As a thermostable esterase with cold adaptation, EstL5 might be of significant industrial interest and value in scientific research. In order to provide new insights into the structure–function relationship of EstL5 and to help better understand the biochemical data, a project to determine the three-dimensional structure of this enzyme was initiated.
Keywords: EstL5, thermostability, GDSL esterases, Geobacillus thermodenitrificans
Abstract
The novel thermostable esterase EstL5 belonging to the GDSL family exhibits a unique cold-adaptation feature at low temperatures. To better understand its biochemical and enzymatic properties, recombinant EstL5 protein was purified and crystallized using the vapour-diffusion method. The EstL5 crystals diffracted X-rays to 2.79 Å resolution using a synchrotron-radiation source, belonged to the tetragonal space group P41212 or P43212, with unit-cell parameters a = b = 101.51, c = 124.22 Å, and are expected to contain two molecules in each asymmetric unit. To obtain initial phases, selenomethionyl-substituted protein was overproduced. Purified SeMet-labelled EstL5 protein was crystallized and formed crystals that diffracted to a resolution of 3.0 Å.
1. Introduction
Esterases (EC 3.1.1.1) and lipases (EC 3.1.1.3) are collectively known as lipolytic enzymes which catalyse the hydrolysis of ester bonds present in acyl alcohols/acylglycerols with the liberation of fatty acids and alcohols/glycerols (Arpigny & Jaeger, 1999 ▶; Sharma et al., 2001 ▶). Although similar in molecular structure and catalytic mechanism, esterases can be distinguished from true lipases by their preference for short-chain acyl derivatives and their lack of requirement for interfacial activation (Sharma et al., 2001 ▶). The biological roles of most lipolytic enzymes still remain unclear, but their involvement in the metabolism of ester components and ubiquitous nature indicate their importance in cell physiology.
Lipolytic enzymes have attracted enormous attention in industrial applications owing to the fact that they not only catalyse enantioselective or regioselective hydrolysis in aqueous emulsions but also catalyse reverse synthetic ester or transesterification reactions in organic solvents (Jaeger & Eggert, 2002 ▶). These enzymes have been widely used to produce fine chemicals and pharmaceuticals, in food processing and as detergent additives (Jaeger & Eggert, 2002 ▶; Bornscheuer, 2002 ▶). A number of lipolytic enzymes from various sources displaying diversified biochemical properties have been isolated and characterized (Panda & Gowrishankar, 2005 ▶). Among them, thermostable lipolytic enzymes from thermophiles display an optimal catalytic activity at high temperatures as well as inherent stability (Haki & Rakshit, 2003 ▶; Hess et al., 2008 ▶; Hotta et al., 2002 ▶). These unique characteristics of thermostable enzymes are valuable in industrial and biotechnology interests owing to the fact that they are better suited to the harsh processes in these applications.
Recently, we have cloned and partially characterized a thermostable lipolytic gene from Geobacillus thermodenitrificans strain T2, denoted estl5 (accession Nos. EU623977 and ACD02023.1; Yang et al., 2013 ▶). Amino-acid sequence comparison showed that EstL5 is a novel member of the GDSL family of serine esterases/lipases with a distinct GDSL sequence motif that differs from the GXSXG motif found in many lipolytic enzymes (Akoh et al., 2004 ▶). Because of the presence of four strictly conserved and functionally important residues Ser, Gly, Asn and His in conserved blocks I, II, III and V, respectively, EstL5 can be further classified as a member of the SGNH hydrolase superfamily (Akoh et al., 2004 ▶). Recombinant EstL5 was identified as an esterase which preferred hydrolysis of short p-nitrophenyl esters of butyrate (C4). Interestingly, the recombinant EstL5 esterase was stable at temperatures of up to 333 K, but it also retained relatively high activity in a low-temperature environment, displaying 24% of its maximum activity even at 273 K (Yang et al., 2013 ▶). As a thermostable esterase with cold adaptation, EstL5 might be of significant industrial interest and value in scientific research. In order to provide new insights into the structure–function relationship of EstL5 and to obtain a better understanding of the biochemical data, we started a project to determine the three-dimensional structure of this enzyme.
2. Materials and methods
2.1. Protein expression and purification
The estl5 gene was cloned into a pET28b vector (Novagen, USA) and recombinant proteins were expressed in Escherichia coli BL21 (DE3) cells as described previously (Yang et al., 2013 ▶). The bacterial culture was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the OD600 reached 0.6–0.8 and grown for a further 12 h at 289 K. The induced cells were harvested by centrifugation and disrupted by sonication. The recombinant EstL5 protein was affinity purified using an Ni–NTA column (Qiagen, USA) and eluted with buffer A (250 mM imidazole, 300 mM NaCl, 100 mM Tris–HCl pH 8.5, 10 mM β-mercaptoethanol). The obtained EstL5 protein was further purified by gel filtration using a Superdex 75 column (GE Healthcare, USA) equilibrated with buffer consisting of 25 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM dithiothreitol (DTT). Cells producing selenomethionine-labelled EstL5 were grown at 298 K in minimal medium supplemented with selenomethionine (SeMet) and the proteins were purified using the same protocol as used for unlabelled EstL5. Finally, the pure EstL5 proteins were concentrated to 20 mg ml−1 using a 10 kDa Vivaspin-20 concentrator (GE Healthcare, USA) for crystallization.
2.2. Crystallization and X-ray data collection
Preliminary crystallization conditions were screened by Crystal Screen Lite and Crystal Screen 2 (Hampton Research, USA) using the hanging-drop vapour-diffusion method at 277 K. Droplets for crystallization were prepared by mixing 1 µl protein solution and 1 µl reservoir solution and were equilibrated against 500 µl reservoir solution. The crystallization conditions were optimized via variation of protein concentration (to a maximum of 20 mg ml−1), precipitant concentration and pH using the hanging-drop vapour-diffusion method at 277 K in 24-well plates. Before data collection, a single crystal was soaked in mother liquor containing 20%(v/v) glycerol as a cryoprotectant and flash-cooled directly in liquid nitrogen. Native and single-wavelength anomalous diffraction (SAD) data sets were collected on beamline BL17U-MX at the Shanghai Synchrotron Radiation Facility (SSRF), China, using a MAR DTB system. The exposure time was 1.0 s per frame. One complete data set was obtained by collecting 360 images with 1° oscillation. The data collected were processed and scaled with the HKL-2000 suite (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
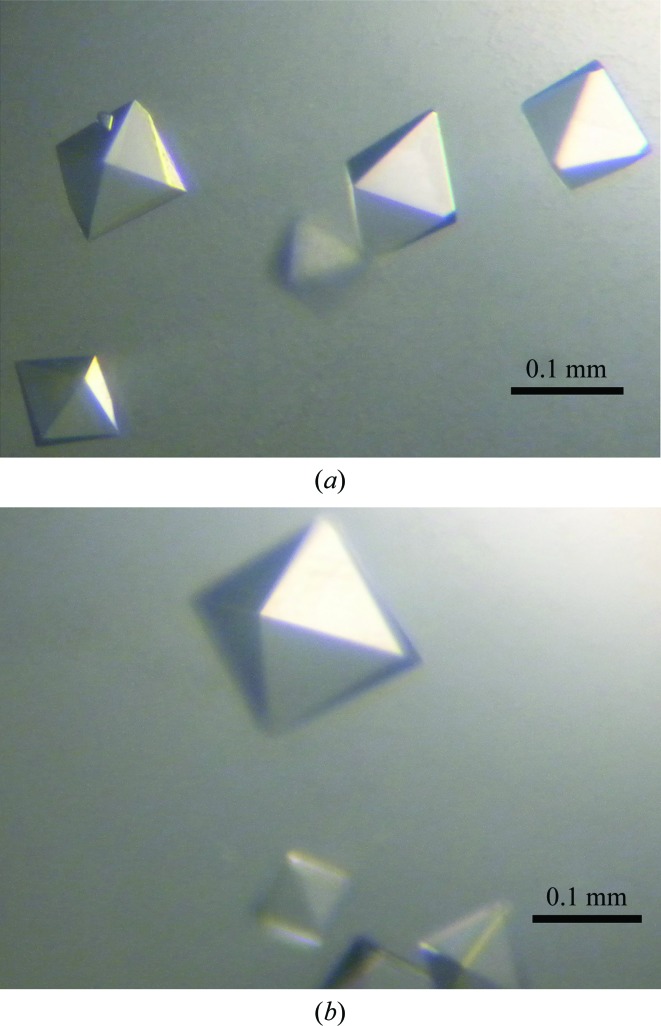
N-terminally hexahistidine-tagged (additional residues MGSSHHHHHHSSGLVPRGSHMAS-) EstL5 protein was purified by Ni2+-affinity chromatography and gel-filtration chromatography, with significant enzyme activity being maintained (Yang et al., 2013 ▶). The purified recombinant EstL5 proteins were subjected to extensive crystallization screening. Small diamond-shaped crystals of EstL5 could only be obtained from the condition 0.1 M sodium acetate pH 4.6, 0.01 M cobalt chloride hexahydrate, 1.0 M 1,6-hexanediol. The crystallization conditions were optimized to produce larger diamond-shaped single crystals. Ultimately, the condition 0.1 M sodium acetate pH 4.6, 0.01 M cobalt chloride hexahydrate, 1.7 M 1,6-hexanediol proved to be most effective for generating large crystals. The dimensions of the crystal were approximately 0.1 × 0.1 × 0.15 mm (Fig. 1 ▶ a). The crystal belonged to the tetragonal space group P41212 or P43212, with unit-cell parameters a = b = 101.51, c = 124.22 Å. The diffraction data set was processed to 2.79 Å resolution with 99.8% completeness. Based on consideration of the Matthews coefficient, the most probable number of EstL5 molecules in the asymmetric unit is two (Matthews, 1968 ▶). In this case, the Matthews coefficient is 2.72 Å3 Da−1, corresponding to a solvent content of 54.79%. Data-collection statistics are summarized in Table 1 ▶.
Figure 1.
Crystals of EstL5 from G. thermodenitrificans T2. (a) EstL5 crystals with approximate dimensions of 0.1 × 0.1 × 0.15 mm grown under the condition 0.1 M sodium acetate pH 4.6, 0.01 M cobalt chloride hexahydrate, 1.7 M 1,6-hexanediol. (b) SeMet-EstL5 crystals grown under the condition 0.1 M sodium acetate pH 4.6, 0.01 M cobalt chloride hexahydrate, 1.4 M 1,6-hexanediol.
Table 1. Summary of native X-ray data collection.
Values in parentheses are for the outermost resolution shell.
| SeMet-EstL5 | EstL5 | |
|---|---|---|
| Detector | MAR 225 CCD | MAR 225 CCD |
| Wavelength (Å) | 0.97885 | 0.97885 |
| Rotation range per frame (°) | 1 | 1 |
| Crystal-to-detector distance (mm) | 300 | 300 |
| Data-collection temperature (K) | 100 | 100 |
| Total rotation range (°) | 360 | 360 |
| Space group | P41212 or P43212 | P41212 or P43212 |
| Unit-cell parameters (Å) | a = b = 101.48, c = 124.56 | a = b = 101.51, c = 124.22 |
| Resolution range (Å) | 50–3.0 (3.11–3.00) | 50–2.79 (2.85–2.79) |
| Unique reflections | 955854 (13542) | 1028867 (15157) |
| Completeness (%) | 99.9 (100) | 99.8 (100) |
| R merge † (%) | 15.6 (70.7) | 14.3 (80.1) |
| Mean I/σ(I) | 27.2 (4.5) | 17.9 (2.8) |
| Multiplicity | 26.5 (21.6) | 14.0 (14.2) |
| V M (Å3 Da−1) | 2.69 | 2.72 |
| Solvent content (%) | 54.19 | 54.79 |
R
merge = 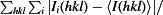
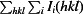 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl, 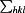 is the sum over all reflections, and
is the sum over all reflections, and 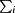 is the sum over i measurements of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
is the sum over i measurements of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
The deduced amino-acid sequence of EstL5 exhibits a relatively low sequence similarity to other structurally known enzymes (less than 20% sequence identity over 265 residues). In order to solve the structure of EstL5, we intend to obtain experimental phases by the SAD approach using SeMet-derivatized crystals. EstL5 protein was overexpressed in the presence of SeMet. Finally, SeMet-substituted EstL5 protein was purified according to the protocol established for the native protein and identified by mass spectrometry. The SeMet-substituted EstL5 could be crystallized under the condition 0.1 M sodium acetate pH 4.6, 0.01 M cobalt chloride hexahydrate, 1.4 M 1,6-hexanediol and the crystal diffracted to a resolution of 3.0 Å (Fig. 1 ▶ b). The diffraction data are also summarized in Table 1 ▶. Six heavy-atom sites (presumably Se) were found in one asymmetric unit using the SHELXC/D/E programs (Sheldrick, 2008 ▶). Ultimate structure determination of the unique GDSL-family thermostable esterase EstL5 is currently underway. Once structural information has been obtained for EstL5, it will be used as a starting point to better understand the catalytic mechanism and stability of the widely distributed GDSL-family enzymes.
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility beamline BL17U for assistance in data collection. This work was supported by the National Basic Research Program of China (973 Program; 2007CB914304 and 2009CB825505), the National Natural Science Foundation of China (30770427 and 31200597), New Century Excellent Talents in University (NCET-06-0356), the Shanghai Leading Academic Discipline Project (B111) and the National Talent Training Fund in Basic Research of China (J0630643).
References
- Akoh, C. C., Lee, G., Liaw, Y., Huang, T. & Shaw, J. (2004). Prog. Lipid Res. 43, 534–552. [DOI] [PubMed]
- Arpigny, J. L. & Jaeger, K.-E. (1999). Biochem. J. 343, 177–183. [PMC free article] [PubMed]
- Bornscheuer, U. T. (2002). FEMS Microbiol. Rev. 26, 73–81. [DOI] [PubMed]
- Haki, G. D. & Rakshit, S. K. (2003). Bioresour. Technol. 89, 17–34. [DOI] [PubMed]
- Hess, M., Katzer, M. & Antranikian, G. (2008). Extremophiles, 12, 351–364. [DOI] [PubMed]
- Hotta, Y., Ezaki, S., Atomi, H. & Imanaka, T. (2002). Appl. Environ. Microbiol. 68, 3925–3931. [DOI] [PMC free article] [PubMed]
- Jaeger, K.-E. & Eggert, T. (2002). Curr. Opin. Biotechnol. 13, 390–397. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Panda, T. & Gowrishankar, B. S. (2005). Appl. Microbiol. Biotechnol. 67, 160–169. [DOI] [PubMed]
- Sharma, R., Chisti, Y. & Banerjee, U. C. (2001). Biotechnol. Adv. 19, 627–662. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Yang, Z., Zhang, Y., Shen, T., Xie, Y., Mao, Y. & Ji, C. (2013). J. Biosci. Bioeng. 115, 133–137. [DOI] [PubMed]