15 Alteration of miR-34a expression may be responsible for important protective mechanisms counteracting radiation cytotoxicity.16 miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells and renders cells hypersensitive to γ-irradiation and genotoxic drugs.17 miR-182 targets BRCA1 to impact homologous recombination-mediated repair and cellular radiosensitivity.18 Recently, it was found that ATM is downregulated by N-Myc-regulated miR-421, and ectopic expression of miR-421 results in S-phase cell cycle checkpoint changes.19 These reports implicate that miRNAs have an important roles in DNA damage responses and signaling pathways. However, no direct interaction between miRNAs and ATR has been reported so far.
In this study, we set out to analyze the downregulated miRNAs in response to genotoxic stresses such as ionizing radiation, and to investigate the feasibility of introducing individual miRNAs into cancer cells to interrupt signaling transduction, therefore enhancing lethal effect or sensitivity of cancer cells during treatment.
Results
miR-185 expression is downregulated in response to ionizing radiation
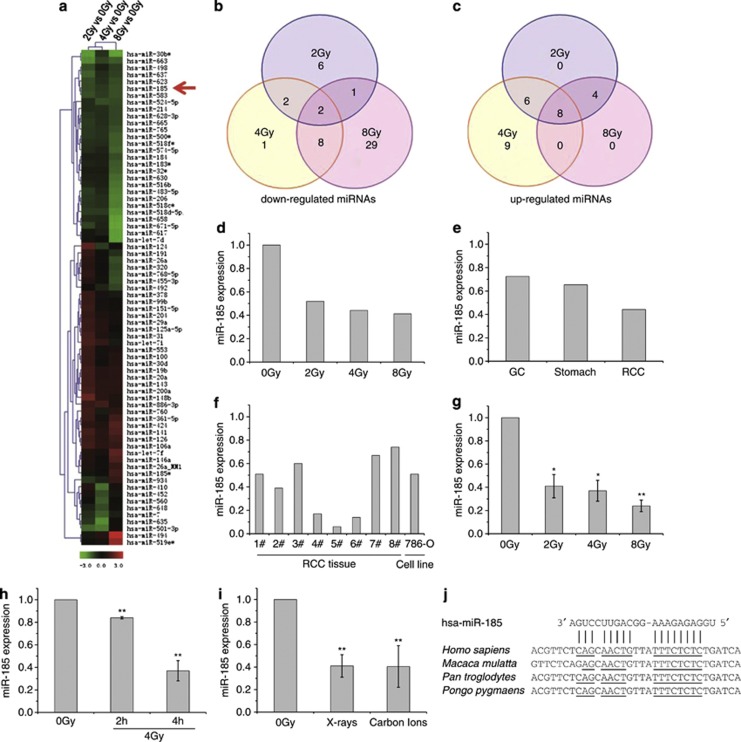
To determine the miRNAs that might be involved in DNA damage responses induced by extrinsic genotoxic stress, clinical renal cell carcinoma (RCC) tissues were treated with various doses of X-rays. The miRNA expression profiles were obtained by miRNA microarray analysis. Among the 588 miRNAs checked, 49 miRNAs were downregulated while 27 were upregulated (Figures 1a–c), indicating alteration of miRNA levels caused by ionizing radiation. Hsa-miR-185 on chromosome 22 was one of the miRNAs that were dramatically downregulated by all radiation doses (Figure 1d). Decreased miR-185 expression was also observed in gastric cancer and in normal stomach tissues, but the downregulation was not as remarkable as in RCC tissues tested using the same miRNA microarray (Figure 1e).
Figure 1.
miR-185 expression is downregulated in response to ionizing radiation. (a) Hierarchical clustering of miRNA expression obtained with Exiqon miRNA microarrays (version 9.2) after exposure of clinical RCC tissues to 2, 4 and 8 Gy of X-rays, respectively. Colored bars represent the differential levels of miRNAs expressed in irradiated samples versus sham-irradiated samples (0 Gy). The red arrow indicates hsa-miR-185. (b) Venn diagram of the miRNA expression profile depicting consistency of downregulated miRNAs between samples. (c) Venn diagram of the miRNA expression profile depicting consistency of upregulated miRNAs between samples. (d) Relative changes of the miR-185 expression level in clinical RCC tissues 4 h after X-ray irradiation. (e) Relative changes of the miR-185 expression level in various clinical tissues 4 h after 4 Gy X-ray irradiation, which was also obtained with a miRNA microarray assay. GC, gastric cancer tissues; Stomach, normal stomach tissues. (f) Verification of miR-185 expression altered by 4 Gy of X-rays in clinical RCC tissues obtained from patients (n=8) and in the cultured RCC cell line 786-O, analyzed by qRT-PCR. The histograms show the relative changes of the miR-185 expression level compared with sham-irradiated samples (0 Gy). (g) Dose-dependent inhibition of miR-185 expression in 786-O cells 4 h after irradiation, obtained by qRT-PCR. (h) Time-dependent inhibition of miR-185 expression in 786-O cells exposed to 4 Gy of X-rays, obtained by qRT-PCR. (i) Radiation-type-dependent inhibition of miR-185 in 786-O cells exposed to 2 Gy X-rays or carbon ions, obtained by qRT-PCR. (j) Predicted binding sites of miR-185 on the 3′-UTR of ATR mRNA in several species. Data with error bars represent the means of at least three independent experiments while others represent the means of two independent experiments. *P<0.05 compared with the sham-irradiated samples (0 Gy); **P<0.01 compared with the sham-irradiated samples (0 Gy)
The downregulation of miR-185 in clinical RCC tissues and the cultured RCC cell line 786-O was verified using quantitative real-time polymerase chain reaction (qRT-PCR) (Figure 1f). The dose-dependent and time-dependent inhibition of miR-185 investigated in 786-O cells can be seen in Figures 1g and h, respectively. Considering radiotherapy, photons such as X-rays and particles such as carbon ions or protons are the major ionizing radiation used for cancer treatment, thus the radiation-type-dependent inhibition of miR-185 is observed and shown in Figure 1i.
Based on the data obtained from open resources, miR-185 is highly conserved and the mature sequence is exactly the same among various mammalian species, furthermore, the binding sites of miR-185 on ATR mRNA of several species are also highly conserved (Figure 1j), which imply that miR-185 may have important biological functions across these species. It was reported that miR-185, miR-16 and miR-22 have strong positive correlation to the appearance of erythroid surface antigens (CD71, CD36 and CD235a) and hemoglobin synthesis.20 miR-185 is highly expressed in brain, kidney, lung, ovary, placenta, prostate, spleen and thyroid.21 Ectopic expression of miR-185 is observed in tumor cells.22 The general downregulation of miR-185 by ionizing radiation suggests that miR-185 may have an important regulatory role in cellular responses to DNA damage stresses. Thus, we carried out further experiments to investigate miR-185 function with the cultured 786-O cells.
Elevation of miR-185 sensitizes RCC cells to X-rays both in vitro and in vivo
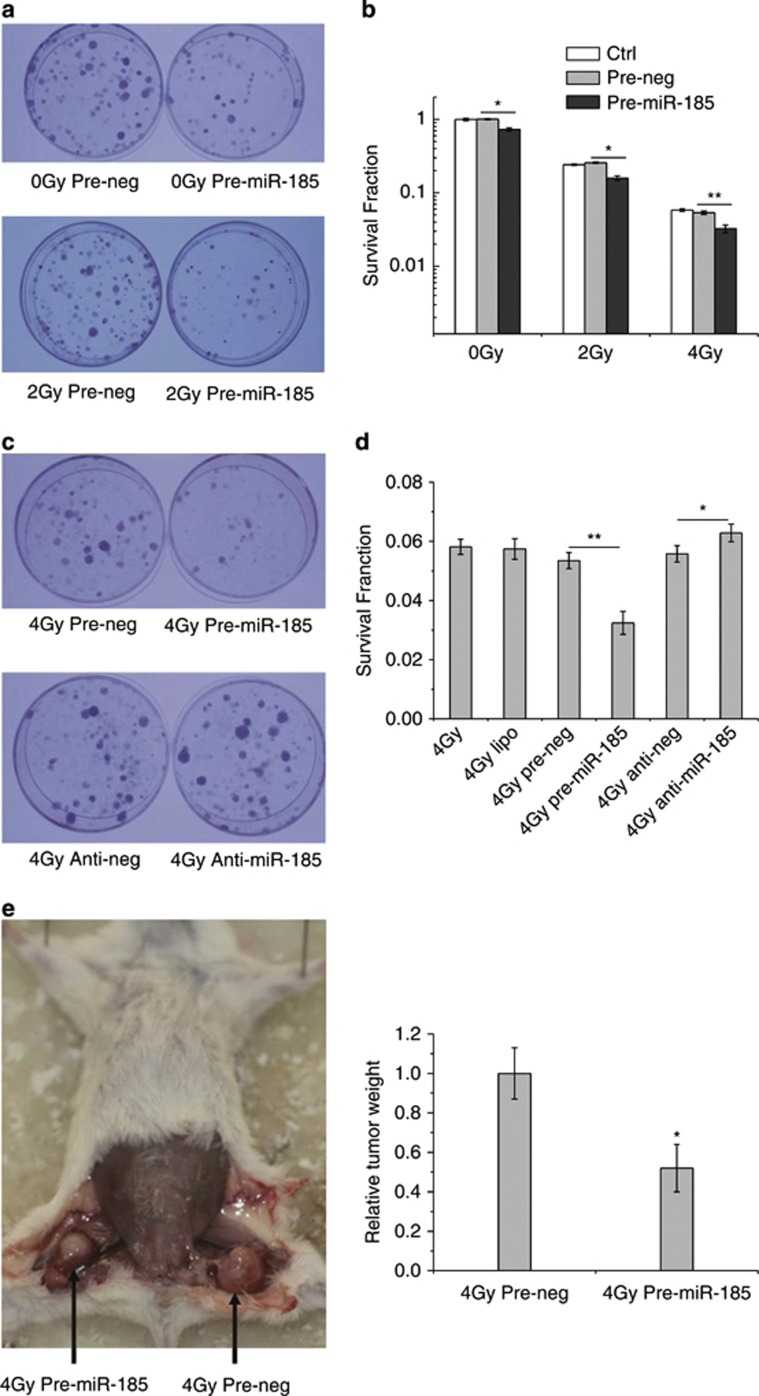
To investigate the impact of miR-185 on radiobiological effects, we elevated miR-185 levels by introducing exogenous pre-miR-185 into RCC cells and tested cell survival by colony formation assay after X-ray irradiation. The survival fraction of 786-O cells transfected with pre-miR-185 was significantly lower than that transfected with pre-neg (negative control of pre-miR-185), indicating reduced cell survival with upregulation of miR-185 expression (Figures 2a and b). To investigate further, we manipulated miR-185 levels in 786-O cells with various treatments and tested the survival fraction after subjecting the samples to 4 Gy of X-rays. As expected, the survival fraction of cells transfected with pre-miR-185 was the lowest among all treatments (Figures 2c and d). The survival fraction of cells transfected with anti-miR-185, an inhibitor that depletes endogenous miR-185, increased slightly but significantly when compared with the survival fraction of cells transfected with its negative control (anti-neg).
Figure 2.
Impact of miR-185 levels on cell survival. (a) Photographs of colonies formed by 786-O cells exposed to 0 and 2 Gy of X-rays. (b) Survival fractions of cells without transfection (Ctrl) and cells transfected with pre-miR-185 or pre-neg (30 nM final) in response to 0, 2 and 4 Gy of X-rays measured by colony formation assay. (c) Photographs of the colony formation with various treatments. (d) Survival fractions of 786-O cells with various treatments. (e) Tumor formation by the RCC cells in NOD/SCID mice subsequently exposed to 4 Gy of X-rays. Tumors formed by 786-O cells transfected with pre-miR-185 or pre-neg were separated from the hind legs of the mice (n=9) and weighed on the 55th day after subcutaneous injection. Each experiment was conducted at least three times independently. *P<0.05 compared with pre-neg; **P<0.01 compared with pre-neg
The role of miR-185 in sensitization was confirmed by NOD/SCID mouse experiments in vivo. As shown in Figure 2e, the weight of tumor formed by 786-O cells transfected with pre-miR-185 and exposed to 4 Gy of X-rays was only half of that transfected with pre-neg and exposed to the same dose of X-rays, supporting the importance of miR-185 in modulating radiobiological effects.
miR-185 targets ATR
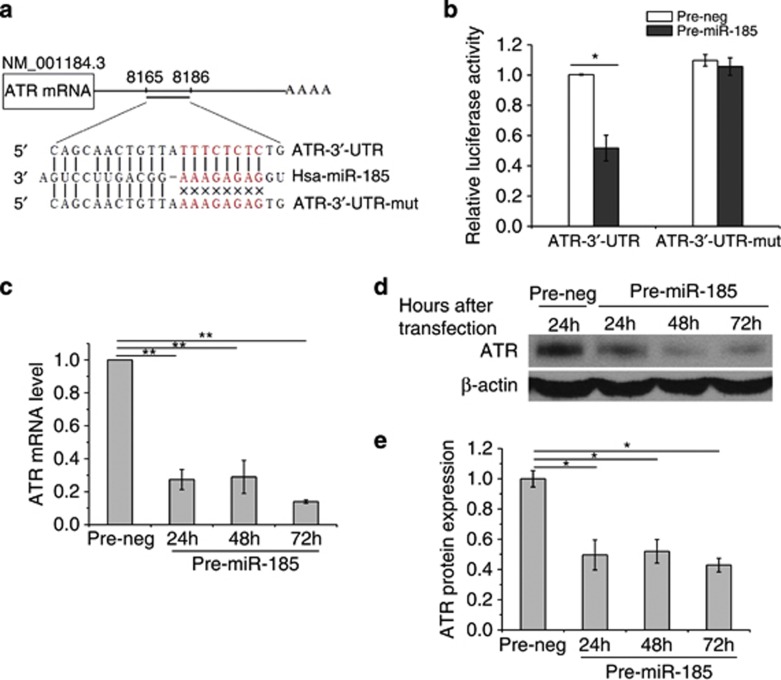
Although Six1, RhoA and Cdc42 have been reported as the targets of miR-185,23, 24, 25 it remains unclear whether any target of miR-185 is related to radiation response. To address this question, we predicted the putative targets of miR-185 by combining several prediction tools available online including MicroCosm Targets version 5 (http://microrna.sanger.ac.uk/targets/v5/) and RNA22 miRNA target detection (http://cbcsrv.watson.ibm.com/rna22.html). Among the predicted targets that showed significant matches with the miR-185 sequence, we focused on ATR because it is a crucial kinase of the signaling pathways for cells to cope with cellular stresses. Above all, there are no reports describing ATR regulation by miRNA up to date. Eight miR-185-binding sites were found on ATR mRNA, one in the 3′-untranslated region (3′-UTR) and seven in the coding region.
Next, we performed luciferase reporter and mutation assays to validate the target prediction. As shown in Figure 3a, we constructed the vector by inserting either the wild-type sequence of the 3′-UTR of ATR mRNA (ATR-3′-UTR) or a mutated seed sequence of the miR-185-binding site (ATR-3′-UTR-mut) into the pMIR-REPORT luciferase reporter. We found that co-transfection of the vector with wild-type ATR-3′-UTR and pre-miR-185 inhibited luciferase activity whereas co-transfection of the vector with ATR-3′-UTR-mut and pre-miR-185 caused no inhibition of luciferase activity in 786-O cells (Figure 3b). These results validated the binding of miR-185 to the 3′-UTR of ATR mRNA. After that, we investigated the impact of miR-185 on the expression of ATR. qRT-PCR revealed that ATR mRNA decreased significantly 24 h after 786-O cells were transfected with pre-miR-185 (Figure 3c). Consistent with these results, western blotting indicated that the protein level of ATR was also remarkably suppressed 24 h after pre-miR-185 transfection (Figures 3d and e). The targeted inhibition of ATR by miR-185 was further confirmed by the downregulation of ATR at both mRNA and protein levels in cells transfected with pre-miR-185.
Figure 3.
miR-185 negatively regulates ATR expression at post-transcriptional level. (a) Construction of a vector with either the wild-type sequence of the 3′-UTR of ATR mRNA (ATR-3′-UTR) or a mutated seed sequence of the miR-185-binding site (ATR-3′-UTR-mut). The seed sequence is shown in red. (b) Luciferase reporter assays. Each constructed vector was co-transfected with exogenous pre-miR-185 or pre-neg into 786-O cells. Luciferase activity was read 24 h after transfection. (c) ATR expression regulated by miR-185 at the mRNA level. qRT-PCR was conducted to quantify the expression level of ATR mRNA at the indicated time points after 786-O cells were transfected with pre-miR-185 or pre-neg. (d) ATR expression regulated by miR-185 at the protein level. Western blotting was performed at the indicated time points after transfection with pre-miR-185 or pre-neg. β-Actin, loading control. (e) Densitometric analysis of ATR protein levels shown in panel d. Each experiment was conducted at least three times independently. *P<0.05 compared with pre-neg; **P<0.01 compared with pre-neg
Taken together, these findings show that miR-185 represses ATR expression by post-transcriptional regulation through binding to the 3′-UTR of ATR mRNA.
miR-185 enhances radiation-induced apoptosis and inhibition of proliferation by repressing ATR pathway
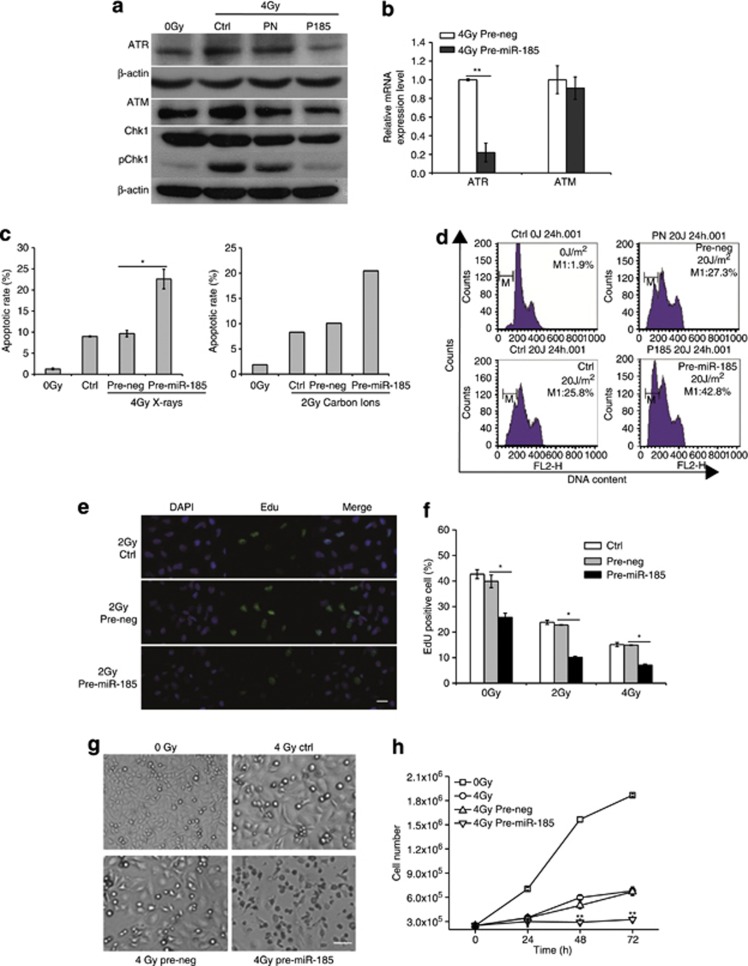
Having established a negative regulatory link between miR-185 and ATR, we were then interested in the mechanisms of sensitization of miR-185 in cancer cells. First, we confirmed the induction of ATR-Chk1 pathway after exposure of 786-O cells to 4 Gy of X-rays. The protein levels of ATR, ATM and pChk1 were increased in the irradiated samples without transfection (4 Gy Ctrl) when compared with the sham-irradiated samples (0 Gy). However, obvious reduction of ATR and pChk1 emerged in the irradiated samples transfected with pre-miR-185 (4 Gy P185). The protein levels of ATM and Chk1 were relatively stable implicating that the repression of ATR and phosphorylation of Chk1 has a central role in the target effects mediated by miR-185 (Figure 4a). In agreement with these results, qRT-PCR analysis revealed that ATR mRNA levels decreased significantly in the irradiated samples transfected with pre-miR-185 while ATM mRNA level did not (Figure 4b). These results indicate the repressive function of exogenous miR-185 on the induction of ATR-Chk1 pathway in response to DNA damage induced by extrinsic stress.
Figure 4.
miR-185 enhances apoptosis and proliferation inhibition by negatively regulating ATR. (a) ATR, ATM, Chk1 and pChk1 expression at the protein level. Western blotting was performed to measure the protein levels of ATR, ATM, Chk1 and pChk1 in cells transfected with pre-miR-185 (P185), pre-neg (PN) or without transfection (Ctrl) 24 h after exposure of 786-O cells to 0 and 4 Gy X-rays. β-Actin, loading control. (b) ATR and ATM expression at the mRNA level. qRT-PCR was conducted to quantify mRNA of ATR and ATM in cells transfected with pre-miR-185 or pre-neg 24 h after exposure of 786-O cells to 4 Gy X-rays. (c) Apoptotic rate of 786-O cells analyzed by flow cytometry 24 h after X-ray or carbon ion irradiation. (d) DNA histograms where the relative cell counts were plotted against DNA content 24 h after cells were exposed to UVB and stained with propidium iodide. Sub-G1 (gate M1) demonstrates the apoptotic cells. One of three independent experiments is shown. (e) Micrographs of EdU-positive 786-O cells 24 h after exposure to 2 Gy of X-rays. Scale bar, 25 μm. (f) Bar charts for the fraction of EdU-positive cells after various treatments. (g) Micrographs of the density and morphology of 786-O cells 48 h after exposure to 4 Gy of X-rays; Scale bar, 100 μm. (h) Growth curves of 786-O cells after various treatments. Each experiment was conducted at least three times independently except that carbon ion irradiation was conducted two times because of the beam time restriction and did not show error bars in the histograms. *P<0.05 compared with pre-neg; **P<0.01 compared with pre-neg
As ATR-Chk1 pathway inhibition promotes apoptosis,26, 27 we tested whether the repression of ATR mediated by miR-185 has a role in establishing apoptosis after exposure of 786-O cells to X-rays or carbon ions. We found that pre-miR-185 transfection enhanced radiation-induced apoptosis dramatically (Figure 4c). DNA damage induced by UVB typically includes the formation of cyclobutane pyrimidine dimers and 6-4 photoproducts, which are effective to activate ATR. As shown by the results of flow cytometry, the apoptotic rate was 25.8% in cells without transfection (Ctrl) while the apoptotic rate was 42.8% in cells transfected with pre-miR-185 after exposure to 20 J/m2 UVB (Figure 4d), indicating that the repression of ATR by elevating miR-185 increased apoptotic levels.
ATR is essential for the sustained survival of proliferating cell.28 Thus, we evaluated the consequence of elevating miR-185 levels by checking the S phase incorporated 5-ethynyl-2′-deoxyuridine (EdU) cells. On 2 and 4 Gy X-ray irradiation, the fraction of EdU-positive cells in samples transfected with pre-miR-185 decreased significantly compared with samples transfected with pre-neg (Figures 4e and f), suggesting the negative regulatory role of miR-185 in cellular proliferation. On the other hand, we also noticed that the fraction of EdU-positive cells in samples transfected with pre-miR-185 decreased significantly even without radiation, supporting the important role of ATR in DNA replication during the unperturbed S phase (Figure 4f). In line with these findings, cell growth curves showed that the cell number in samples transfected with pre-miR-185 was significantly lower than the cell number in samples transfected with pre-neg after 4 Gy X-ray irradiation, indicating that the repression of ATR by miR-185 involved in the inhibition of cellular proliferation (Figures 4g and h).
Altogether, these data demonstrate that elevated miR-185 enhances apoptosis and proliferation inhibition by repressing the ATR pathway, which can be induced by intrinsic and extrinsic cellular stresses, and results in increased lethal effect or sensitivity of cancer cells.
Discussion
As both a sensor and a transducer, ATR is crucial in cellular responses to DNA damage and DNA replication stresses. It mediates DNA damage signals, regulates cell cycle checkpoints, stabilizes replication forks, and promotes DNA repair and replication restart.29, 30 It is essential for the maintenance of genome integrity and cell survival31, 32 as well as the suppression of telomere fragility and recombination.33 Its activation requires not only replication protein A and ATR-interacting protein for ssDNA, but also DNA topoisomerase II-binding protein 1, RAD17 and the 9-1-1 complex for dsDNA adjacent to ssDNA. Activated ATR phosphorylates hundreds of proteins, including Chk1. Dysregulation of ATR disturbs a wide range of cellular processes. However, little was known about the regulatory mechanisms for ATR expression. Moreover, the targets of miR-185 that are tightly linked to radiation responses remain unclear. Here, we demonstrate that miR-185 targets ATR directly by binding to the 3′-UTR of ATR mRNA. ATR expression is downregulated by miR-185 at both mRNA and protein levels. It has been reported that miR-185 induces G1 cell cycle arrest and apoptosis, inhibiting proliferation of cancer cells by targeting Six1,23 RhoA and Cdc42.24 Our results also indicate that miR-185 regulates apoptosis and cellular proliferation although the target is ATR. On the basis of our own data, which demonstrate that miR-185 represses the expression of ATR and the activation of its downstream effector Chk1, we favor a model in which elevation of miR-185 enhances apoptosis and inhibition of proliferation by downregulating the ATR-Chk1 pathway, which is induced by ionizing radiation (Figure 5). To the best of our knowledge, this is the first report on the regulation of ATR expression mediated by a miRNA.
Figure 5.

A schematic diagram illustrating miR-185 and ATR function in radiation response. ATR is activated by DNA damage resulting from exposure to ionizing radiation in combination with a series proteins such as replication protein A (RPA), ATR-interacting protein (ATRIP), DNA topoisomerase II-binding protein 1 (TOPBP1), RAD17 and the 9-1-1 complex (Flynn and Zou30). Activated ATR phosphorylates its effector kinase Chk1. Consequently, the ATR-Chk1 pathway has an important role in detecting DNA damage, initiating DNA repair and coordinating with other cellular processes, and serves as a central safeguard of the genome. However, elevation of miR-185 represses the ATR-Chk1 pathway and sensitizes cancer cells to ionizing radiation by promoting apoptosis and inhibition of proliferation
miR-185 is inferred to be connected with cancers because of its localization in frequently altered chromosomal regions34 and its suppressive effect on tumor growth,23, 24 metastasis,35 DNA methylation36 and invasion.25 We found that the level of miR-185 was downregulated in response to ionizing radiation, suggesting that its ectopic expression might be connected with cell survival or radioresistance. Our results show that introduction of exogenous pre-miR-185 into RCC cells reduce cell survival by negatively regulating ATR. But what might be the underlying mechanisms for the radiation-initiated downregulation of miR-185 itself? It has been reported that miRNA expression can be regulated by Dicer, a cytoplasmic RNase III, which produces mature miRNAs from their precursors.37, 38 Dicer is downregulated when cells are subjected to extrinsic stresses.39, 40 Indeed, our preliminary data indicated that miR-3928 expression increased in 786-O cells after exposure to X-rays and verified that miR-3928 targeted Dicer. Dicer expression was suppressed in cells overexpressing miR-3928 and the maturation of some miRNAs including miR-185, miR-300 and miR-663 was inhibited.41 Downregulation of miR-185 may be due to the suppression of Dicer induced by ionizing radiation. In the case of pre-miR-185 transfection, miR-185 was already matured and overexpressed before irradiation. Thus, the suppression of Dicer induced by ionizing radiation could not apparently impact the miR-185 levels at this stage. On the other hand, an oscillation of Dicer expression was also observed in irradiated cells. The mechanisms involved in maintaining the proper balance between miRNAs biogenesis and Dicer expression need to be further addressed in the future.
Taken together, our results provide strong evidence that miR-185 has an important role in regulating cellular processes by repressing the ATR pathway, promoting radiation-induced apoptosis and inhibition of proliferation. Elucidation of this mechanism offers opportunities for application of miR-185 in therapeutic intervention and radiosensitization.
Materials and Methods
Clinical tissues, cell culture and irradiation
Clinical tissue samples were collected at Institute of Urology at the Second Hospital of Lanzhou University and the First Hospital of Lanzhou City, with informed consent in accordance with the relevant ethical standards and approval by the internal review board. The human RCC cell line 786-O was purchased from Shanghai Institutes for Biological Sciences (Shanghai, China), and cultured at 37 °C with 5% CO2 in RPMI-1640 medium (GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA). Irradiation was carried out with a Faxitron RX-650 (Faxitron Bioptics, Lincolnshire, IL, USA) producing 100 kVp X-rays, or with a regular UV light monitored by a UV radiometer (UV-B, Beijing Normal University, Beijing, China) on a clean bench. Carbon ion irradiation was performed at the HIRFL (Heavy Ion Research Facility of Lanzhou, Institute of Modern Physics, Lanzhou, China).
miRNA microarray hybridization and analysis
Fresh tissues were irradiated with various doses of X-rays (2/4/8 Gy) immediately after the surgical operation. Total RNAs were isolated using TRIzol Reagent (Invitrogen, Eugene, OR, USA). Then, the isolated miRNAs were labeled with Hy3 using the miRCURY array labeling kit (Exiqon, Vedbaek, Denmark) and hybridized to a miRCURY LNA microRNA array (version 9.2, Exiqon). Microarray images were acquired using a Genepix 4000B scanner (Axon Instruments, Union City, CA, USA) and analyzed with Genepix Pro 6.0 software (Axon Instruments).
RT-PCR and qRT-PCR
Total RNAs were extracted from the clinical tissues and cultured cells using TRIzol Reagent (Invitrogen). For miR-185 detection, reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA); qRT-PCR was performed using TaqMan Universal PCR Master Mix, and U6 was analyzed as an internal control. For mRNA expression, reverse transcription was performed using a Q-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA). The qRT-PCR reaction was performed using SYBR Green PCR master mix (GeneCopoeia). The primer sequences for ATR were, forward: 5′-TGCAGTAATGTCAATGGTTGG-3′ and reverse: 5′-CTGGAACTTCAAAGGTTTCTCC-3′. GAPDH was used as an internal control. The primer sequences were, forward: 5′-GTGGACCTGACCTGCCGTCT-3′ and reverse: 5′-GGAGGAGTGGGTGTCGCTGT-3′. The primer of ATM was purchased from GeneCopoeia (catalog #: HQP011736). Samples were run on a Bio-Rad Chromo4 System Real-Time PCR Detector (Bio-Rad Laboratories, Hercules, CA, USA). The relative fold change of miRNA or mRNA was determined using the equation 2-▵▵Ct, in which ▵▵Ct=(CtmiRNA − CtU6) sample − (CtmiRNA − CtU6) control or ▵▵Ct=(CtmRNA − CtGAPDH) sample − (CtmRNA - CtGAPDH) control.
Transfection
miRNA duplexes including miR-185 precursor (pre-miR-185) and its negative control (pre-neg), miR-185 antisense oligonucleotide (anti-miR-185) and its negative control (anti-neg) were purchased from Ambion (Foster City, CA, USA). Transfection of the miRNA duplexes was performed with 40–60% confluent cells using Lipofectamine 2000 (Invitrogen). The medium was replaced with new culture medium 5 h after transfection.
Colony formation assay
The clonogenic survival assay was conducted as described previously.42 In brief, cells were harvested by trypsinization and resuspended in RPMI-1640 medium supplemented with 10% fetal bovine serum. An appropriate number of cells were plated into each 60 mm dish to produce 20–100 colonies. After incubating for 10 days, cells were fixed with 70% ethanol for 5 min and then stained with 0.5% crystal violet for 3 min. Colonies containing >50 cells were counted as survivors. A total of 3–6 parallel dishes were scored for each treatment.
Luciferase reporter assay
ATR-3′-UTR was amplified by PCR from cDNA of 786-O cells. ATR-3′-UTR-mut was synthesized by GenScript Incorporation (Nanjing, China). These segments were inserted into the pMIR-REPORT miRNA Expression Reporter (Ambion). In all, 1 × 105 786-O cells in 12-well plates were co-transfected with 300 ng constructed pMIR-REPORT and pre-miR-185 or pre-neg (30 nM final), along with 300 ng pRL Renilla Luciferase Control Reporter Vector (Promega, Madison, WI, USA). Assays were performed 24 h after transfection using the Dural Luciferase Reporter Assay Kit (Promega). Renilla luciferase activity was used to normalize the firefly luciferase activity. The primers for the 3′-UTR of human ATR gene used for PCR were 5′-TGTTCTTGACATTGAGCAG-3′ and 5′-GAAAGCAGTTTATTTGTT-3′. Three independent experiments were performed in triplicate.
Western blots
Cells were lysed in RIPA buffer (Beyotime, Nanjing, China). Proteins were separated by 10% SDS-PAGE and transferred to a methanol-activated PVDF membrane (GE Healthcare, Piscataway, NJ, USA). The membrane was blocked for 1 h in PBST containing 5% milk and subsequently probed with anti-ATR antibody (Santa Cruz Biotechnologies, Santa Cruz, CA, USA), anti-Chk1 antibody(Santa Cruz Biotechnologies), anti-pChk1 antibody (Cell Signaling Technology, Danvers, MA, USA), anti-ATM antibody (Abcam, Cambridge, UK) and anti-β-actin antibody (Santa Cruz Biotechnologies) for 2 h. After 1-h incubation with goat-anti-mouse HRP-conjugated secondary antibody (Santa Cruz Biotechnologies), the protein bands were detected with luminal reagent (GE Healthcare) and their relative intensities were quantified using Adobe Photoshop software (Adobe Systems Incorporated, San Jose, CA, USA).
Flow cytometry assay
Cells were fixed in −20 °C pre-chilled 70% ethanol overnight, and then stained with propidium iodide.42 A FACScalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) and Modfit software (Version 3.0, Verity Software House, Topsham, ME, USA) were used to analyze the level of apoptosis in each sample.
EdU incorporation assay
The EdU incorporation assay was carried out using a Cell-Light EdU In Vitro Imaging Kit (RiboBio, Guangzhou, China) according to the manufacturer's instructions. In brief, cells were cultured for 2 h in EdU medium after X-ray irradiation, then fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were stained with Apollo mixture for 30 min after treatment with 0.5% Triton X-100 for 10 min.
Cell growth curves
In all, 1 × 105 786-O cells were plated into 12 well-plates for 24 h. After pre-miR-185 or pre-neg transfection, the cell number was counted at 0, 24, 48 and 72 h after 4 Gy X-ray irradiation using a Coulter Counter (Beckman, Brea, CA, USA).
In vivo tumorigenesis assay
NOD/SCID mice were purchased from Institute of Laboratory Animal Sciences, CAMS and PUMC (Beijing, China). Immediately after transfection with pre-miR-185 or pre-neg, 1 × 105 786-O cells were injected subcutaneously into the hind legs of 8-week-old mice (n=9). Tumors formed by the RCC cells were subsequently exposed to 4 Gy of X-rays. The mice were killed on the 55th day after injection for evaluating tumor formation. This research was conducted in accordance with the Guide for Care and Use of Laboratory Animals as adopted and promulgated by the United National Institutes of Health.
Statistical analysis
Results are expressed as mean±S.D. The statistical significance of the results was determined by Student's t-tests using Microsoft Excel software (Microsoft Campus, Redmond, WA, USA).
Acknowledgments
This work was supported by grants from the Major State Basic Research Development Program of China (973 Program, no. 2010CB834201) and the National Natural Science Foundation of China (no. 10979062, 31270895 and U1232125).
Glossary
- ATM
ataxia telangiectasia mutated
- ATR
ATM- and Rad3-related
- Chk
checkpoint kinase
- RCC
renal cell carcinoma
- 3′-UTR
3′-untranslated region
- UVB
ultraviolet B
- PCR
polymerase chain reaction
The authors declare no conflict of interest.
Footnotes
Author contributions
JW designed most of the experiments and co-wrote the paper. JH performed qRT-PCR, western blot, EdU incorporation and colony formation assay. FS conducted the animal and clinic tissue experiments and apoptosis assay. ND collaborated in the miRNA profiling and experimental design. WH performed the luciferase report assay. WW collected clinic tissue and helped with qRT-PCR. GZ conceived and supervised the work and co-wrote the paper.
Edited by E Candi
References
- Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme. Curr Opin Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rew Mol Cell Bio. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Saito T. Radiation-induced response of microRNA expression in murine embryonic stem cells. Med Chem. 2006;2:555–563. doi: 10.2174/1573406410602060555. [DOI] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wu H, Wang E. Changes in microRNA expression patterns in human fibroblasts after low-LET radiation. J Cell Biochem. 2008;105:824–834. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, et al. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35:81–86. [PubMed] [Google Scholar]
- Ilnytskyy Y, Zemp FJ, Koturbash I, Kovalchuk O. Altered microRNA expression patterns in irradiated hematopoietic tissues suggest a sex-specific protective mechanism. Biochem Biophys Res Co. 2008;377:41–45. doi: 10.1016/j.bbrc.2008.09.080. [DOI] [PubMed] [Google Scholar]
- Lal A, Pan Y, Navarro F, Dykxhoorn MD, Moreau L, Meire E, et al. miR-24-mediated downregulation of H2 AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Du L, Nagabayashi G, Seeger R, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci USA. 2010;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, Chen Y, et al. Find all citations by this author (default)Or filter your current search Find all citations by this author (default)Or filter your current search Find all citations by this author (defaulOfilter your current search. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers Oncogene 2010294971–4979. [DOI] [PubMed] [Google Scholar]
- Liu M, Lang N, Chen X, Tang Q, Liu S, Huang J, et al. miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett. 2011;301:151–160. doi: 10.1016/j.canlet.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Tang H, Wang Z, Liu X, Liu Q, Xu G, Li G, et al. LRRC4 inhibits glioma cell growth and invasion through a miR-185-dependent pathway. Curr Cancer Drug Targ. 2012;12:1032–1042. doi: 10.2174/156800912803251180. [DOI] [PubMed] [Google Scholar]
- Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan TP, Kawasumi M, Blasina A, Anderes K, Conney AH, Nghiem P. ATR-Chk1 pathway inhibition promotes apoptosis after UV treatment in primary human keratinocytes: potential basis for the UV protective effects of caffeine. J Invest Dermatol. 2009;129:1805–1815. doi: 10.1038/jid.2008.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees CJ, Tejera AM, Martinez P, Murga M, Mulero F, Fernandez-Capetillo O, et al. ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J Cell Biol. 2010;188:639–652. doi: 10.1083/jcb.200908136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akcakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, et al. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39:311–318. doi: 10.3892/ijo.2011.1043. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tang H, Wang Z, Zhang B, Liu W, Lu H, et al. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol Cancer. 2011;10:124. doi: 10.1186/1476-4598-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane DR, Cittelly DM, Howe EN, Spoelstra NS, McKinsey EL, LaPara K, et al. MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Horm Cancer. 2010;1:306–319. doi: 10.1007/s12672-010-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligori M, Izzotti A, Pulliero A, Arrigo P. Mutagens interfere with microRNA maturation by inhibiting DICER. An in silico biology analysis. Mutat Res. 2011;717:116–128. doi: 10.1016/j.mrfmmm.2011.07.020. [DOI] [PubMed] [Google Scholar]
- Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada S, Takahashi T, Isodono K, Adachi A, Imoto H, Ogata T. Downregulation of Dicer expression by serum withdrawal sensitizes human endothelial cells to apoptosis. Am J Physiol Heart Circ Physiol. 2008;295:H2512–H2521. doi: 10.1152/ajpheart.00233.2008. [DOI] [PubMed] [Google Scholar]
- Chang L, Hu W, Ye C, Yao B, Song L, Wu X, et al. miR-3928 activates ATR pathway by targeting Dicer. RNA Biol. 2012;9:1247–1254. doi: 10.4161/rna.21821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DM, Rogers RA, Sepulveda R, Donaldson K, Schuler D, Gaering S, et al. Quantification of the pathological response and fate in the lung and pleura of chrysotile in combination with fine particles compared to amosite-asbestos following short-term inhalation exposure. Inhal Toxicol. 2011;23:372–391. doi: 10.3109/08958378.2011.575413. [DOI] [PubMed] [Google Scholar]