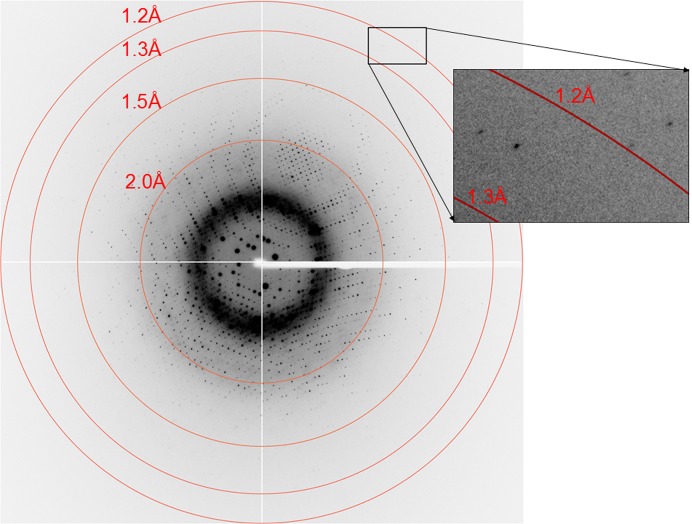
Rice l-galactose dehydrogenase was crystallized and X-ray diffraction data were collected to 1.2 Å resolution.
Keywords: l-galactose dehydrogenase, ascorbic acid biosynthesis, Oryza sativa L.
Abstract
In plants, l-galactose dehydrogenase (l-GalDH) is a key enzyme in the biosynthesis of ascorbic acid (AsA), which is well known as a unique antioxidant compound and a cofactor for many enzymes. l-GalDH catalyses the oxidation of l-galactose to l-galactono-1,4-lactone. Rice l-GalDH was overexpressed in Escherichia coli, purified and crystallized. Diffraction-quality rod-shaped crystals were grown using a sitting-drop vapour-diffusion method. The l-GalDH crystals exhibited the symmetry of space group P21 and diffracted to a resolution of 1.2 Å. The crystals had unit-cell parameters a = 46.8, b = 54.9, c = 56.9 Å, β = 102.3°. On the basis of the Matthews coefficient (V M = 2.1 Å3 Da−1, solvent content of 42.3%), it was estimated that one peptide was present in the asymmetric unit.
1. Introduction
l-Ascorbic acid (AsA), also known by its popular name vitamin C, is an important antioxidant (Du et al., 2012 ▶) and is also a cofactor for many enzymes such as haem oxygenases (Muramoto et al., 2002 ▶; Wang et al., 2005 ▶). AsA is synthesized by plants and animals, with the exceptions of primates (including humans), guinea pigs, bats and some birds. A pathway for AsA formation has been well characterized in animal systems (Chatterjee et al., 1960 ▶). AsA is synthesized in liver and kidney from d-glucose as an initial precursor through the intermediates d-glucuronate and l-gulono-1,4-lactone (l-GulL). At the final stage, l-GulL dehydrogenase oxidizes l-GulL to AsA.
In plants, several pathways different from the pathway in animals have been suggested. A novel d-mannose/l-galactose (d-Man/l-Gal) pathway was proposed as the main process of AsA biosynthesis (Wheeler et al., 1998 ▶). In addition to the d-Man/l-Gal pathway, at least three other pathways have been suggested based on genetic and biochemical studies: the d-galacturonic acid pathway, the l-gulose pathway and the myo-inositol pathway (Li et al., 2010 ▶). These pathways cooperate or alternatively work under various environmental, physiological and special (tissue-specific) conditions (Ishikawa et al., 2006 ▶). Most of the enzymes related to the pathway have been identified and studied in detail. In the d-Man/l-Gal pathway, AsA can be synthesized from d-mannose-1-phosphate via GDP-mannose and GDP-l-galactose (GDP-l-Gal), and free l-Gal is produced from GDP-l-Gal through the action of GDP-l-Gal phosphorylase and l-galactose-1-phosphate phosphatase (Conklin et al., 1999 ▶; Dowdle et al., 2007 ▶). l-Gal is then oxidized by l-galactose dehydrogenase (l-GalDH) to form l-galactono-1,4-lactone (Wheeler et al., 1998 ▶; Gatzek et al., 2002 ▶). Finally, it is oxidized to AsA by l-galactono-1,4-lactone dehydrogenase (Mapson & Breslow, 1958 ▶).
Because of their lack of ability to synthesize AsA (Nishikimi et al., 1994 ▶), humans have to depend on plant-derived AsA sources. AsA production by plants is very important for eliminating oxidants that threaten human health. There have been many genetic and biochemical investigations that have helped to elucidate the enzymatic events involved in the AsA biosynthetic pathways. Although no structures of any of the enzymes in the AsA pathway have been reported, structure determination would still be a powerful tool to elucidate the enzymatic activities involved in the AsA synthetic pathways. Structural information on the enzymes should allow the breeding of plants with high AsA production and the production of foods with high nutritional value.
We report the structural determination of rice (Oryza sativa L. japonica cultivar group) l-GalDH (EC 1.1.1.117; OsLGalDH), which is one of the key enzymes in the d-Man/l-Gal AsA biosynthesis pathway. OsLGalDH shares less than 29% amino-acid sequence homology with NAD(P)-dependent oxidoreductases for which structures are known. Here, we report the expression, purification, crystallization and preliminary X-ray analysis of OsLGalDH.
2. Materials and methods
The DNA fragment corresponding to the OsLGalDH protein (316 amino acids) was amplified using PCR with forward primer 5′-ATGGAGCTCGCGAGCTCGGC-3′ and reverse primer 5′-AGCTCTAGATCAGGCTTGCTCAATGCCACT-3′ (gene-specific sequences are shown in bold) using the cDNA clone AK102223 (KOME Database; http://cdna01.dna.affrc.go.jp/cDNA/; Kikuchi et al., 2003 ▶) as a template. The fragment was digested by XbaI and cloned into the pET-45b expression vector (Merck–Novagen, Madison, Wisconsin, USA) using PmlI and AvrII sites. The protein was overexpressed using Escherichia coli BL21 (DE3) cells in LB medium by induction with 0.25 mM isopropyl β-d-1-thiogalactopyranoside overnight at 298 K. The protein was purified from the soluble fraction of the bacterial cell lysate by Ni2+-charged HiTrap chelating FF column chromatography (GE Healthcare, Buckinghamshire, England) in a buffer consisting of 20 mM sodium phosphate pH 7.2, 500 mM NaCl, 200 mM imidazole. The protein was further purified by gel-filtration column chromatography using Superdex 200 pg 16/60 (GE Healthcare) with a buffer consisting of 0.2 M NaCl, 20 mM Tris–HCl pH 8.0.
The protein solution was desalted and concentrated to 10 mg ml−1 by ultrafiltration using a YM-10 membrane (Merck–Millipore, Billerica, Massachusetts, USA) and filtered through a 0.1 µm membrane (Merck–Millipore). The pooled solution without buffer reagents was used in crystallization trials. Sparse-matrix crystal screening was performed using Crystal Screen HT, Index HT (Hampton Research, Aliso Viejo, California, USA), Wizard I and II and Cryo I and II (Emerald BioSystems, Bainbridge Island, Washington, USA). Sitting-drop vapour-diffusion trials were set up using the Thermo Scientific Matrix Hydra II (Thermo Fisher Scientific Inc., Hudson, New Hampshire, USA) automated liquid-handling system in 96-well Intelli-Plates (Art Robbins Instruments, Sunnyvale, California, USA) at 293 K using 50 µl reservoir solution. Each drop consisted of 0.3 µl protein solution and 0.3 µl reservoir solution. Within 2 weeks, several tiny rectangular plate-shaped crystals were observed under several conditions using polyethylene glycol 3350 or polyethylene glycol 6000 as a precipitant.
The diffraction data were collected from a single crystal on beamline BL-1A of the Photon Factory (PF), High Energy Accelerator Research Organization, Tsukuba, Japan. The crystal was scooped up in a nylon CryoLoop (Hampton Research) and then flash-cooled in a nitrogen-gas stream at 95 K. Diffraction data were collected with 0.75 s exposures for 1° oscillations over a total of 360° at a wavelength of 1.000 Å using a Quantum 270 CCD detector (ADSC, Poway, California, USA). Data were integrated and scaled using the programs DENZO and SCALEPACK from the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
OsLGalDH could be overexpressed in a stable soluble form in E. coli, with a yield of 5 mg per litre culture broth. The purified protein showed a single band in SDS–PAGE (Fig. 1 ▶) and was eluted as a single peak from the gel-filtration column chromatography. The apparent molecular weight was 35 000 Da from SDS–PAGE and 34 000 Da from gel-filtration chromatography. Considering the calculated molecular weight of 33 500 from the amino-acid sequence, OsLGalDH should be a monomeric protein in solution.
Figure 1.
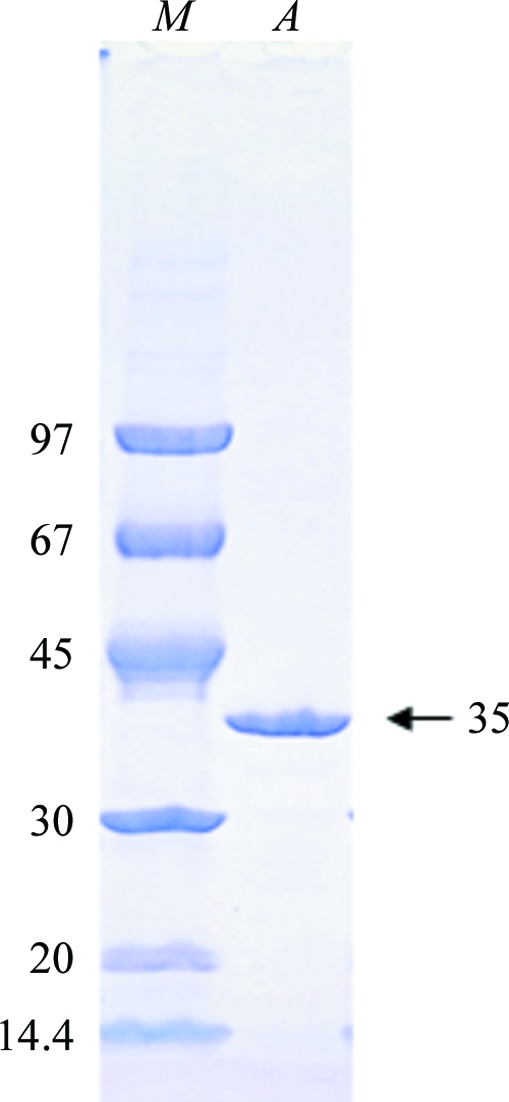
SDS–PAGE of purified OsLGalDH. Lane M, molecular-weight markers (labelled in kDa); lane A, OsLGalDH after gel-filtration column chromatography.
Diffraction-quality crystals were obtained from a crystallization solution consisting of 0.1 M bis-tris pH 6.5, 0.2 M MgCl2, 25% polyethylene glycol 3350 (Fig. 2 ▶). The crystal diffracted to a maximum resolution of 1.2 Å using synchrotron radiation at PF (Fig. 3 ▶). The crystal belonged to space group P21, with unit-cell parameters a = 46.8, b = 54.9, c = 56.9 Å, β = 102.3°. The processing statistics of the collected data are summarized in Table 1 ▶. Assuming that the asymmetric unit of the crystal contained one OsLGalDH molecule, the Matthews coefficient was calculated to be 2.1 Å3 Da−1 and the solvent content was 42.3% (Matthews, 1968 ▶).
Figure 2.
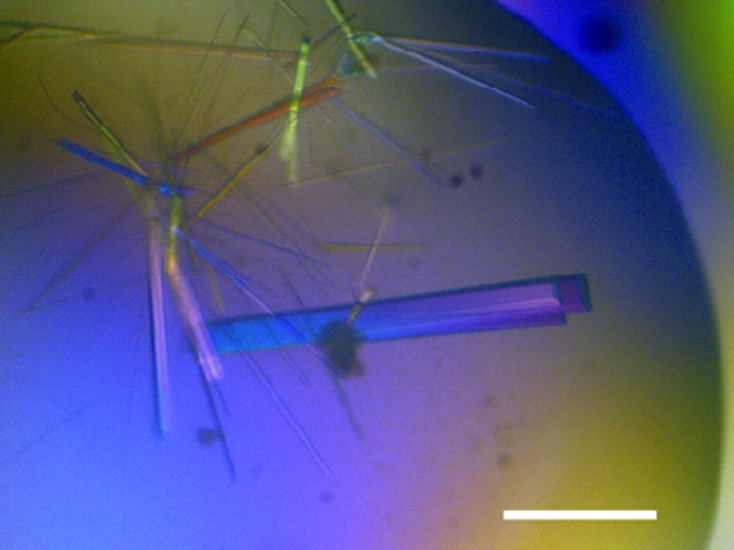
Crystals of OsLGalDH. The scale bar represents 300 µm.
Figure 3.
Diffraction image of an OsLGalDH crystal.
Table 1. Data-collection statistics for the OsLGalDH crystal.
Values in parentheses are for the outermost resolution shell.
| Beamline | BL-1A, PF |
| Detector | ADSC Quantum 270 CCD |
| Wavelength (Å) | 1.000 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 46.8, b = 54.9, c = 56.9, β = 102.3 |
| Resolution range (Å) | 50.0–1.2 (1.22–1.20) |
| R merge † | 0.073 (0.560) |
| Completeness (%) | 99.1 (89.6) |
| Multiplicity | 7.0 (5.8) |
| Unique reflections | 87015 (3919) |
| Total reflections | 608744 |
| Average I/σ(I) | 17.4 (4.1) |
| Mosaicity range (°) | 0.36–1.36 |
| Overall B factor from Wilson plot (Å2) | 12.0 |
R
merge = 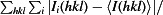
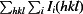 , where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all i observations of reflection hkl.
, where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all i observations of reflection hkl.
Crystallization and structure determination of OsLGalDH are the first step towards identifying the substrate-binding residues of this enzyme and gaining insight into its mechanism of action. As its amino-acid sequence homology to NAD/NADP-dependent oxidoreductases with known structures is less than 29%, it appeared that it would be difficult to determine the phase by the molecular-replacement (MR) method. The Phaser MR (McCoy et al., 2007 ▶) method with a reference model constructed from bacterial oxidoreductase (PDB entry 3lut; Chen et al., 2010 ▶) using the program CHAINSAW (Stein, 2008 ▶) in the CCP4 program suite (Winn et al., 2011 ▶) gave a poor initial model with an R factor of 0.522. Model building and refinement of the model are currently in progress.
Acknowledgments
We would like to thank the beamline researchers and staff at PF for the diffraction data collection.
References
- Chatterjee, I. B., Chatterjee, G. C., Ghosh, N. C., Ghosh, J. J. & Guha, B. C. (1960). Biochem. J. 74, 193–203. [DOI] [PMC free article] [PubMed]
- Chen, X., Wang, Q., Ni, F. & Ma, J. (2010). Proc. Natl Acad. Sci. USA, 107, 11352–11357. [DOI] [PMC free article] [PubMed]
- Conklin, P. L., Norris, S. R., Wheeler, G. L., Williams, E. H., Smirnoff, N. & Last, R. L. (1999). Proc. Natl Acad. Sci. USA, 96, 4198–4203. [DOI] [PMC free article] [PubMed]
- Dowdle, J., Ishikawa, T., Gatzek, S., Rolinski, S. & Smirnoff, N. (2007). Plant J. 52, 673–689. [DOI] [PubMed]
- Du, J., Cullen, J. J. & Buettner, G. R. (2012). Biochim. Biophys. Acta, 1826, 443–457. [DOI] [PMC free article] [PubMed]
- Gatzek, S., Wheeler, G. L. & Smirnoff, N. (2002). Plant J. 30, 541–553. [DOI] [PubMed]
- Ishikawa, T., Dowdle, J. & Smirnoff, N. (2006). Physiol. Plant. 126, 343–355.
- Kikuchi, S. et al. (2003). Science, 301, 376–379.
- Li, M., Ma, F., Liang, D., Li, J. & Wang, Y. (2010). PLoS One, 5, e14281. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mapson, L. W. & Breslow, E. (1958). Biochem. J. 68, 395–406. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Muramoto, T., Tsurui, N., Terry, M. J., Yokota, A. & Kohchi, T. (2002). Plant Physiol. 130, 1958–1966. [DOI] [PMC free article] [PubMed]
- Nishikimi, M., Fukuyama, R., Minoshima, S., Shimizu, N. & Yagi, K. (1994). J. Biol. Chem. 269, 13685–13688. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Stein, N. (2008). J. Appl. Cryst. 41, 641–643.
- Wang, J., Lad, L., Poulos, T. L. & Ortiz de Montellano, P. R. (2005). J. Biol. Chem. 280, 2797–2806. [DOI] [PubMed]
- Wheeler, G. L., Jones, M. A. & Smirnoff, N. (1998). Nature (London), 393, 365–369. [DOI] [PubMed]
- Winn, M. D. et al (2011). Acta Cryst. D67, 235–242.