The purification and crystallization of the isolated toxin VapC (MvpT) from the VapBC toxin–antitoxin complex of the pathogen S. flexneri are reported. The crystals belonged to space group H3 and diffracted to 1.9 Å resolution.
Keywords: toxin–antitoxin, ribonucleases, tRNA, PIN domain, Shigella flexneri, VapC
Abstract
Upon release from the stable complex formed with its antitoxin VapB, the toxin VapC (MvpT) of the Gram-negative pathogen Shigella flexneri is capable of globally down-regulating translation by specifically cleaving initiator tRNAfMet in the anticodon region. Recombinant Shigella flexneri VapCD7A harbouring an active-site mutation was overexpressed in Escherichia coli, purified to homogeneity and crystallized by the vapour-diffusion technique. A preliminary X-ray crystallographic analysis shows that the crystals diffracted to at least 1.9 Å resolution at a synchrotron X-ray source and belonged to the trigonal space group in the hexagonal setting, H3, with unit-cell parameters a = b = 120.1, c = 52.5 Å, α = β = 90, γ = 120°. The Matthews coefficient is 2.46 Å3 Da−1, suggesting two molecules per asymmetric unit and corresponding to a solvent content of 50.0%.
1. Introduction
Toxin–antitoxin (TA) loci have been found in all prokaryotic genomes that have been sequenced to date and typically encode two proteins, a toxin and an antitoxin, that associate to form a tight complex in which binding of the antitoxin inhibits the cellular action of the toxin (Makarova et al., 2009 ▶; Pandey & Gerdes, 2005 ▶). Upon changes in the cellular environment, such as during oxidative stress or nutrient deprivation, the antitoxin undergoes degradation and the more stable toxin is freed from the inhibitory TA complex. Once released, many toxins are active as nucleases that are able to cleave specific cellular RNAs and thus induce growth arrest by down-regulating the overall translational rate (Ahidjo et al., 2011 ▶; Yamaguchi & Inouye, 2009 ▶). The antitoxins, on the other hand, contain a DNA-binding domain conferring high affinity for the TA promoter, enabling auto-regulation of the level of transcription from the TA operon (Gerdes et al., 2005 ▶). TA gene pairs can be subdivided into several families, of which the vapBC (Virulence-Associated Proteins) loci are the most abundant (Pandey & Gerdes, 2005 ▶). Interestingly, vapBC loci are particularly common in pathogenic bacteria (Ramage et al., 2009 ▶), where they are involved in bacterial persister cell formation, and thus are highly relevant in disease control (Maisonneuve et al., 2011 ▶).
VapC toxins are characterized by an approximately 100 amino-acid N-terminal ribonuclease motif belonging to the PilT N-terminus (PIN) domain type. This domain harbours four highly conserved acidic residues that have been shown to be essential for metal-ion coordination in other PIN-domain-containing ribonucleases, but the exact enzymatic mechanism used by the VapC toxins is as yet unresolved (Bäckbro et al., 2004 ▶; Fatica et al., 2004 ▶). The VapB antitoxins are highly specific towards their cognate toxins and inhibit the ribonucleolytic activity through direct interaction (Ramage et al., 2009 ▶). Crystal structures have been determined of both VapBC-like complexes and isolated VapC-like toxins from archaea and pathogenic bacteria, which show a great diversity in both sequence and structure, as well as the cellular target of the toxin (Bunker et al., 2008 ▶; Mattison et al., 2006 ▶; Miallau et al., 2009 ▶). As in most other TA systems, VapBC complexes bind to the operator sequences of their own promoter region and down-regulate transcription from the operon (Bodogai et al., 2006 ▶; Wilbur et al., 2005 ▶). A recent crystal structure of the intact VapBC complex from the Gram-negative pathogen Shigella flexneri revealed the DNA-binding complex is a compact hetero-octameric assembly with the two DNA-binding domains juxtaposed in a manner compatible with adjacent major-groove binding (Dienemann et al., 2011 ▶). This structure also showed that VapB inhibits VapC through displacement of divalent metal ions at the active site.
Once activated, S. flexneri VapC (MvpT) has been found to specifically target the anticodon region of tRNAfMet through endoribonucleolytic cleavage by a mechanism involving Asp7 of VapC (Winther & Gerdes, 2011 ▶). In order to understand the basis of target RNA recognition and the enzymatic mechanism employed by the Shigella VapC toxin in degrading tRNAfMet, we introduced the D7A mutation with the aim of determining the structure of the VapCD7A in isolation as well as bound to the anticodon region of tRNAfMet. Here, we report the expression, purification and crystallization of the isolated VapCD7A (MvpT) toxin as well as initial crystallographic analysis of diffraction data collected to 1.9 Å resolution. The results provide a basis for determining the crystal structure of the isolated toxin and thus understanding both the principle of toxin activation as well as the mechanism of the ribonucleolytic activity of VapC.
2. Materials and methods
2.1. Cloning
The vapCD7A gene with codon 7 encoding aspartate substituted by an alanine codon was amplified from plasmid pKW254813 (Winther & Gerdes, 2011 ▶) using primers H6-VapC_SF_Down (5′-CCCCCGGTAC CGGATCCAAA ATAAGGAGGA AAAAAAAATG CATCACCATC ACCATCACCT GAAGTTTATG CTC-3′) and VAPC#SF-UP (5′-CCCCCAAGCT TGAATTCGAT TTCTGATGAA CAGGTCAGC-3′). The resulting PCR product was digested with BamHI and HindIII restriction nucleases and ligated into the pMG25 expression plasmid. This plasmid, pKW2583HN, expresses N-terminally polyhistidine-tagged VapCD7A upon induction by IPTG (isopropyl β-d-1-thiogalactopyranoside).
2.2. Expression and purification
pKW2583HN was transformed into Escherichia coli C41 (DE3) cells for expression (Winther & Gerdes, 2011 ▶). The cells were grown in LB medium containing 100 mg ml−1 ampicillin at 310 K until the OD600 reached 0.5 and were subsequently induced with 1 mM IPTG overnight at 298 K with shaking at 120 rev min−1. Cells were harvested by centrifugation at 12 000g for 15 min and resuspended in lysis buffer consisting of 50 mM Tris pH 8.0, 500 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, 10 mM imidazole and protease-inhibitor tablets (Sigma). The cells were opened by a combination of sonication and high-pressure homogenization at 103 MPa and the lysate was cleared by centrifugation at 15 000 rev min−1 for 45 min. The resulting supernatant was loaded onto a pre-packed 5 ml HiTrap column (GE Healthcare) charged with Ni2+ followed by extensive washing of the column (20 column volumes) in a buffer identical to the lysis buffer but including 35 mM imidazole. Elution was finally achieved with the same buffer only containing 250 mM imidazole. Next, the protein pool was concentrated to 3.37 mg ml−1 by ammonium sulfate precipitation (65% saturation) and resuspended in 25 mM MES pH 6.0, 5 mM MgCl2, 5 mM β-mercaptoethanol. As a final step, a monodisperse protein sample was obtained using a Superdex 75 10/300 HR size-exclusion column (GE Healthcare) running in 25 mM MES pH 6.0, 50 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol.
2.3. Crystallization
Prior to crystallization, the monodisperse sample of VapCD7A was concentrated to 4 mg ml−1 using a Vivaspin 6 filter (5 kDa cutoff, GE Healthcare) pre-washed in size-exclusion buffer. Initial screening for crystallization conditions was carried out using an Oryx4 crystallization robot (Douglas Instruments) by mixing 250 nl protein solution with 250 nl reservoir solution (using the PEG/Ion HT and Index commercial screens; Hampton Research) for sitting-drop vapour-diffusion drops placed in 96-well Swissci plates (Hampton Research) at 292 K. Optimization of the crystallization conditions was performed in standard 24-well crystallization plates (Molecular Dimensions) using a 1:1 protein:reservoir ratio.
2.4. X-ray data collection and processing
Single crystals were transferred into a solution containing initially 25%(w/v) followed by 30%(w/v) PEG 3350 in 0.05 M citric acid, 0.05 M bis-tris propane pH 5.0 using a mounted CryoLoop (Hampton Research) and then flash-cooled in liquid nitrogen. All data were collected at 100 K on beamline I911-2 (wavelength 1.04 Å) at MAX-lab (Lund, Sweden) and processed with XDS (Kabsch, 2010 ▶) through xia2 (Winter, 2010 ▶). Matthews parameters were calculated using MATTHEWS_COEF as implemented in CCP4 (Matthews, 1968 ▶; Winn et al., 2011 ▶).
3. Results and discussion
The D7A active-site mutant of VapC, which has been shown to be inactive in vivo, can readily be expressed in E. coli and purified directly in the absence of the antitoxin VapB (Winther & Gerdes, 2011 ▶). Essentially pure VapCD7A was obtained by Ni–NTA chromatography followed by size-exclusion chromatography (Fig. 1 ▶). The protein elutes after 13.5 ml from a standard 24 ml Superdex 75 10/300 HR column as a monodisperse peak corresponding to a molecular weight of 24.5 kDa (Fig. 1 ▶), thus suggesting that the free toxin (theoretical molecular weight 14.8 kDa) exists as a dimer in solution. The initial crystallization screening identified a condition consisting of 16%(w/v) PEG 3350, 0.05 M citric acid, 0.05 M bis-tris propane pH 5.0 that yielded large single crystals with a hexagonal morphology and did not require optimization (Fig. 2 ▶). The crystals could readily be reproduced and appeared after 1 d at 292 K and grew to maximum size (0.4 × 0.2 × 0.1 mm) within a week.
Figure 1.
Purification and oligomeric state of isolated S. flexneri VapCD7A toxin. (a) Fractions of the purified protein were analysed on 15% SDS–PAGE. Lane 1, molecular-weight marker (labelled in kDa); lane 2, elution from the Ni–NTA affinity column; lane 3, final protein sample after size-exclusion chromatography. The theoretical molecular mass of the VapC monomer is 14.8 kDa. (b) Analysis of the oligomeric state of isolated VapC using size-exclusion chromatography. The arrows at the top indicate the elution volumes of proteins of known mass (given in kDa). The elution volume of VapCD7A (13.5 ml) suggests that the molecule is present as a dimer in solution (∼30 kDa).
Figure 2.
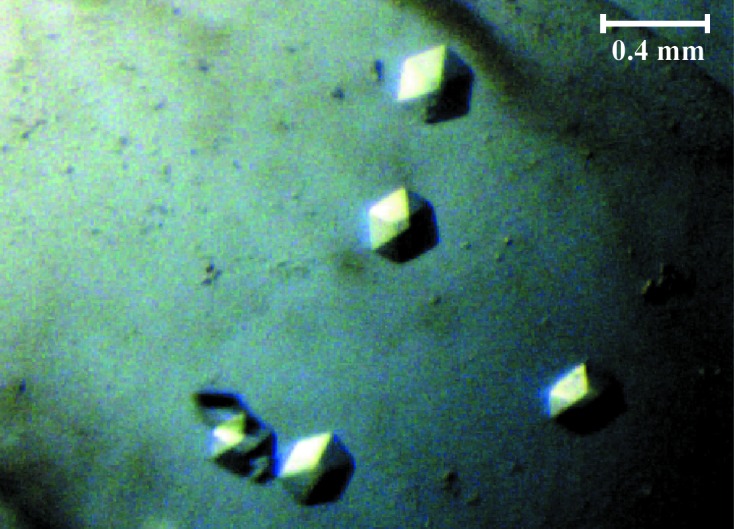
Crystals of isolated S. flexneri VapCD7A toxin. Single crystals of hexagonal appearance; the scale bar indicates the approximate crystal size.
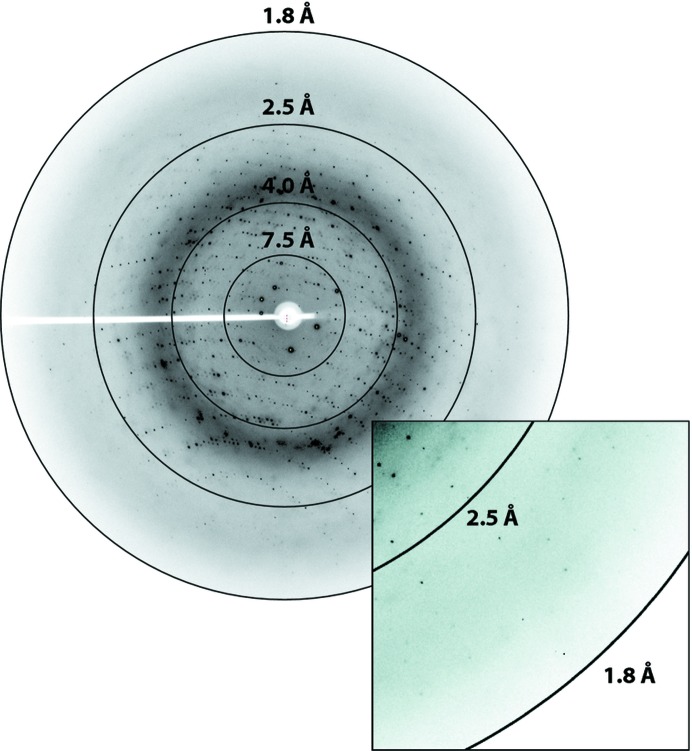
The flash-cooled VapCD7A crystals diffracted to beyond 1.9 Å resolution at a synchrotron X-ray source (Fig. 3 ▶) and a full data set was collected at MAX-lab, Lund, and processed by XDS (Kabsch, 2010 ▶) through xia2 (Winter, 2010 ▶). A summary of the data-processing statistics is shown in Table 1 ▶. The crystals belonged to the trigonal space group in the hexagonal setting, H3 (space group No. 146), with unit-cell parameters a = b = 120.1, c = 52.5 Å, α = β = 90, γ = 120°. The Matthews coefficient and solvent content were estimated using CCP4 (Winn et al., 2011 ▶) as 2.46 Å3 Da−1 and 50.0%, respectively, with an estimated two copies of the monomer per asymmetric unit, suggesting that a single VapC dimer could form the crystallographic asymmetric unit. The self-rotation function, however, did not reveal any nontrivial peaks suggestive of twofold noncrystallographic symmetry. Structure solution by molecular replacement and model building are in progress.
Figure 3.
Synchrotron X-ray diffraction pattern collected from a single cryocooled VapCD7A crystal. Diffraction was observed beyond 1.9 Å resolution. The inset shows a close-up view of the reflections near the edge of the detector.
Table 1. Crystallographic data statistics.
Values in parentheses are for the outermost resolution shell.
| Space group | H3 (No. 146) |
| Unit-cell parameters (Å, °) | a = b = 120.1, c = 52.5, α = β = 90, γ = 120 |
| Wavelength (Å) | 1.041 |
| Matthews coefficient (Å3 Da−1) | 2.46 |
| Likely No. of protomer copies in the asymmetric unit | 2 |
| Solvent content (%) | 50.1 |
| Resolution (Å) | 23.4–1.92 (1.97–1.92) |
| No. of reflections | 132504 |
| No. of unique reflections | 21158 |
| Multiplicity | 6.4 (6.3) |
| Completeness (%) | 98.1 (86.3) |
| R meas † (%) | 3.9 (40.3) |
| CC1/2 (%) | 100.0 (96.7) |
| 〈I/σ(I)〉 | 29.9 (5.3) |
Redundancy-independent R factor calculated on intensities (Diederichs & Karplus, 1997 ▶). R
meas = 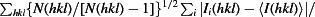
 .
.
In summary, the purification, crystallization and preliminary crystallographic analysis of the S. flexneri VapCD7A toxin reported here form the basis for determining the structure of the active conformation of the isolated toxin upon release of the antitoxin VapB. This structure will allow us to better understand the mechanism of toxin activation for the VapBC toxin–antitoxin family as well as the catalytic mechanism of the toxin. In the longer term, cocrystallization with tRNAfMet will allow a full understanding of the toxin target specificity and the cleavage mechanism.
Acknowledgments
We are grateful to the staff at MAX-lab, Lund, for help with data collection. This work was supported by grants from the Novo Nordisk Foundation, the Danish National Research Council and the Danish Natural Research Foundation Centre for mRNP Biogenesis and Metabolism to DEB.
References
- Ahidjo, B. A., Kuhnert, D., McKenzie, J. L., Machowski, E. E., Gordhan, B. G., Arcus, V., Abrahams, G. L. & Mizrahi, V. (2011). PLoS One, 6, e21738. [DOI] [PMC free article] [PubMed]
- Bäckbro, K., Roos, A., Baker, E. N. & Arcus, V. L. (2004). Acta Cryst. D60, 733–735. [DOI] [PubMed]
- Bodogai, M., Ferenczi, S., Bashtovyy, D., Miclea, P., Papp, P. & Dusha, I. (2006). Mol. Plant Microbe Interact. 19, 811–822. [DOI] [PubMed]
- Bunker, R. D., McKenzie, J. L., Baker, E. N. & Arcus, V. L. (2008). Proteins, 72, 510–518. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Dienemann, C., Bøggild, A., Winther, K. S., Gerdes, K. & Brodersen, D. E. (2011). J. Mol. Biol. 414, 713–722. [DOI] [PMC free article] [PubMed]
- Fatica, A., Tollervey, D. & Dlakić, M. (2004). RNA, 10, 1698–1701. [DOI] [PMC free article] [PubMed]
- Gerdes, K., Christensen, S. K. & Løbner-Olesen, A. (2005). Nature Rev. Microbiol. 3, 371–382. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Maisonneuve, E., Shakespeare, L. J., Jørgensen, M. G. & Gerdes, K. (2011). Proc. Natl Acad. Sci. USA, 108, 13206–13211. [DOI] [PMC free article] [PubMed] [Retracted]
- Makarova, K. S., Wolf, Y. I. & Koonin, E. V. (2009). Biol. Direct, 4, 19. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mattison, K., Wilbur, J. S., So, M. & Brennan, R. G. (2006). J. Biol. Chem. 281, 37942–37951. [DOI] [PubMed]
- Miallau, L., Faller, M., Chiang, J., Arbing, M., Guo, F., Cascio, D. & Eisenberg, D. (2009). J. Biol. Chem. 284, 276–283. [DOI] [PMC free article] [PubMed]
- Pandey, D. P. & Gerdes, K. (2005). Nucleic Acids Res. 33, 966–976. [DOI] [PMC free article] [PubMed]
- Ramage, H. R., Connolly, L. E. & Cox, J. S. (2009). PLoS Genet. 5, e1000767. [DOI] [PMC free article] [PubMed]
- Wilbur, J. S., Chivers, P. T., Mattison, K., Potter, L., Brennan, R. G. & So, M. (2005). Biochemistry, 44, 12515–12524. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Winther, K. S. & Gerdes, K. (2011). Proc. Natl Acad. Sci. USA, 108, 7403–7407. [DOI] [PMC free article] [PubMed]
- Yamaguchi, Y. & Inouye, M. (2009). Prog. Mol. Biol. Transl. Sci. 85, 467–500. [DOI] [PubMed]