Abstract
Epigenetic mechanisms have important roles in carcinogenesis. We certified that the mRNA translation-related gene cytoplasmic polyadenylation element-binding protein 1 (CPEB1) is hypomethylated and overexpressed in glioma cells and tissues. The knockdown of CPEB1 reduced cell senescence by regulating the expression or distribution of p53 in glioma cells. CPEB1 is also regulated directly by the tumor suppressor miR-101, a potential marker of glioma. It is known that the histone methyltransferase enhancer of zeste homolog 2 (EZH2) and embryonic ectoderm development (EED) are direct targets of miR-101. We demonstrated that miR-101 downregulated the expression of CPEB1 through reversing the methylation status of the CPEB1 promoter by regulating the presence on the promoter of the methylation-related histones H3K4me2, H3K27me3, H3K9me3 and H4K20me3. The epigenetic regulation of H3K27me3 on CPEB1 promoter is mediated by EZH2 and EED. EZH2 has a role in the regulation of H3K4me2. Furthermore, the downregulation of CPEB1 induced senescence in a p53-dependent manner.
Keywords: hypomethylation, histone-modifition, miR-101, CPEB1, p53, senescence
Gliomas are the most common type of central nervous system tumors. Patients with high-grade glioma have a poor prognosis. The median survival period of the patients with malignant gliomas is no more than 15 months after diagnosis.1 Unfortunately, high-grade glioma tumor cells are highly mobile and infiltrate the surrounding, otherwise healthy, brain tissue;2 this characteristic is one of the primary reasons for the lack of a successful treatment for this type of tumor. There is an urgent need to develop mechanism-based approaches for the management of glioma. In glioma, silencing gene expression by DNA hypermethylation affects the genes involved in the key cellular functions, such as DNA repair and genome integrity (MGMT and MLH1), tumor suppression (RB1, VHL, RASSF1A and BLU), cell cycle (p16INK4a and p15INK4b) and tumor invasion and apoptosis (DAPK, TIMP3, CDH1 and PCDH-gamma-A11).3, 4, 5, 6, 7 The hypomethylated genes DNMT3B and Sox2 are involved in the cell cycle and metastasis, respectively.8, 9 Thus, DNA methylation has a significant role in glioma.
In our previous research study, the DNA methylome in glioma was constructed by high-throughput methylated DNA IP combined with the use of a promoter and CpG islands microarray. We then certified two new hypomethylated genes, F10 and POTEH, and confirmed the prognostic value of these genes in glioma.10, 11 It has been shown that the hypomethylation of single-copy genes has an important role in glioma.
Cytoplasmic polyadenylation element-binding (CPEB) protein is the key factor that controls mRNA translation. CPEB binds the cytoplasmic polyadenylation element in the 3′-UTRs of responsive mRNAs.12 In Xenopus oocytes and early-stage embryos, CPEB protein 1 (CPEB1) is a dual-function protein. In unstimulated conditions, CPEB1 maintains the mRNA in a repressed state. Although in response to developmental cues or stimulation, CPEB1 dissociates from the repression complex and activates translation.13 Recent data have demonstrated a correlation between CPEB1 and senescence.14 Sasayama T et al15 examined CPEB1 mRNA levels in several tumor cell lines, such as hepatoma, glioma, lung cancer, colon cancer and pancreatic cancer, and demonstrated that 50% of these cell lines had high expression levels of CPEB1.
Cellular senescence is a largely irreversible growth arrest, in which the cells are arrested in the G0/G1 phase of the cell cycle and do not enter the S phase in response to growth factors.16 However, tumor cells can be induced to undergo senescence by treatment with chemotherapeutic drugs, radiation or differentiation agents. The importance of cellular senescence is increasingly being recognized as a mechanism of glioma suppression. Although several genes influence senescence, perhaps the most central genes are the tumor suppressors p53, Rb, p19ARF and p16INK4A.15
In this study, we confirmed the promoter methylation status and the expression of CPEB1 in glioma cell lines and tissues and elucidated the phenotype and mechanism of CPEB1 dysregulation.
Results
CPEB1 was overexpressed because of promoter hypomethylation and correlated with a poor outcome in glioma
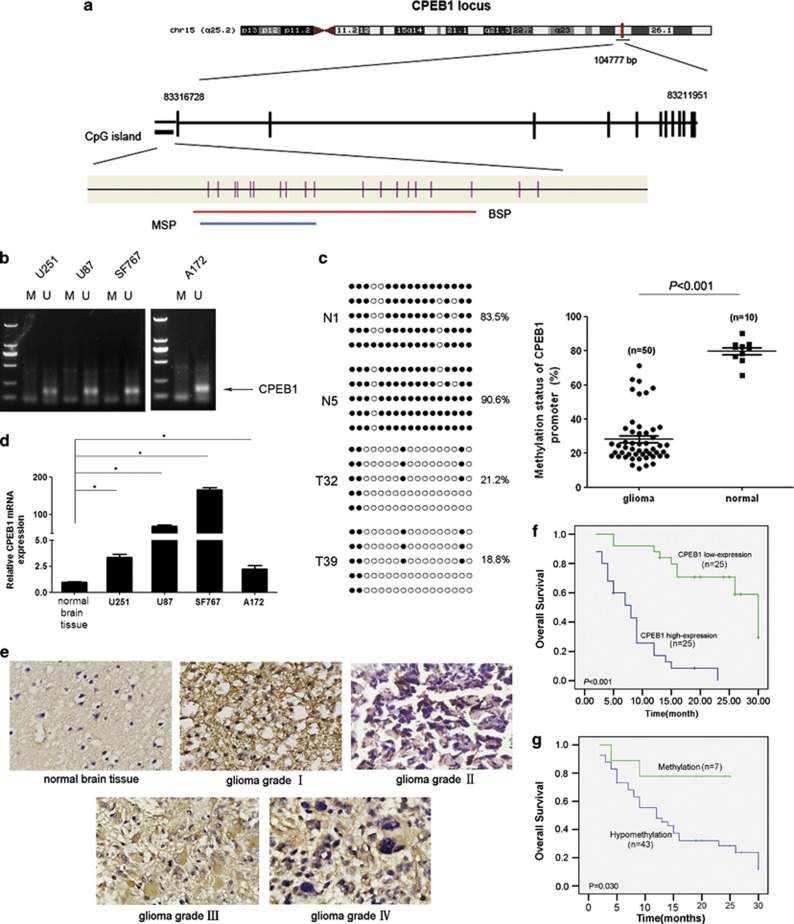
We demonstrated previously that the CPEB1 gene located at 15q25.2 was hypomethylated in glioma tissues.17 To further confirm the hypomethylation of CPEB1 in glioma, we designed and validated bisulfite sequencing PCR (BSP) and methylation-specific PCR (MSP) methods (Figures 1a and b). CpG dinucleotides were heavily methylated in normal brain samples, and the decrease in methylation observed in the tumor cell lines had a mean of 20.1%. In contrast, the decrease in methylation that was observed in the glioma samples had a mean of 71.8% (P<0.001) (Figure 1c). A methylation decrease level of >45.3% was regarded as hypomethylation. CPEB1 was hypomethylated in 43 (86.0%) of 50 tumors. There was no statistically significant correlation between sex, age or histological grade and CPEB1 hypomethylation (Table 1). The increased expression of the CPEB1 protein was found in the glioma cell lines and in 25 of 50 glioma tissues (Figures 1d and e). The expression of CPEB1 in the normal brain tissues was found to be lower compared with the glioma cell lines. There was no statistical correlation between sex, age or histological grade and CPEB1 expression (Table 2). A comparison of the methylation status with protein expression revealed that all of the 25 tumors that had a high level of CPEB1 expression harbored hypomethylation. CPEB1 methylation was observed in 18 of 25 (72.0%) samples with a reduced or total loss of CPEB1 expression (Table 1). There was a significant relationship between CPEB1 promoter hypomethylation and CPEB1 protein overexpression (P=0.001, Table 2). The correlation between CPEB1 expression, methylation status and overall survival (OS) was statistically significant (Figures 1f and g). These results suggest that CPEB1 overexpression and hypomethylation may be involved in glioma carcinogenesis.
Figure 1.
CPEB1 is overexpressed due to promoter hypomethylation and correlates with a poor outcome in glioma. (a) Schematic diagram of CpG dinucleotides within the CPEB1 promoter. The nucleotide number is relative to the transcription start site of CPEB1. The red line indicates the region tested with BSP; the blue line indicates the region detected with MSP. (b) The methylation status of CPEB1 in glioma cell lines was detected with MSP. U, unmethylated primer, M, methylated primer. (c) BSP of the upstream regulatory region of CPEB1 was performed for representative tissues (N, normal brain tissue, T, glioma sample). For each sample, at least five separate clones were sequenced, and the results are shown here. Unmethylated CpG sites are shown as open circles, whereas methylated CpG sites are indicated by closed circles. For each row of circles, the sequence results for an individual clone of the bisulfite-PCR product are given. The number of methylated CpGs divided by the total number of true CpGs analyzed is given as a percentage on the right of each BSP result. *P<0.05. (d) Real-time PCR was used to detect the expression of CPEB1. The expression level of CPEB1 in the normal brain tissue was much lower than that in the four glioma cell lines. *P<0.05. (e) The expression of CPEB1 in normal brain tissues and glioma tissues was tested by ISH. (f) The correlation between CPEB1 protein expression in the tumor and the OS of the glioma patients. The patients with a high level of CPEB1 expression had a poor outcome. (g) The correlation between CPEB1 methylation in the tumor tissue and the OS of the glioma patients. The patients with hypomethylation of CPEB1 had a shorter OS than those with normal levels of CPEB1 methylation
Table 1. Relationship of CPEB1 expression to clinical parameters in glioma patients.
| Variable | CPEB1 |
P | |
|---|---|---|---|
| <8 | ≥8 | ||
| Total (N=50) | 25 | 25 | |
| Sex | 1.000 | ||
| Male (34) | 17 | 17 | |
| Female (16) | 8 | 8 | |
| Age(years)a | 0.145 | ||
| <42 (23) | 14 | 9 | |
| ≥42 (27) | 11 | 16 | |
| Grade | 0.868 | ||
| Low grade (i+ii) (26) | 14 | 12 | |
| High grade (iii+iv) (24) | 11 | 13 | |
Abbreviation: CPEB1, cytoplasmic polyadenylation element-binding protein 1
Table 2. Correlation between CPEB1 methylation status, protein expression and clinical parameters in glioma patients.
| Variable | CPEB1 |
P | |
|---|---|---|---|
| hypomethylation | methylation | ||
| Total (N=50) | 43 | 7 | |
| Expression | 0.004 | ||
| <8 (25) | 18 | 7 | |
| ≥8 (25) | 25 | 0 | |
| Sex | 1.000 | ||
| Male (34) | 29 | 5 | |
| Female (16) | 14 | 2 | |
| Age(years)a | 0.145 | ||
| <42 (23) | 18 | 5 | |
| ≥42 (27) | 25 | 2 | |
| Grade | 0.697 | ||
| Low grade (i+ii) (26) | 23 | 3 | |
| High grade (iii+iv) (24) | 20 | 4 | |
Abbreviation: CPEB1, cytoplasmic polyadenylation element-binding protein 1
The knockdown of CPEB1 reduced senescence by regulating p53 distribution in glioma cells
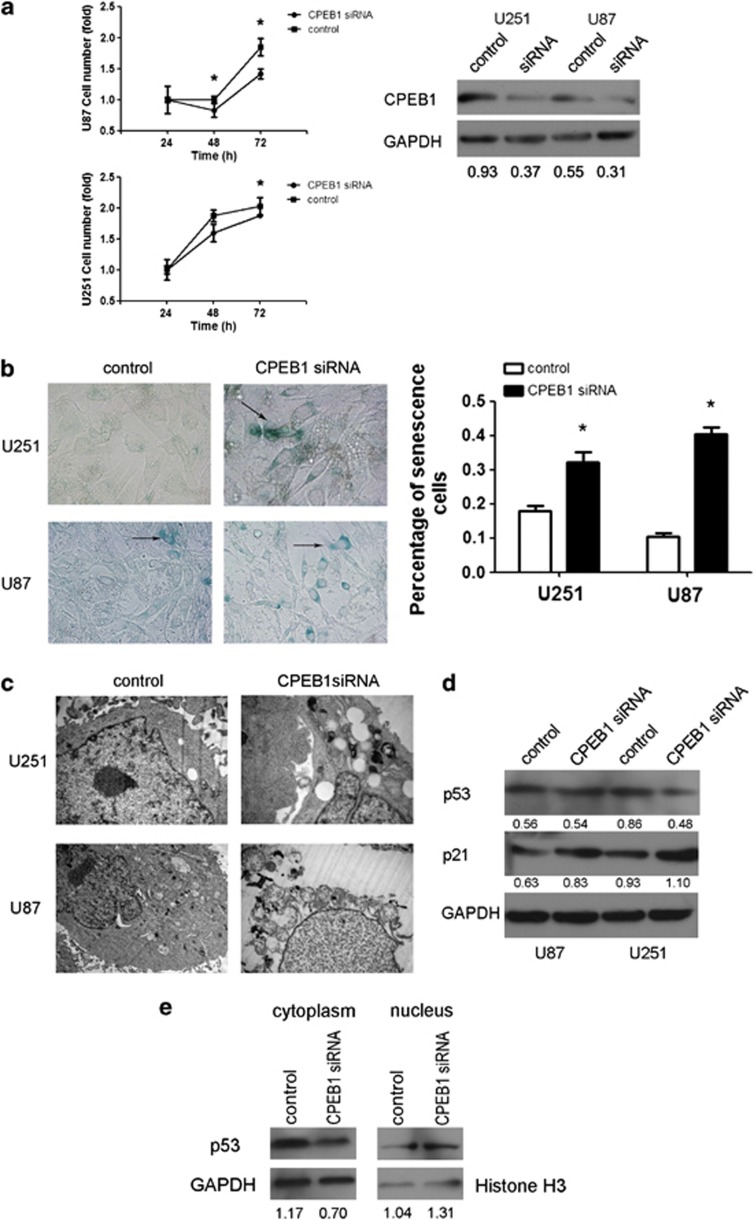
Burns DM et al.18 demonstrated that CPEB1 was repressed by miR-122, and the inhibition of CPEB1 activated p53 translation in human skin fibroblasts. p53 is one of the hallmarks of cellular senescence;19 therefore, the p53-mutated cell line U251 and the p53 wild-type cell line U87 were selected for use in the following experiments. U251 and U87 cell growth was inhibited by CPEB1 knockdown. The inhibition was observed at 72 h in U251 cells and at 48 and 72 h in U87 cells (Figure 2a). Senescent cells can be detected with an assay that detects senescence-associated β-gal (SA-β-gal) activity.20 Whether in p53-mutated or in wild-type glioma cells, CPEB1 knockdown initiated senescence (Figure 2b). Next, the morphology of U251 and U87 cells treated with a CPEB1 small interfering RNA (siRNA) was observed with transmission electron microscopy. The results revealed vacuolar degeneration in the cytoplasm of the U251 cells. However, there were two different lesions in the U87 cells: one was cellular edema, including cytoplasm, nucleus and mitochondria; another lesion was mitochondrial vacuolar degeneration (Figure 2c). After the knockdown of CPEB1, the level of expression of p53 was decreased in the U251 cells but not changed in the U87 cells (Figure 2d). Furthermore, it was discovered that the knockdown of CPEB1 induced the overexpression of p21, which is downstream of p53 (Figure 2d). Even though the CPEB1 knockdown did not influence the expression of p53 in the U87 cell lysate, the loss of CPEB1 expression still enhanced the expression of the downstream p53 effector p21. The data indicate that the knockdown of CPEB1 impeded wild-type p53 transport to the cytoplasm in U87 cells (Figure 2e). This observation supports the hypothesis that the knockdown of CPEB1 induces cell senescence in glioma in a p53-dependent manner.
Figure 2.
CPEB1 reduces senescence by regulating p53 distribution in glioma cells. (a) A cell growth assay was used to analyze the function of CPEB1 siRNA in glioma cell proliferation. *P<0.05. (b) The effect of CPEB1 siRNA on senescence in glioma cell lines as determined with SA-β-gal staining. Arrows indicate the green senescent cells. *P<0.05. (c) Transmission electron micrographs of the morphology of glioma cells transfected with CPEB1 siRNA. (d) The regulation of p53 and p21 by CPEB1 siRNA. (e) The changes in the cellular distribution of p53 induced by CPEB1 siRNA in the U87 cell line
CPEB1 was a direct and epigenetic target of miR-101
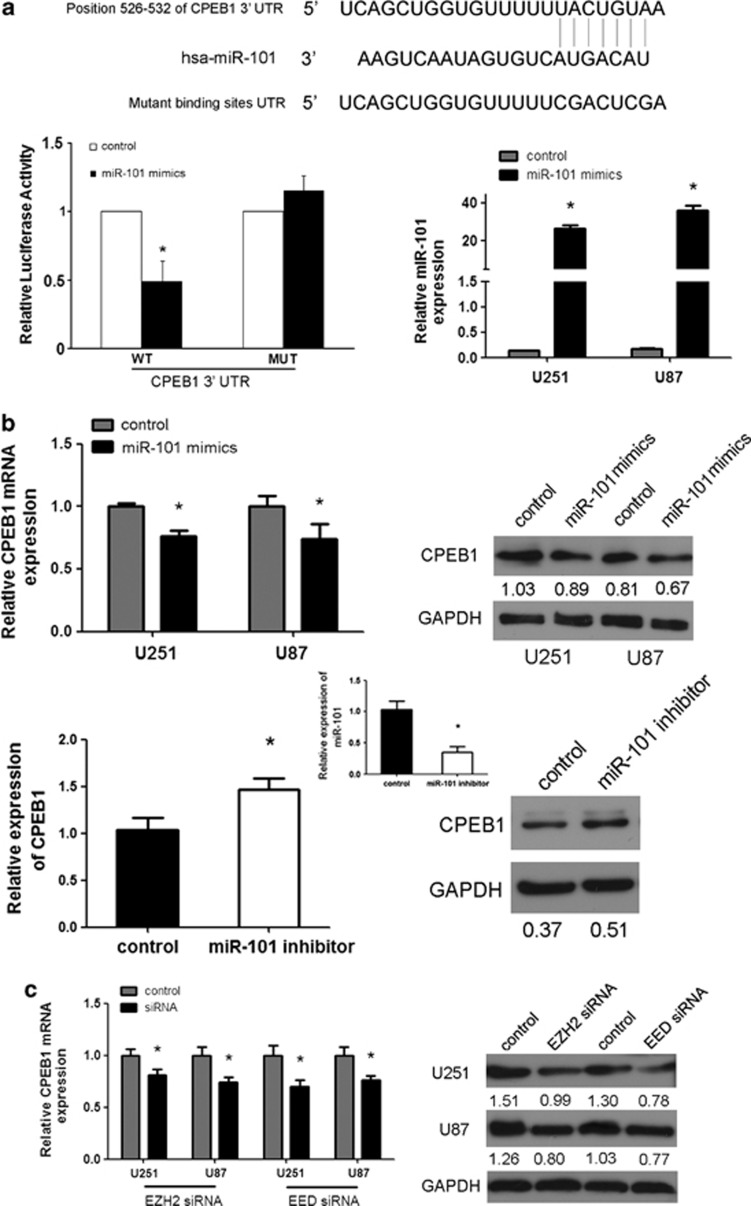
Having established a role for CPEB1 in glioma cell senescence, we next aimed to clarify the mechanism of CPEB1 regulation. To date, there has been only one published study that demonstrated the repression of CPEB1 by miR-122.18 We used the online software TargetScan 5.1 (Cambridge, MA, USA) to predict potential miRNA-binding sites in the 3′-UTR sequence of CPEB1. CPEB1 was predicted to be a target of miR-101. Recent data have demonstrated the suppressor role of miR-101 in tumors.21 The predicted binding sites were cloned downstream of the firefly luciferase gene in the pMIR-REPORT vector (Figure 3a). When the cells were individually cotransfected with miR-101 and pMIR-CPEB1-3′-UTR-WT, there was a significant reduction in luciferase activity compared with cells transfected with the negative control (Figure 3a), thereby confirming that miR-101 directly interacted with the 3′-UTR of CPEB1 in glioma cells. It was gratifying to observe that miR-101 could inhibit the expression of CPEB1, and that the suppression of miR-101 could enhance the expression of CPEB1 (Figure 3b). CPEB1 can be considered to be a new target of miR-101 in glioma.
Figure 3.
CPEB1 is a direct and epigenetic target of miR-101. (a) miR-101 binds to the 3′-UTR of CPEB1 as shown by the predicted binding between miR-101 and the 7mer seed matches in the CPEB1 3′-UTR. miR-101 regulates the expression of the CPEB1 3′-UTR reporter constructs. The luciferase reporter assays were performed 48 h after transfection with the indicated pMIR-REPORT plasmids and a renilla transfection control plasmid that were cotransfected with miR-101 or a relevant scrambled control. The data shown are the mean±S.D. of six replicates and are representative of three-independent experiments. *P<0.05. (b) miR-101 regulates CPEB1 expression. A real-time PCR analysis was performed 48 h after transfection with miR-101 mimics, an inhibitor of miR-101 or a scrambled control. A western blot analysis was performed 72 h after transfection with miR-101 mimics, an inhibitor or a scrambled control. The gain of miR-101 function assay was performed in U251 and U87 cells. The loss of miR-101 function assay was performed in U251 cells. *P<0.05. (c) The expression of CPEB1 is regulated by EZH2 siRNA and EED siRNA. A real-time PCR analysis was performed 48 h after transfection with EZH2 siRNA, EED siRNA or a scrambled control. A western blot analysis was performed 72 h after transfection with EZH2 siRNA, EED siRNA or a scrambled control. *P<0.05. (d) A luciferase reporter assay defines the position of the CPEB1 core promoter. 0 is the TSS. The core promoter ranged from −307 to −207. pGL3-control is the positive control, pGL3-enhancer is the negative control. The relative activity as compared with the pRL-TK plasmid is shown. Where the relative luciferase activity is higher than the pGL3-enhancer is considered to embody the core promoter region. *P<0.05. (e) A ChIP assay was performed to detect the H3K4me2, H3K27me3, H3K9me3 and H4K20me3 occupancy of the CPEB1 core promoter. U251 and U87 cells transfected with miR-101 mimics, EZH2 siRNA and EED siRNA were analyzed. *P<0.05. (f) The methylation status of CPEB1 is affected by miR-101. The unmethylated CpG sites are shown as open circles, whereas the methylated CpG sites are indicated by closed circles. For each row of circles, the sequence results for an individual clone of the bisulfite-PCR product are given. The number of methylated CpGs divided by the total number of true CpGs analyzed is given as a percentage on the right of each BSP result. The methylation level of CPEB1 is increased by miR-101 in the U251 and U87 cells as determined with BSP. MiR-101 mimics reverse the methylation status of CPEB1 as determined with MSP in U251 cells
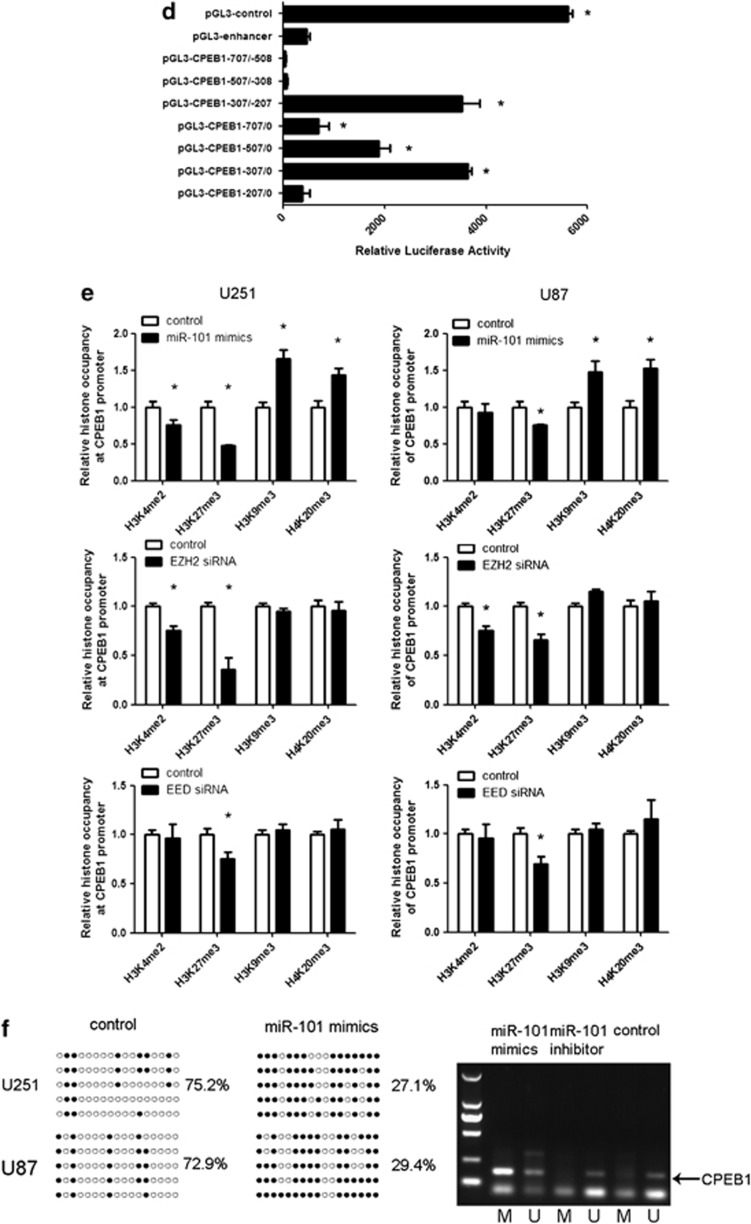
Enhancer of zeste homolog 2 (EZH2) and embryonic ectoderm development (EED) are the core subunits of polycomb repressive complex 2 (PRC2) and also the targets of miR-101.22, 23 The polycomb group proteins form macromolecular complexes that function as epigenetic gene silencers.24 PRC2 has a signature activity to methylate histone H3 at lysine 27 (H3K27), adding up to three methyl groups to this target Lys residue. Trimethylated H3K27 (H3K27me3) is thought to be the main form of H3K27 that mediated PRC2 silencing. DNA methylation is correlated with histone modification. We hypothesized that the EZH2 and EED, which are the targets of miR-101, regulate the promoter methylation status of CPEB1 by modification of H3K27me3. Then, we determined that the expression of CPEB1 was downregulated by EZH2 siRNA and EED siRNA in the glioma cell lines U251 and U87 (Figure 3c). As EZH2 and EED are gene silencers, their siRNA was thought to upregulate the expression of CPEB1. However, the result was in contrast to what was expected. In order to verify the mechanism how EZH2 and EED regulate the expression of CPEB1, we determined the core promoter region of CPEB1. The luciferase reporter assay demonstrated that the core promoter ranged from −307 to −207(Figure 3d), and a possible repressive element resides between positions −707 and −508. In addition, we determined the presence of H3K27me3, a marker for DNA methylation, at the core CPEB1 promoter locus upon the treatment of U251 cells with miR-101. As shown in Figure 3e, we found H3K27me3 occupancy was decreased at the CPEB1 core promoter in miR-101-treated cells compared with the control. EZH2 is a histone methyltransferase that predominantly methylates histone H3 lysine 27. Consistently, we observed that both in U251 and U87 cells H3K27me3 was downregulated in CPEB1 promoter when the expression of EZH2 and EED was knocked down. If miR-101 targeted EZH2 for degradation and EZH2/EED is important for CPEB1 expression, one would expect H3K27me3 to be upregulated in the cells expressing miR-101 mimics. Instead, our data showed a H3K27me3 downregulation when miR-101 mimics and EZH2/EED siRNA were expressed. These results were not in accordance with what we supposed previously. Therefore, we considered that there may exist other mechanisms in the epigenetical regulation of miR-101 on CPEB1.
In general, permissive regions of DNA exhibit an open chromatin structure marked by the hyperacetylation of histones H3 and H4 and the dimethylation and trimethylation of histone H3 at lysine 4 (H3K4me2/3). In contrast, repressed regions exhibit a compact chromatin structure that is enriched in ‘repressive' modifications: dimethylation and trimethylation of H3K9 (H3K9me2/3) and H3K27me3 and the trimethylation of H4K20 (H4K20me3).25, 26 Although the code has not yet been fully deciphered, it is apparent that DNA methylation can both influence and be influenced by histone modifications. There are also three other markers correlated with the gene methylation status, the activated gene expression marker H3K4 and the repressed gene expression markers H3K9 and H4K20.27, 28, 29 The occupancy of the CPEB1 core promoter by H3K9me3 and H4K20me3 was upregulated by miR-101 compared with the control. H3K4me2 occupancy can be used as a tracer to mark a locus that has undergone DNA demethylation. The H3K4me2 occupancy at the core promoter of CPEB1 was reduced by miR-101 only in the U251 cells (Figure 3e).
As EZH2 and EED are also shown to correlate with H3K4, H3K9 and H4K20, we analyzed the effects of EZH2 and EED on histone H3K4, H3K9 and H4K20 modification at the core promoter of CPEB1. The downregulation of EZH2 decreased the occupancy by H3K4me2 at the core promoter of CPEB1 but had no effect on H3K9me3 and H4K20me3 occupancy; the knockdown of EED could not regulate H3K4me2, H3K9me3 and H4K20me3 occupancy at the core promoter of CPEB1. So we postulated that miR-101 reversed the methylation status of CPEB1 by downregulating the H3K4me2 occupancy and upregulating the H3K9me3 and H4K20me3 occupancy at the core promoter of CPEB1 by targeting EZH2. A reduction in the methylation level of CPEB1 was induced by miR-101 mimics in both U251 and U87 cells as determined by BSP. MiR-101 mimics could also reduce the level of methylated CPEB1 in U251 cells as determined by MSP, whereas miR-101 inhibitor could maintain the hypomethylation of CPEB1 in U251 cells (Figure 3f). Taken together, the above results indicated that miR-101 epigenetically downregulated the expression of CPEB1 by inhibiting EZH2.
miR-101 was frequently downregulated in human glioma cell lines and tissues and was associated with poor prognosis
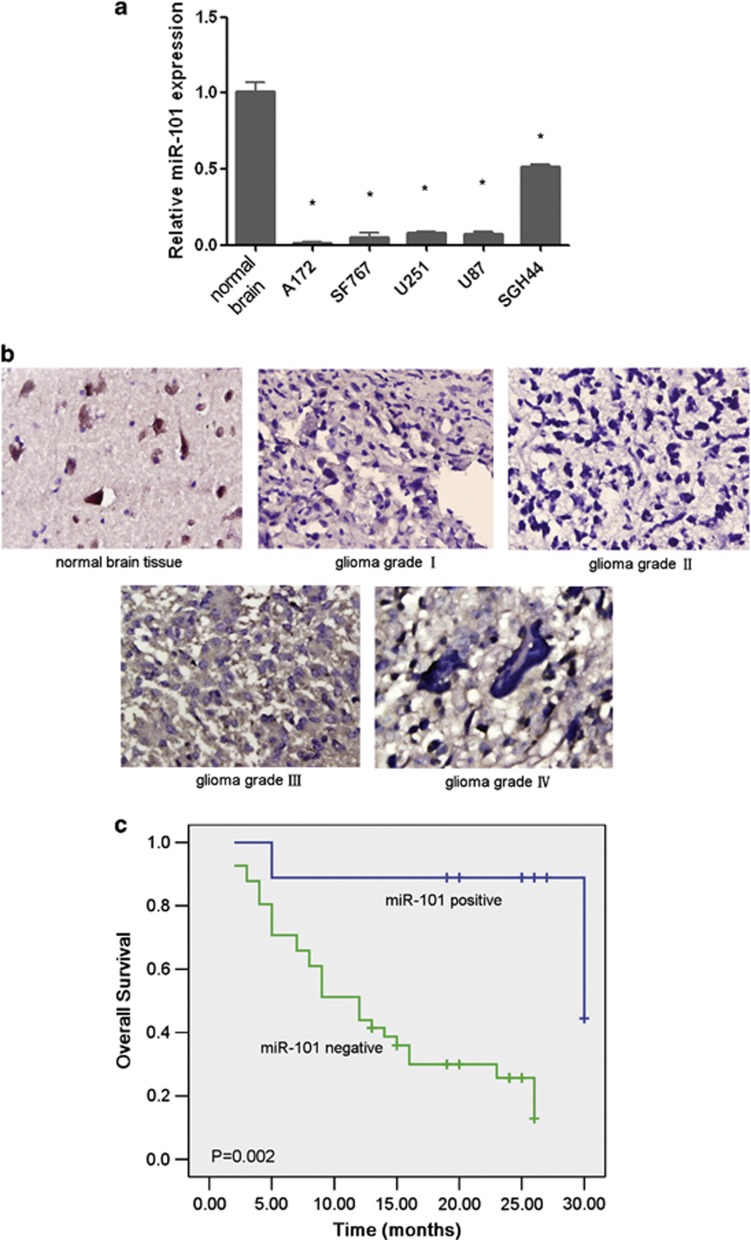
Whether MiR-101 is reported to be downregulated in multiple tumors.21, 30, 31, 32, 33 In the present study, we detected the expression of miR-101 in glioma cell lines and tissues. In-situ hybridization was performed to analyze the expression level of miR-101 in human normal brain tissues and glioma tissues, and real-time PCR was performed for normal brain tissue and human glioma cell lines, including U251, U87, A172, SF767 and SHG44. The mature form of miR-101 was at an obviously lower level in normal brain tissues compared with that in glioma cells and tissues (Figures 4a and b). The expression level of miR-101 was further examined in 50 samples of glioma tissue and 10 normal brain tissue samples. In comparison with the cell lines, miR-101 was downregulated in 82% of the tumor samples: 2 grade I, 18 grade II, 10 grade III and 11 grade IV. The reduction in miR-101 expression was not correlated with tumor grade, sex or age (Figure 4b, Table 3). However, the expression level in grade I was much lower than that found in grades II, III and IV (P<0.05). As can be seen, a markedly reduced OS was observed in the glioma patients who had lost miR-101 expression compared with the patients who exhibited normal expression levels (Figure 4c). These results suggest that reduced miR-101 expression is a frequent event in human glioma tissues and may be involved in glioma carcinogenesis. Furthermore, miR-101 may function as a potential marker for the initiation of a glioma.
Figure 4.
miR-101 is frequently downregulated in human glioma cell lines and tissues and is associated with a poor prognosis. (a) The expression of miR-101 in normal brain tissue and glioma cell lines. The real-time PCR data indicate that miR-101 is downregulated in glioma cell lines. *P<0.05. (b) The expression of miR-101 in glioma tissues and normal brain tissues. The ISH assay indicates that miR-101 is expressed at high levels in normal brain tissue, whereas the expression is lost in glioma tissues. (c) The correlation between miR-101 expression in the tumor and the OS of glioma patients
Table 3. Relationship of miR-101 to clinical parameters in glioma patients.
| Variable | miR-101 |
P | |
|---|---|---|---|
| <8 | ≥8 | ||
| Total (N=50) | 41 | 9 | |
| Sex | 0.942 | ||
| Male (34) | 29 | 5 | |
| Female (26) | 22 | 4 | |
| Age(years)a | 0.525 | ||
| <42 (23) | 18 | 5 | |
| ≥42 (27) | 23 | 4 | |
| Grade | 0.467 | ||
| Low grade (i+ii) (26) | 20 | 6 | |
| High grade (iii+iv) (24) | 21 | 3 | |
Abbreviation: CPEB1, cytoplasmic polyadenylation element-binding protein 1.
Median of age is 42 years
miR-101 had a role as a tumor suppressor and induced senescence in glioma cell lines
Although it has been reported that miR-101 can suppress carcinogenesis in many tumors,34, 35, 36, 37 the role of miR-101 in glioma formation and the precise mechanism by which this miRNA acts have been unclear. The exogenous expression of miR-101 could inhibit the growth of the glioma cells at 48 and/or 72 h (Figure 5a). It was found that miR-101 could abrogate the migration abilities of the glioma cell lines U251 and U87 (Figure 5b). We performed SA-β-gal staining of these cell lines and found that there was an increased number of SA-β-gal-positive cells in the cell lines that were transfected with miR-101 (Figure 5c). In addition, transmission electron microscopy revealed cytoplasmic and nuclear condensation, the aggregation of chromatin at the periphery of the nucleus and mitochondrial vacuolar degeneration in the U251 cells. However, there were two different lesions in the U87 cells: cellular edema, including cytoplasm, nucleus and mitochondria; cellular condensation, including cytoplasmic, nuclear and mitochondrial vacuolar degeneration (Figure 5d).
Figure 5.
miR-101 acts as a tumor suppressor and induces senescence in glioma cell lines. (a) The inhibition of proliferation by miR-101 in glioma cells. (b) A wound-healing assay analyzes the effect of miR-101 on glioma cells. *P<0.05. (c) The effect of miR-101 on senescence in glioma cells was measured with SA-β-gal staining. Arrows point to the green senescent cells. *P<0.05. (d) Transmission electron micrographs of the morphology of the glioma cells transfected with miR-101 mimics. The U251 cells exhibited cytoplasmic and nuclear concentration, chromatin aggregation at the periphery of the nucleus and mitochondrial vacuolar degeneration after transfection with miR-101. Two different lesions were observed in the treated U87 cells: one was cellular edema, including in the cytoplasm, nucleus and mitochondria; the other was cellular condensation, including cytoplasmic, nuclear and mitochondrial vacuolar degeneration
miR-101 induced glioma cell senescence in a p53-dependent manner
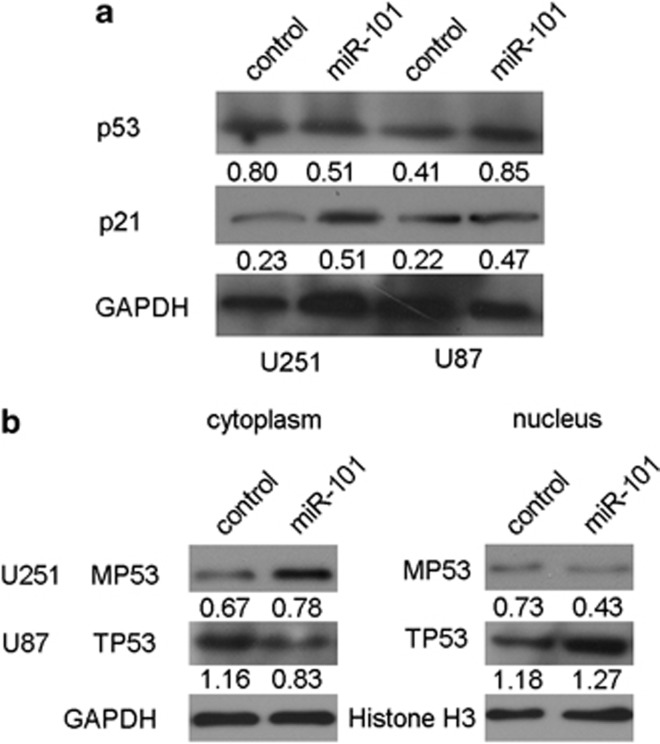
Whether miR-101 regulated CPEB1 and induced glioma cell senescence in a p53-dependent manner. The western blot analysis revealed that the expression of p53 was downregulated in the U251 cells and was upregulated in the U87 cells that had been transfected with miR-101 mimics, but the level of p21 expression was increased in both U251 and U87 cells (Figure 6a). In this experiment, we postulated that miR-101 regulates the distribution of p53 in U87 and U251 cells. As was expected, the western blot analysis indicated that miR-101 facilitated the transport of mutant p53 to the cytoplasm in the U251 cells and retained the wild-type p53 in the nucleus of the U87 cells (Figure 6b). These results indicate that miR-101 not only suppresses the expression of mutant p53 and enhances the expression of wild-type p53 but also regulates the distribution of p53 in glioma cells. The findings are in accordance with the tumor suppressor role of miR-101 in glioma cells.
Figure 6.
miR-101 induces glioma cell senescence in a p53-dependent manner. (a) Western blot analysis of the expression of p53 and p21. (b) Western blot analyzes the distribution changes in U87 cells
Discussion
Hypomethylation is a common mechanism of dysregulation in cancer. We identified that hypomethylation was one of the mechanisms upregulating CPEB1. Using the Kaplan–Meier method, we found that the glioma patients who harbored the overexpression or hypomethylation of CPEB1 had poor outcomes. Therefore, CPEB1 overexpression and hypomethylation can be considered to be potential markers in the prognosis of glioma.
A recent study showed that exogenous CPEB1 induces senescence in CPEB1-knockout MEFs.14 In this study, we certified the positive role of CPEB1 in the proliferation and senescence of glioma cells. p53 is an important gene in cellular senescence, and p53expression is regulated by CPEB1.18 Studies have shown that CPEB1 regulates the distribution of β-catenin in astrocytes.38 In this study, we demonstrated that the knockdown of CPEB1 could also result in the regulation of p53 in glioma cells. These results demonstrate a role for CPEB1 in the p53-mediated senescence of glioma cells.
We proved that CPEB1 was hypomethylated and overexpressed in glioma. However, the mechanism of CPEB1 dysregulation is unclear. miRNAs have important roles in the process of glioma formation.39, 40 With bioinformatics, we predicted candidate miRNAs that may control the expression of CPEB1 and selected the tumor suppressor miR-101 for further validation. The results obtained from the luciferase reporter assay, the real-time PCR and the western blot indicated that CPEB1 was the direct target of miR-101. However, the mechanism of hypomethylation remained unclear.
Most of the polycomb group protein targets are transcription factors and components of signaling pathways.41 The polycomb group protein proteins comprise two groups: PRC1 and PRC2. PRC2 proteins participate in histone modification and have an inhibitory role at the level of transcript initiation.42 PRC2 has four core subunits: EZH1, EZH2, EED and SUZ12. The most remarkable feature is the methylation of H3K27. H3K27me3 is considered to be the main form of H3K27 in the polycomb group protein protein that inhibits gene expression.43 Additional functions of the PRC include DNA methylation, histone methylation and histone acetylation.44 Surprisingly, the histone methyltransferases EZH2 and EED are the targets of miR-101.23, 45 We postulated that the hypomethylation of CPEB1 was due to the modification of histone methylation that was regulated by miR-101. First, we verified that the CPEB1 methylation status was regulated by miR-101, EZH2 and EED. To determine whether miR-101 modulates the histone modification at the CPEB1 promoter through EZH2 and EED, we identified the core promoter of CPEB1 and tested whether the occupancy of the promoter by the DNA methylation-related modified histone H3K27me3 was affected by miR-101, EZH and EED. According to the published references, EZH2 and EED act as expression suppressors, and their knockdown could upregulate the expression of gene. However, in our study, it is in contrast to what was expected. So we supposed that there were other mechanisms of EZH2 and EED in modulating CPEB1. DNA methylation was shown to be closely correlated with histone modification, especially the demethylation of H3K4 and the methylation of H3K9.46 Next, we detected the occupancy by H3K4me2, H3K9me3 and H4K20me3 with the knockdown of EZH2 and EED. The results from a chromatin immunoprecipitation (ChIP) assay suggest that the occupancy by H3K4me2 was decreased with EZH2 siRNA transfection. H3K4me2 occupancy can be used as a tracer to mark a locus that has undergone DNA demethylation. H3K4 demethylation is a gene-silencing marker.47 H3K9me2/3 is a marker of the inhibition of gene expression. In mouse myoblast cells, CDC12, EZH2 and DNMT3A are recruited to the promoter of myogenin, the occupancy by H3K9me2/3 and H3K9Ac at the promoter is increased, and the expression of myogenin is then suppressed.27 In our study, the downregulation of EZH2 decreased the occupancy by H3K4me2 at the core promoter of CPEB1 but had no effect on H3K9me3 and H4K20me3 occupancy; the knockdown of EED could not regulate H3K4me2, H3K9me3 and H4K20me3 occupancy at the core promoter of CPEB1. Further, in 2007, Hoffmann MJ et al.48 found it was interesting that the expression of EZH2 was elevated in prostate cancers exhibiting significantly increased LINE-1 hypomethylation. In normal cells, DNA methyltransferases would be directed by repressor protein complexes to inactive parts of the genome, of which LINE-1 sequences constitute a major fraction. Chromatin disorganization in cancer cells would involve misconduction of repressor complexes and, consequentially, DNA methyltransferases to aberrantly silenced regions of the genome, away from their normal location.48 This might explain why EZH2 has a positive role in controlling the expression of CPEB1. In a brief, we inferred that miR-101 could suppress the expression of CPEB1 by reversing promoter methylation status through decreasing the demethylation-related H3K4me2 and methylation-related H3K9me3 and H4K20me3. Our results indicated that miR-101 may modulate the occupancy by H3K4me2, H3K9me3 and H4K20me3 via the regulation of other histone methyltransferases, such as H3K9-specific SUV39H1/2,49 G9a50 or H4K20-specific SUV4−20H.51 However, we did not conduct further study.
MiR-101 is considered to be a tumor suppressor. In this study, we certified the repressive nature of miR-101 in glioma. The results indicated that miR-101 inhibits proliferation and migration and induces senescence in glioma cells. As CPEB1, a target of miR-101, regulated glioma cell senescence in a p53-dependent manner, we inferred that the miR-101-induced senescence is also dependent on a p53 pathway. The western blot analysis revealed that the level of expression of mutant p53 in U251 cells was decreased and the expression level of wild-type p53 in U87 cells was increased by miR-101, but the expression of p21 was increased in both U251 and U87 cells. In other words, miR-101 facilitated the transport of mutant p53 to the cytoplasm in U251 cells and resulted in the nuclear retention of wild-type p53 in U87 cells.
In summary, the hypomethylated gene CPEB1 can be directly and epigenetically targeted by miR-101, a potential marker for glioma, and epigenetically regulated by the methylation of histones by the miR-101 targets EZH2, EED and DNMT3A. CPEB1 also induces senescence in a p53-dependent manner.
Materials and Methods
Tissue specimens
We obtained frozen tissue samples of 50 gliomas and 10 normal brain tissues from the Xiangya Hospital of the Central South University, Hunan, China, between January 2009 and July 2011. The study was approved by the Ethical Committee of the Faculty of Medicine, the Central South University, and informed consent was obtained from all patients. Tumor samples were diagnosed by two pathologists who were blinded to patient data using the World Health Organization system. Clinical data, including gender, age, initial presentation, postoperative irradiation, chemotherapy, follow-up and outcome, were obtained from the medical records. They included 16 female and 34 male patients who were in the age range from 16 to 65 years, with a mean age of 41 years and a median age of 42 years (Supplementary Table S1).
Cell lines and treatments
The following human glioma cell lines obtained from the Cell Center of Peking Union Medical College (Beijing, China) were used: U251, A172, SHG44 and U87. U251, A172 and U87 cells were maintained in the Dulbecco's Modified Eagle medium (Gibco, Grand Island, NY, USA) and SHG44 cells were maintained in RPMI-1640 (Gibco), with 10% FCS, 100 units/ml penicillin and 100 μg/ml streptomycin, at 37 °C in a humidified atmosphere of 5% CO2 and 95% air.
Isolation of genomic DNA from cell lines and tissues and Bisulfite DNA
Genomic DNA was isolated from cell lines, glioma tissues and normal brain tissues, by the Universal Genomic DNA Extraction Kit Ver.3.0 (Takara, Dalian, China) according to the manufacturer's instructions. The quality and integrity of DNA from tissues and cells were checked by electrophoresis on 1% agarose gel and quantified spectrophotometrically. Genomic DNA (0.5 μg) extracted from the cells, tumor and the normal tissue specimens was subjected to bisulfite treatment using an Epitect Bisulfite Kit (Qiagen, Hilden, Germany) and stored at −20 °C until further use.
Bisulfite sequencing PCR and methylation-specific PCR
BSP and MSP were conducted as described previously (Reed K), commencing with the amplification of the bisulfite-treated CPEB1 promoter containing 17 CpG sites. For PCR, 2.5 U of Taq mix (Takara), 0.5 μl of 1 μM forward (5′-GAGGGGTAGGAGGGTAGAGTTATA-3′) and reverse primers (5′-AACAAAAACAATTACCATACAAACC-3′), was used in a 50 μl of total reaction volume. Here, 100 ng of bisulfite-treated DNA was used as the template of the PCR. The PCR cycles were as follows: at 95 °C for 5 min, followed by 38 cycles at 95 °C for 0.5 min, at 61.7 °C for 2 min and at 72 °C for 2.5 min, followed by a final extension at 72 °C for 10 min. The PCR products were purified by gel extraction from a 1% agarose gel and ligated into the pGEM-T vector (Promega, Madison, WI, USA) in a 3:1 vector/PCR product ratio. The ligation products were used to transform competent Escherichia coli cells (strain JM109) using standard procedures, and blue/white screening was used to select a minimum of five bacterial transformants (clones). The CPEB1 promoter of positive clones was sequenced by the Genscript company (Nanjing, China) and Invitrogen (Guangzhou, China). The methylation decrease for each sample was calculated as the percentage of unmethylated CpG dinucleotides from the total number of CpG dinucleotides analyzed. For MSP, 2.5 U of Taq mix (Takara), 0.5 μl of 1 μM forward and reverse primers (forward methylated primer: 5′-GGAGGGGTAGGAGGGTAGAGTTATAC-3′, reverse methylated primer: 5′-AAAATAATCCCGATACAATACCGTT-3′, forward unmethylated primer: 5′-GGGGTAGGAGGGTAGAGTTATATGA-3′, reverse unmethylated primer: 5′-TAAAATAATCCCAATACAATACCATT-3′), was used in a 25 μl of total reaction volume. Here, 50 ng of bisulfite-treated DNA was used as the template of the PCR. The PCR cycles were as follows: at 95 °C for 5 min, followed by 35 cycles at 95 °C for 0.5 min, at 60 °C for 2 min and at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. PCR products were separated on 1% agarose gels and analyzed by ethidium bromide staining.
MicroRNAs, siRNAs, DNA plasmids and transfection
MiR-101 mimics (5′-UACAGUACUGUGAUAACUGAA-3′) and scramble control (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by GenePharma CO, Ltd (Shanghai, China). The sequence of EZH2 siRNA is: 5′-GCTCTAGACAACAAACCTT-3′ the sequence of EED siRNA is: 5′-GCAGTGTATAAAGCACTAT-3′ the sequence of CPEB1 siRNA is: 5′-GCUCUGCAUGGAAUGCUAATT-3′. To generate a luciferase reporter construct, we synthesized 54 bp of the 3′-UTR of CPEB1 mRNA from human genomic DNA (Invitrogen). This 3′-UTR region of CPEB1 containing the predicted target sites of miR-101 was then subcloned downstream of pMIR-REPORT miRNA expression reporter vector (Ambion, Shanghai, China). We also constructed plasmids with mutated target sites of miR-101. miR-101 mimics, siRNA and DNA plasmids were transfected by LipofectAMINE 2000 (Life Technologies, Gaithersburg, MD, USA).
Cloning of the CPEB1 promoter, plasmid construction and transfection
Different upstream regulatory regions of the CPEB1 gene were amplified from U251 cells DNA by PCR with UltraPF DNA polymerase (GeneCopoeia Inc, Rockville, MD, USA). Sequences of primers used are listed in Supplementary Table S2. PCR fragments were digested with MluI/XhoI and linked to the luciferase-based promoter-less plasmid-pGL3-Enhancer Vector (Promega) to create plasmids pGL3-CPEB1-707/0, pGL3-CPEB1-507/0, pGL3-CPEB1-307/0, pGL3-CPEB1-207/0, pGL3-CPEB1-507/-308 and pGL3-CPEB1-707/-508 (primers were shown in Supplementary Table S2). The sequences and the orientation of the cloned fragments were confirmed by direct DNA sequencing. Plasmids used for transfection were isolated and purified using the PURELINK PLASMID MINI 25 REACTIONS (Invitrogen–Life Technologies, Carlsbad, CA, USA). The promoter activities of these fragments were tested by transient transfection of 1 μg of plasmid DNA into U251 cell lines using LipofectAMINE 2000. For the luciferase-based assay, results are normalized against Renilla luciferase activity. At least three-independent assays were performed.
Luciferase reporter assay
For luciferase reporter assay, U251 cells were plated in a 24-well plate and then cotransfected with 0.5 nmol of either miR-101 mimics or scramble control, 20 ng of either pMIR-CPEB1-3′-UTR-WT or pMIR-CPEB1-3′-UTR-MUT and 2 ng of pRL-TK (Promega). Cells were collected 48 h after transfection and analyzed using the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was detected by M200 microplate fluorescence reader (Tecan, Beijing, China). The pMIR-REPORT-β-gal control vector was cotransfected as an internal control to correct the differences in both transfection and harvest efficiencies. Transfection experiments were performed in duplicates and repeated at least thrice in independent experiments.
RNA extraction, reverse transcription and real-time quantification PCR
Total RNA was extracted using Trizol (Life Technologies, Rockville, MD, USA) from glioma cells. For real-time, PCR 2 μg of total RNA was reverse-transcribed using the cDNA synthesis KIT (Fermentas, Burlington, ON, Canada). Amplification of β-actin was used as a control for this reaction. Primers sequence of CPEB1 was: 5′-TGGGTATTAGCCGACAGT-3′ (forward) and 5′-CTCCGTTGGTTATTGAAAGT-3′ (reverse). Data were normalized for β-actin levels by the 2−ΔΔCt method and the level of CPEB1 mRNA in glioma cell lines by the 2−ΔCt method. The level of miR-101 was performed using a SYBR-green-containing PCR kit (GenePharma CO), and RNA input was normalized to the level of human U6 snRNA.
In-situ hybridization (ISH)
We performed nonradioactive ISH on paraffin tissue sections with a double DIG-labeled probe according to the manufacturer's instructions (Exiqon, Copenhagen, Denmark). In brief, the sections were deparaffinized in xylene and then gradually rehydrated in decreasing concentrations of ethanol. Sections were treated with proteinase K and then fixed with paraformaldehyde. Prehybridization was then carried out for 3 h in hybridization buffer in a humidified chamber at 42 °C. Slides were incubated in the DIG-labeled probe (containing miR-101 with the following sequence: 5′-TCAGGAACTGCCTTTCTCTCCA-3′) diluted to 25 nM in hybridization buffer overnight at 55 °C. A stringency wash was then performed by rinsing slides in 2 × SCC, and the slides were placed in blocking buffer for 30 min in a humidified chamber at 37 °C. Immunological detection was performed using the DIG Nucleic Acid Detection Kit (Boshide, Wuhan, China) according to the manufacturer's directions. Slides were incubated with blocking buffer in a humidified chamber at room temperature for 2 h. 3,3′-Diaminobenzidine hydrochloride (DAB) staining Kit and hematoxylin were used for staining purpose. The slides were then examined under a transmission light microscope. Staining intensity was scored as 0 (negative), 1+(weak), 2+(medium) or 3+(strong). Low expression was defined as an intensity of 0, 1, 2 or 3 and <10% stained cells or an intensity of 0 or 1 and <50% stained cells. High expression was defined as an intensity of 2 or 3 and >10% stained cells, or an intensity of 1, 2 or 3 and >50% stained cells.
Cell number counting assay
A total of 1 × 105 cells were cultured in each well in a 24-well plate, transfected with miR-101 mimics, CPEB1 siRNA or scramble sequence. Count the cell number at 24, 48 and 72 h after transfection. Compare the growing fold of each group.
Wound-healing assays
Glioma cells were used to study the effects of miR-101 inhibition on long-term migration owing to their higher transfection efficiency. Glioma cells were cultured in a six-well plate. Cells were transfected with Lipofectamine 2000 with 0.1 nm of either miR-101 mimcs or scramble sequence. Cultures were scratched 6 h post transfection and cell migration was assessed 12, 24 and 36 h later. Fields containing scratches were visualized and selected at × 20 magnification under bright-field phase optics to ensure a non-biased sample. For each field, the distance of the wound boarders was compared with the control.
SA-beta-gal staining
At day 4 after transfection with miR-101, CPEB1 siRNA (5′-GCUCUGCAUGGAAUGCUAATT-3′) or scramble control, glioma cells were fixed and stained by using an SA-beta-gal kit (Beyotime, Beijing, China) according to the manufacturer's instructions. The blue senescent cells were distinguished from viable cells under the microscope. Four high-power fields per sample were counted in three-independent samples to quantify the number of senescent cells.
Transmission electron microscopy
U251 and U87 cells were transfected as indicated for 72 h before fixation. Samples were fixed in 1% w/v OsO4 in 0.12 M sodium cacodylate buffer (pH 7.2) for 24 h. The specimens were dehydrated in a graded series of ethanol, transferred to propylene oxide and embedded in Epon according to standard procedures. Sections, B 80 nm thick, were cut with a Reichert-Jung Ultracut E microtome and collected on copper grids with Formvar-supporting membranes. Sections were stained with uranyl acetate and lead citrate. Imaging was carried out on a HITACHI-7500 (HITACHI, Tokyo, Japan) transmission electron microscope (magnifications: × 20 000).
Western blot
Cell protein lysate, cytosol protein or nuclear protein was separated in 10% SDS-polyacrylamide gels, electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Danvers, MA, USA), then detected with goat polyclonal antibody for CPEB1 (Santa Cruz Biotechnology, Santa Cruz County, CA, USA), mouse monoclonal antibody for p53 (Santa Cruz Biotechnology) and GAPDH (Millipore) and the commercial ECL kit (Pierce, Rockford, IL, USA). The intensity of protein fragments was quantified using ChemicalDocTM XRS+ (Bio-Rad, Berkeley, CA, USA).
Immunohistochemical staining
Immunohistochemical studies were performed using the UltraSensitive S-P Kit (Maixin Biotechnology Company, Fuzhou, China) and color was developed using DAB as chromogen, counterstained with Mayer's hematoxylin and mounted for evaluation using microscope (OLYMPUS BX-51, Osaka, Japan). Two independent pathologists blinded to the clinicopathological information performed scoring. Staining was scored for determining the intensity (1, 0+ 2, 1+ 3, 2+ 4, 3+) and percentage of membranous and cytoplasmic staining in malignant cells (1, 0–25% 2, 26–50% 3, 51–75% 4, 76–100%). The score of intensity multiplied by the score of percentage counts was used as the final score. A score of <8 was considered as a low-level expression and that of ≥8 was considered as a high-level expression.52
ChIP assay
A total of 2 × 107 cells of each parental U251 and U87 were used for the ChIP assay. Chromatin was cross-linked with formaldehyde and sonicated to an average size of 200–1000 bp. ChIP assay was performed with the EZ chromatin immunoprecipitation kit according to the manufacturer's protocol (Upstate biotechnology, Lake Placid, NY, USA). One milligram of rabbit antibody against dimethylated H3K4, trimethylated H3K9, trimethylated H3K27 and trimethyl-H4K20 (Millipore), respectively, were mixed with sheared chromatin and incubated at 4 °C overnight. Chromatin–antibody complexes were then precipitated with Salmon Sperm DNA/protein A agarose. Reagents and antibodies used in the ChIP assay were obtained from Upstate. Real-time PCR amplification was carried out with Power SYBR Green PCR Master Mix according to manufacturer's instructions (Takara). PCR cycles were identical to the quantitative RT-PCR used for gene expression analysis described above. Primer sequences were: 5′-TGGGTATTAGCCGACAGT-3′ (forward), 5′-CTCCGTTGGTTATTGAAAGT-3′ (reverse). Data are presented as ‘Relative Occupancy' by the equation 2[ΔCt (No antibody−Input)−ΔCt (Target−Input)].
Follow-up study
In this prospective study, patients were followed up for a period of up to 2.5 years. None of the patients received radiation or chemotherapy before surgery. Patients were subjected to a physical examination every 3 months for 1 year postoperatively and thereafter a brain CT examination every 6 months. Overall survival , defined as the time from the date of surgery to the date of death or last contact if the patient was still alive, ranged from 2 to 30 months (median, 13 months). Among the 50 glioma patients included in this study, complete follow-up was available for 38 patients; 15 patients who did not show recurrence were alive until the end of the follow-up period. Thirty-two patients died due to disease recurrence.
Statistical analysis
Data are presented as mean±S.D. from at least three separate experiments. Multiple group comparisons were performed using ANOVA with a post hoc test for subsequent individual group comparisons. The difference in CPEB1 promoter methylation status between normal brain tissues and glioma tissues was examined by an independent sample t-test. The relationships between CPEB1 methylation status, protein expression and clinicopathological parameters were examined by the χ2-test. OS curves were calculated by the Kaplan–Meier method, and the log-rank test was used to determine the difference in OS rates between the two groups. Results were considered significant when P≤0.05 was obtained. All the statistical analyses were performed using SPSS13.0 for Windows (SPSS Inc., Chicago, IL, USA).
Acknowledgments
This study was supported by Grants from The 111 project (111-2-12), the National Science Foundation of China (81171932; 30901539; 30901718) and the Hunan Province Natural Sciences Foundations of China (11JJ1013). We thank Bo Xiang Ph.D in Cancer Research Institute of Central South University for supplying p21 antibody.
Glossary
- CPEB1
cytoplasmic polyadenylation element-binding protein 1
- EZH2
enhancer of zeste homolog 2
- EED
embryonic ectoderm development
- H3K4me2
histone H3 lysine 4 dimethylation
- H3K27me3
histone H3 lysine 27 trimethylation
- H3K9me3
histone H3 lysine 9 trimethylation
- H4K20me3
histone H4 lysine 20 trimethylation
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Candi
Supplementary Material
References
- Starkweather AR, Sherwood P, Lyon DE, McCain NL, Bovbjerg DH, Broaddus WC. A biobehavioral perspective on depressive symptoms in patients with cerebral astrocytoma. J Neurosci Nurs. 2011;43:17–28. doi: 10.1097/jnn.0b013e3182029859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler F, Kienast Y, Fuhrmann M, Von Baumgarten L, Burgold S, Mitteregger G, et al. Imaging glioma cell invasion in vivo reveals mechanisms of dissemination and peritumoral angiogenesis. Glia. 2009;57:1306–1315. doi: 10.1002/glia.20850. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Horiguchi K, Tomizawa Y, Tosaka M, Ishiuchi S, Kurihara H, Mori M, et al. Epigenetic inactivation of RASSF1A candidate tumor suppressor gene at 3p21.3 in brain tumors. Oncogene. 2003;22:7862–7865. doi: 10.1038/sj.onc.1207082. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Watanabe T, Klangby U, Asker C, Wiman K, Yonekawa Y, et al. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001;11:159–168. doi: 10.1111/j.1750-3639.2001.tb00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waha A, Guntner S, Huang TH, Yan PS, Arslan B, Pietsch T, et al. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005;7:193–199. doi: 10.1593/neo.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N, et al. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran G, Shanmuganandam K, Bendre A, Muzumdar D, Goel A, Shiras A. Epigenetic regulation of DNA methyltransferases: DNMT1 and DNMT3B in gliomas. J Neurooncol. 2011;104:483–494. doi: 10.1007/s11060-010-0520-2. [DOI] [PubMed] [Google Scholar]
- Liu X, Tang H, Wang Z, Huang C, Zhang Z, She X, et al. F10 gene hypomethylation, a putative biomarker for glioma prognosis. J Neurooncol. 2012;107:479–485. doi: 10.1007/s11060-011-0775-2. [DOI] [PubMed] [Google Scholar]
- Liu X, Tang H, Zhang Z, Li W, Wang Z, Zheng Y, et al. POTEH hypomethylation, a new epigenetic biomarker for glioma prognosis. Brain Res. 2011;1391:125–131. doi: 10.1016/j.brainres.2011.03.042. [DOI] [PubMed] [Google Scholar]
- Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB. Genes Dev. 2006;20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasayama T, Marumoto T, Kunitoku N, Zhang D, Tamaki N, Kohmura E, et al. Over-expression of Aurora-A targets cytoplasmic polyadenylation element binding protein and promotes mRNA polyadenylation of Cdk1 and cyclin B1. Genes Cells. 2005;10:627–638. doi: 10.1111/j.1365-2443.2005.00870.x. [DOI] [PubMed] [Google Scholar]
- Braig M, Schmitt CA. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 2006;66:2881–2884. doi: 10.1158/0008-5472.CAN-05-4006. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tang H, Wang Z, Zhang B, Liu W, Lu H, et al. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol Cancer. 2011;10:124. doi: 10.1186/1476-4598-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DM, D'Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- Saito Y, Suzuki H, Matsuura M, Sato A, Kasai Y, Yamada K, et al. MicroRNAs in Hepatobiliary and Pancreatic Cancers. Front Genet. 2012;2:66. doi: 10.3389/fgene.2011.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CW, Huang Y, Leong KW, Chen LC, Chen HC, Chen SJ, et al. PKCalpha mediated induction of miR-101 in human hepatoma HepG2 cells. J Biomed Sci. 2010;17:35. doi: 10.1186/1423-0127-17-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V, et al. Synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA. 2007;13:223–239. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15:3927–3937. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong GM, Bain LJ. Sodium arsenite represses the expression of myogenin in C2C12 mouse myoblast cells through histone modifications and altered expression of Ezh2, Glp, and Igf-1. Toxicol Appl Pharmacol. 2012;260:250–259. doi: 10.1016/j.taap.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Kondo Y, Sugimoto A, Kawashima D, Saito S, Isomura H, et al. Epigenetic histone modification of Epstein-Barr virus BZLF1 promoter during latency and reactivation in Raji cells. J Virol. 2012;86:4752–4761. doi: 10.1128/JVI.06768-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson E, Webb R, Weisbrod A, Bian B, He M, Zhang L, et al. The microRNA expression changes associated with malignancy and SDHB mutation in pheochromocytoma. Endocr Relat Cancer. 2012;19:157–166. doi: 10.1530/ERC-11-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Zhang T, Liu H, Lv T, Yuan D, Yao Y, et al. MiR-101 and Mcl-1 in non-small-cell lung cancer: expression profile and clinical significance. Med Oncol. 2012;29:1681–1686. doi: 10.1007/s12032-011-0085-8. [DOI] [PubMed] [Google Scholar]
- Chandramouli A, Onyeagucha BC, Mercado-Pimentel ME, Stankova L, Shahin NA, LaFleur BJ, et al. MicroRNA-101 (miR-101) post-transcriptionally regulates the expression of EP4 receptor in colon cancers. Cancer Biol Ther. 2012;13:175–183. doi: 10.4161/cbt.13.3.18874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J, van Grieken NC, Pereira PM, Sousa S, Tijssen M, Buffart TE, et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J Pathol. 2012;228:31–44. doi: 10.1002/path.4032. [DOI] [PubMed] [Google Scholar]
- Semaan A, Qazi AM, Seward S, Chamala S, Bryant CS, Kumar S, et al. MicroRNA-101 inhibits growth of epithelial ovarian cancer by relieving chromatin-mediated transcriptional repression of p21(waf(1)/cip(1)) Pharm Res. 2011;28:3079–3090. doi: 10.1007/s11095-011-0547-x. [DOI] [PubMed] [Google Scholar]
- Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- Pang Y, Young CY, Yuan H. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin. 2010;42:363–369. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, et al. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46:2295–2303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Jones KJ, Korb E, Kundel MA, Kochanek AR, Kabraji S, McEvoy M, et al. CPEB1 regulates beta-catenin mRNA translation and cell migration in astrocytes. Glia. 2008;56:1401–1413. doi: 10.1002/glia.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Wang P, Xu J, Zhu Y, Sun G, Pang Q, et al. MicroRNA-205 functions as a tumor suppressor in human glioblastoma cells by targeting VEGF-A. Oncol Rep. 2012;27:1200–1206. doi: 10.3892/or.2011.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Liu X, Wang Z, She X, Zeng X, Deng M, et al. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res. 2011;1390:21–32. doi: 10.1016/j.brainres.2011.03.034. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Otte AP, Kwaks TH. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion. Curr Opin Genet Dev. 2003;13:448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–631. doi: 10.1002/hep.25679. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2011;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann MJ, Engers R, Florl AR, Otte AP, Muller M, Schulz WA. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer. Cancer Biol Ther. 2007;6:1403–1412. doi: 10.4161/cbt.6.9.4542. [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Sharma G, Mirza S, Parshad R, Srivastava A, Datta Gupta S, Pandya P, et al. CpG hypomethylation of MDR1 gene in tumor and serum of invasive ductal breast carcinoma patients. Clin Biochem. 2010;43:373–379. doi: 10.1016/j.clinbiochem.2009.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.