Robo receptors participate in the orchestration of several developmental responses, most notably axonal guidance in the central nervous system (CNS). Here, the expression, derivatization, crystallization, and experimental phasing of the extracellular juxtamembrane segment of the human Robo1 receptor are described.
Keywords: Robo1, Slit, selenomethionine, SAD
Abstract
Robo receptors participate in the orchestration of several developmental responses, most notably axonal guidance in the central nervous system. Robo1 contains five tandem Ig-like and three fibronectin type-III (FnIII) domains in its ectodomain, followed by a single-pass transmembrane segment and an intracellular region. A human Robo1 construct that includes the two extracellular membrane-proximal fibronectin (Fn) domains and the juxtamembrane linker was overexpressed in Escherichia coli and purified. Crystals were obtained using the vapour-diffusion method at 293 K and X-ray diffraction data were collected. Molecular-replacement attempts using related Fn domains as search models did not result in a solution. After introducing two additional methionine residues using PCR site-directed mutagenesis, selenomethionine-derivative crystals were produced. These crystals belonged to the primitive orthorhombic space group P212121, with unit-cell parameters a = 27.24, b = 77.64, c = 91.91 Å. Assuming the presence of a monomer in the asymmetric unit gave a crystal volume per protein weight (V M) of 1.97 Å3 Da−1 and a solvent content of 37.6%. Anisotropic diffraction data and a fragmented single-wavelength anomalous dispersion electron-density map, to which homology-modelled domains were docked, were obtained.
1. Introduction
In the developing nervous system, axons extend themselves over long distances to form proper synaptic connections. This process of neuronal pathfinding/axon guidance is controlled by diffusible and membrane-bound extracellular guidance factors that either attract axons towards a designated location or repel them from it. Slits are a family of large secreted guidance factors that are directly detected by their cognate transmembrane Robo receptors. Slit stimulation of Robo is thought to trigger intracellular signalling cascades that result in cytoskeleton remodelling, thereby affecting the direction of the Robo-presenting navigating axon (Kolodkin & Tessier-Lavigne, 2010 ▶). Several cytoplasmic effectors and transducers are implicated in Robo downstream signalling, including the actin-binding proteins Ena (Bashaw et al., 2000 ▶) and Canoe/AF-6 (Slováková et al., 2012 ▶), the tyrosine kinase Abelson (Abl), the Ras/Rho GEF Son of Sevenless (SOS) and the Rac1/RhoA/CDC42-activating protein srGAP (Hohenester, 2008 ▶). However, their direct interaction and response to Robo are not fully understood, particularly in vertebrate systems. The Slit/Robo pathway also participates in cellular navigation outside the central nervous system (CNS) and plays a role in the development of organs such as heart, kidney, lungs and liver (Ypsilanti et al., 2010 ▶). Deregulation of the Slit–Robo pathway occurs in a wide variety of cancer types (Ballard & Hinck, 2012 ▶); for instance, approximately 23% of pancreatic cancer cases show genetic aberrations in the Robo1/2 and Slit2 genes (Biankin et al., 2012 ▶). Robo1 genetic aberrations are also associated with melanoma, breast cancer, small-cell lung cancer and bladder cancer (Ballard & Hinck, 2012 ▶). Consistent with these observations, the inhibition of Robo1 with monoclonal antibodies and decoy Robo ectodomains has been shown to reduce tumour mass and vasculature density (Wang et al., 2008 ▶).
Previous biochemical and structural studies identified and mapped the major Slit–Robo interacting domains (Liu et al., 2004 ▶; Howitt et al., 2004 ▶; Morlot et al., 2007 ▶); however, the Robo activation mechanism remains obscure. Recently, it was observed that cleavage of the entire Robo ectodomain by the membrane metalloproteinase Kuz/ADAM10 is vital for Robo activation (Coleman et al., 2010 ▶). The proteolysis site is located at the juxtamembrane (JM) linker connecting the membrane-proximal FnIII (fibronectin type-III) domain to the transmembrane segment and it was suggested that Slit binding to Robo somehow induces the proteolytic event (Coleman et al., 2010 ▶; Seki et al., 2010 ▶).
In order to learn more about the Robo activation mechanism and to advance our understanding of the forces that control CNS circuitry, we aim to investigate the Robo structure. Here, we describe the expression, crystallization and preliminary X-ray structure of the extracellular juxtamembrane domains of human Robo1.
2. Experimental procedures
2.1. Design of methionine-substitution mutant
A Robo1 insert (Robo1Fn23), containing the two amino-terminal FnIII domains and the JM linker, was generated by PCR amplification from the complete cDNA clone of human Robo1 (accession No. BC157861, spanning residues 660TSQGVD…DVVKQP897). The insert was ligated into modified pET43 containing an N-terminal Nus-A and His tag followed by a TEV restriction site. A detailed description of the oligonucleotide primers used in this work is included in Table 1 ▶. Native protein expression was performed in the Escherichia coli BL21Tuner strain supplemented with the RIL Codon Plus plasmid in 2×YT medium. Following protein purification and screening for crystallization conditions, the best diffracting crystals of native Robo1Fn23 were obtained from hanging-drop vapour-diffusion plates with a reservoir consisting of 31–35% PEG 400, 0.1 M sodium acetate trihydrate pH 4.25–4.75 supplemented with 5% glycerol at 293 K. A native data set was collected to 1.8 Å resolution on beamline BM14 at the European Synchrotron Radiation Facility (ESRF) at 100 K. As molecular-replacement (MR) trials using structures of FnIII homologous domains as search models failed, we pursued an experimental phasing course through selenomethionine derivatization. Robo1Fn23 bears one methionine (Met809) out of a total of 238 residues, a ratio that might fall short of providing sufficient phasing power. Moreover, Met809 is located in a position predicted to be solvent-exposed and hence has a significant likelihood of having a relatively high B factor. We therefore decided to introduce two additional methionines using site-directed mutagenesis.
Table 1. Primers used for insert subcloning and for methionine substitutions by assembly PCR.
Bold underlined letters mark the sites of mutagenesis.
| Reverse (5′–3′) | Forward (5′–3′) | |
|---|---|---|
| Mutagenesis | TCCAGATGGCCGATACATCATTTTATATCCTTGTAT | ATACAAGGATATAAAATGATGTATCGGCCATCTGGA |
| Subcloning | TATAAAGCTTCTACGGCTGCTTCACCACATCTGA | TATAGGATCCACAAGTCAGGGGGTGGACCAC |
Choosing the locations of the designated methionine substitutions was based on homology models for the Fn2 and Fn3 domains of Robo1 that we produced. Using a BLAST (Altschul et al., 1997 ▶) search of the PDB, we found that the sequence of Fn2 from human Robo2 (PDB entry 1ujt; RIKEN Structural Genomics/Proteomics Initiative, unpublished work) was 48% identical to the Robo1 Fn2 and that Fn domains from the netrin receptors neogenin/DCC (PDB entries 1x5h and 2ed9; RIKEN Structural Genomics/Proteomics Initiative, unpublished work) were 35% identical to the membrane-proximal Fn3 of Robo1. Next, we produced homology models for Robo1 Fn2 and Fn3 using Sculptor (Bunkóczi & Read, 2011 ▶) and evaluated candidate substitution sites while taking into account several factors such as predicted flexibility, chemical similarity and structural compatibility. As Met809 of native Robo1 is located in Fn3, we predicted that substitutions in Fn2 could be useful for positioning of the Fn2 and Fn3 domains by triangulating the locations of the three selenium sites. Given these considerations, we substituted the consecutive Robo1 Ile709 and Leu710, located on strand C of Fn2, to methionines (hereafter referred to as Robo1Fn23SeMet) by a single-assembly PCR mutagenesis (see Table 1 ▶) and ligated the insert to the modified pET43 vector.
2.2. Robo1Fn23SeMet expression and purification
Selenomethionine-substituted Robo1Fn23SeMet was prepared and crystallized for anomalous diffraction experiments. Protein was produced in the same bacterial strain as the native protein by inhibiting the de novo methionine-synthesis pathway (Van Duyne et al., 1993 ▶) as reported previously (Opatowsky et al., 2004 ▶).
An overnight starter culture was grown at 310 K from a single transformed colony in 10% LB medium and 90% new minimal medium (NMM; 22 mM KH2PO4, 50 mM K2HPO4, 7.5 mM ammonium sulfate, 8.5 mM NaCl, 1 mM MgSO4) containing 100 µg ml−1 ampicillin and 34 µg ml−1 chloramphenicol. LB–NMM medium was removed prior to the introduction of 2 l pre-warmed NMM containing 7.2 g glucose, 100 µg ml−1 ampicillin, 34 µg ml−1 chloramphenicol and 100 mg of all amino acids except methionine and fortified with 5 ml Kao and Michayluk vitamin solution (Sigma) per litre. Cells were grown at 310 K to an OD600 of 0.35, whereupon the temperature was reduced to 289 K and lysine, phenylalanine and threonine (100 mg l−1), isoleucine, leucine and valine (50 mg l−1) and dl-selenomethionine (50 mg l−1) were added. 45 min later an OD600 of 0.6 was reached and expression was induced with 200 µM IPTG over a 14 h period before the cells were centrifuged and frozen.
Cells were suspended at a 1:10(w:v) ratio with lysis buffer (0.5 M NaCl, 10% glycerol, 50 mM phosphate buffer pH 7.4, 0.1% Triton) and lysed using a microfluidizer (Microfluidics). Cell debris was removed by 20 min centrifugation (11 742g) at 277 K. The soluble fraction was loaded onto a metal-chelate column (HisTrap, GE Healthcare) pre-equilibrated with buffer A (0.5 M NaCl, 10% glycerol, 50 mM phosphate buffer pH 7.4) at a flow rate of 3 ml min−1. The column was washed with buffer A until a stable baseline was achieved. After applying a gradient elution with buffer A supplemented with 70–200 mM imidazole, Robo1Fn23SeMet-containing fractions were pooled and diluted at a ratio of 1:15(v:v) with 500 mM Tris pH 8.5 (the high concentration of the Tris was a mistake) and 5% glycerol and loaded onto an ion-exchange column (Q Sepharose, GE Healthcare) pre-equilibrated with buffer B (50 mM NaCl, 50 mM Tris pH 8.8, 5% glycerol) at a flow rate of 5 ml min−1. Proteins were eluted using a gradient of buffer B containing 320–544 mM NaCl.
Robo1Fn23SeMet-containing fractions were pooled and subjected to TEV protease with a protein:protease ratio of 50:1(w:w) for 12–14 h at 277 K. After TEV proteolysis was completed, the pH of the sample was adjusted to 7.6 using NaH2PO4 and it was once again loaded onto a metal-chelating column. Robo1Fn23SeMet did not appear in the unbound fractions and was eluted with 30 mM imidazole. The eluted fractions were analysed using SDS–PAGE, pooled and concentrated to a volume of 15 ml using spin concentrators (Sartorius Stedim Biotech).
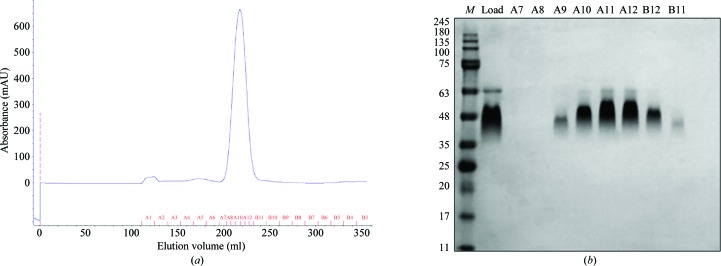
Gel-filtration chromatography was then carried out using a pre-equilibrated Superdex 200 HiLoad 26/60 column (GE Healthcare) (see Fig. 1 ▶). The protein was eluted with buffer C (10 mM Tris pH 8, 20 mM NaCl). Pooled fractions were concentrated to 20 mg ml−1, divided into aliquots and flash-frozen in liquid N2.
Figure 1.
Purification of Robo1Fn23SeMet. (a) Superdex 200 26/60 size-exclusion chromatography elution profile of Robo1Fn23SeMet. The protein peak elutes at a volume of 217 ml, corresponding to ∼30 kDa, which is close to the calculated weight of the protein (26 kDa). (b) SDS–PAGE analysis of (a). Lane M contains molecular-mass markers (labelled in kDa).
2.3. Crystallization
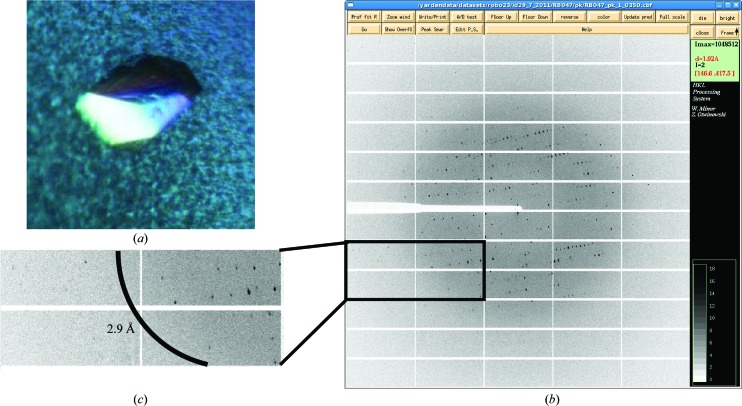
Robo1FN23SeMet did not crystallize under the same conditions as the native protein and was therefore screened using Index HT (Hampton Research, Aliso Viejo, California, USA), ProPlex, Morpheus and JCSG (Molecular Dimensions) screens at 293 K in 96-well hanging-drop clear polystyrene microplates (TTP LabTech) using a Mosquito robot for crystallography (TTP LabTech). The drop size was 0.2 µl with a 1:1 sample:reservoir ratio. After 2 weeks, small crystal clusters appeared in condition E2 of JCSG (0.2 M NaCl, 0.1 M sodium cacodylate pH 6.5, 2 M ammonium sulfate). Crystallization conditions were further refined using 24-well hanging-drop vapour-diffusion plates by varying pH values against ammonium sulfate concentrations and adding 4% glycerol, 2% PEG 400, 2% MPD and 2% dioxan. Optimal crystal-growth conditions were a reservoir consisting of 0.2 M NaCl, 0.1 M sodium cacodylate pH 5.25–5.5, 1.4–1.5 M ammonium sulfate, 2% MPD. Crystals appeared after 3–5 d in 2 µl drops (protein:reservoir ratio of 1:1) and were harvested from 3 d to 1 week after appearance (Fig. 2 ▶ a). Prior to flash-cooling in liquid nitrogen, crystals were transferred to a cryoprotectant solution consisting of 0.2 M NaCl, 0.1 M sodium cacodylate pH 5.5, 1.57 M ammonium sulfate, 18% glycerol.
Figure 2.
X-ray diffraction of a human Robo1Fn23SeMet crystal. (a) Crystals of Robo1Fn23SeMet grown in 1.5 M ammonium sulfate pH 5.5. Crystal dimensions were approximately 0.1 × 0.1 × 0.05 mm. (b) Diffraction image collected on beamline ID29 at the ESRF using a PILATUS 6M detector. (c) Close-up of high-resolution shells; the arc marks the 2.9 Å resolution line.
2.4. SAD data collection and analysis
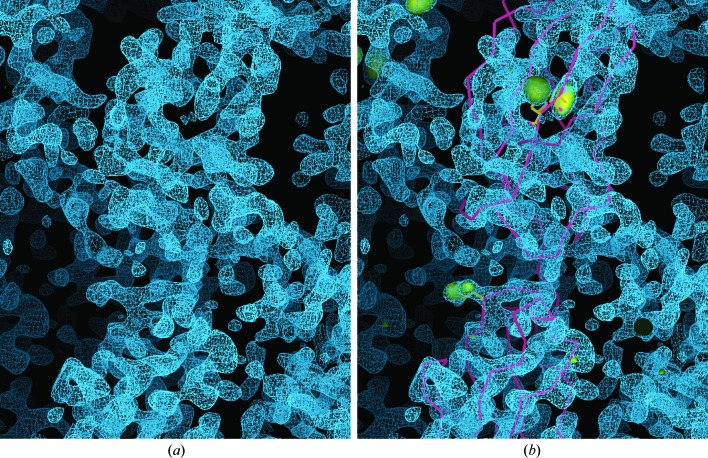
Diffraction data for the Robo1Fn23SeMet crystals were measured on the tunable beamline ID29 at the ESRF (de Sanctis et al., 2012 ▶) under standard cryogenic conditions (Figs. 2 ▶ b and 2 ▶ c), processed with iMOSFLM (Battye et al., 2011 ▶) and scaled with SCALA (Evans, 2006 ▶). A single-wavelength (at the anomalous absorption peak) SAD experiment was performed on a single crystal (Table 2 ▶). AutoSol in PHENIX (Adams et al., 2010 ▶) was used to identify the three selenium sites, calculate the phases and perform density modification. The initial density-modified electron-density map obtained by AutoSol/RESOLVE was fragmented and hardly interpretable; however, the AutoSol/Phaser map contoured to 4σ revealed the locations of the three methionine residues. These sites were used to dock the two homology-modelled Fn2 and Fn3 domains (Fig. 3 ▶) and were further used in SAD–MR (McCoy et al., 2007 ▶) and refinement cycles using PHENIX.
Table 2. Data-collection statistics for Robo1Fn23SeMet.
Values in parentheses are for the outermost resolution shell.
| Beamline | ID29, ESRF |
| Wavelength (Å) | 0.97934 |
| No. of images | 720 |
| Oscillation range (°) | 0.5 |
| Space group | P212121 |
| Unit-cell parameters | |
| a (Å) | 27.24 |
| b (Å) | 77.64 |
| c (Å) | 91.91 |
| Resolution (Å) | 59.3–2.9 (3.06–2.90) |
| Total No. of reflections | 36169 |
| Unique reflections | 4723 |
| Completeness (%) | 99.7 (99.7) |
| 〈I/σ(I)〉 | 24.3 (13.8) |
| R meas (all I+ and I−)† (%) | 7.2 (14.5) |
| Multiplicity | 7.7 (8.2) |
| DelAnom correlation between half-sets | 0.447 (0.071) |
R
meas = 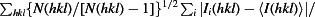
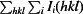 , where Ii(hkl) is the ith observation of reflection hkl, 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl and N(hkl) is the number of observations of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl, 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl and N(hkl) is the number of observations of reflection hkl.
Figure 3.
Preliminary structure of Robo1Fn23. (a) Selenomethionine SAD experimental electron-density map (presented as blue mesh) after solvent-flattening by AutoSol/RESOLVE. Note the clear protein–solvent boundaries and the fragmented electron density. (b) As (a) but with the selenium substructure locations presented as yellow volumes contoured to 4σ (from the AutoSol/Phaser map). Using the three selenomethionine sites as anchors, the Fn2 and Fn3 homology models (shown as a yellow backbone trace) were manually docked into the RESOLVE map.
3. Results and discussion
The extracellular JM region of Robo regulates the activity of the Robo–Slit signalling pathway. Here, we present the design and production of selenomethionine-derived crystals of Robo1Fn23, which includes two FnIII domains and the JM linker. X-ray SAD data were collected at the selenium absorption edge to 2.9 Å resolution on beamline ID29 at the ESRF. The crystals belonged to space group P212121 and exhibited strong diffraction anisotropy of 43.38 Å2, as estimated by the UCLA MBI Diffraction Anisotropy Server (Strong et al., 2006 ▶), resulting in a fragmented electron-density map that could not be well traced. We therefore used the locations of the three selenium substructures to dock homology-modelled FnIII domains into the electron-density map. Subsequent SAD–MR and rigid-body refinement cycles resulted in an interpretable map and a reliable starting backbone model. Next, we will refine this model against the isotropic native data set for further structural interpretation.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Ballard, M. S. & Hinck, L. (2012). Adv. Cancer Res. 114, 187–235. [DOI] [PMC free article] [PubMed]
- Bashaw, G. J., Kidd, T., Murray, D., Pawson, T. & Goodman, C. S. (2000). Cell, 101, 703–715. [DOI] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Biankin, A. V. et al. (2012). Nature (London), 491, 399–405.
- Bunkóczi, G. & Read, R. J. (2011). Acta Cryst. D67, 303–312. [DOI] [PMC free article] [PubMed]
- Coleman, H. A., Labrador, J. P., Chance, R. K. & Bashaw, G. J. (2010). Development, 137, 2417–2426. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Hohenester, E. (2008). Biochem. Soc. Trans. 36, 251–256. [DOI] [PubMed]
- Howitt, J. A., Clout, N. J. & Hohenester, E. (2004). EMBO J. 23, 4406–4412. [DOI] [PMC free article] [PubMed]
- Kolodkin, A. L. & Tessier-Lavigne, M. (2010). Cold Spring Harb. Perspect. Biol. 3, a001727. [DOI] [PMC free article] [PubMed]
- Liu, Z., Patel, K., Schmidt, H., Andrews, W., Pini, A. & Sundaresan, V. (2004). Mol. Cell. Neurosci. 26, 232–240. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Morlot, C., Thielens, N. M., Ravelli, R. B., Hemrika, W., Romijn, R. A., Gros, P., Cusack, S. & McCarthy, A. A. (2007). Proc. Natl Acad. Sci. USA, 104, 14923–14928. [DOI] [PMC free article] [PubMed]
- Opatowsky, Y., Chomsky-Hecht, O. & Hirsch, J. A. (2004). Acta Cryst. D60, 1301–1303. [DOI] [PubMed]
- Sanctis, D. de et al.(2012). J. Synchrotron Rad. 19, 455–461.
- Seki, M., Watanabe, A., Enomoto, S., Kawamura, T., Ito, H., Kodama, T., Hamakubo, T. & Aburatani, H. (2010). FEBS Lett. 584, 2909–2915. [DOI] [PubMed]
- Slováková, J., Speicher, S., Sánchez-Soriano, N., Prokop, A. & Carmena, A. (2012). J. Neurosci. 32, 10035–10044. [DOI] [PMC free article] [PubMed]
- Strong, M., Sawaya, M. R., Wang, S., Phillips, M., Cascio, D. & Eisenberg, D. (2006). Proc. Natl Acad. Sci. USA, 103, 8060–8065. [DOI] [PMC free article] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol. 229, 105–124. [DOI] [PubMed]
- Wang, L.-J., Zhao, Y., Han, B., Ma, Y.-G., Zhang, J., Yang, D.-M., Mao, J.-W., Tang, F.-T., Li, W.-D., Yang, Y., Wang, R. & Geng, J.-G. (2008). Cancer Sci. 99, 510–517. [DOI] [PMC free article] [PubMed]
- Ypsilanti, A. R., Zagar, Y. & Chédotal, A. (2010). Development, 137, 1939–1952. [DOI] [PubMed]