Abstract
This study aimed at characterizing the phenotypic and toxigenic status of circulating strains of cholera during outbreaks in Nigeria, employing molecular typing techniques. Two hundred and one samples of rectal swabs, stool, vomitus, water (from the well, borehole, sachet, stream, and tap) and disinfectants (sodium hypochlorite) were collected from three states in the country. The samples were inoculated on thiosulphate-citrate bile salt-sucrose (TCBS), Cary-Blair transport medium and smeared on glass slides for direct examination. The Vibrio cholerae isolates were serotyped, biotyped, and characterized using PCR of the cytotoxin gene A (ctxA), wbeO1, and wbfO139 gene primer. Of the 201 samples screened, 96 were positive for V. cholerae O1 (48%), with 69 (72%) positive for ctxA gene. The results from this study showed that the circulating strains of cholera in Nigeria were of Ogawa serotype, also observed in other outbreaks in Nigeria (1991, 1992, and 1996). However, the strains were of the Classical biotype and were mainly (72%) ctxA gene-positive. This current investigation has confirmed the production of cholera toxin by the circulating strains, and this could be harnessed for possible cholera vaccine production in Nigeria.
Key words: Cholera toxin gene A, Classical, Ogawa, V. cholerae, Nigeria
INTRODUCTION
Vibrio cholerae is the causative agent of cholera. It is acquired by consuming food or water contaminated with V. cholerae; strains belonging to O1 and O139 serogroups are agents of endemic and pandemic cholera, a potentially life-threatening diarrhoea that produces characteristic rice-water stool. It is a major problem in the developing countries and has been linked to poverty and poor sanitation (1,2).
Epidemiologically, cholera most often occurs in explosive outbreaks throughout several regions simultaneously. Pandemics of cholera have followed progressive patterns, affecting many countries across the continent and over many years (3,4).
Epidemic V. cholerae are classified into two serogroups O1 and O139 as well as two biotypes El Tor and Classical. The ongoing seventh pandemic is due to V. cholerae serogroup O1 biotype El Tor. There are also two serotypes of epidemic cholera, Ogawa and Inaba (5,6).
Between 1995 and 2005, a total of 632 cholera outbreaks were reported worldwide; 66.0% of the total cases and 87.6% of fatal cases were reported from sub-Saharan Africa (7). The World Health Organization (WHO) annual figures on global cholera incidence, which are based on official cases reported by affected countries, are believed to be under-estimated due to limitations relating to a lack of adequate surveillance systems (8). In addition, the actual number of cholera cases globally is estimated to be much higher than officially reported (8) because outbreaks are often not reported to avoid the risk of travel and trade embargos on the affected country. According to WHO, only five countries (Burundi, Cameroon, the Democratic Republic of Congo [DRC], Ghana, and Tanzania) have reported cases of cholera every year since 1990 (9). Prompt and accurate diagnosis of V. cholerae, therefore, remains a key step in cholera outbreak surveillance, which can greatly influence rapid intervention and prevention to minimize spread of disease and mortality.
Due to inadequate disease notification system in the country, it is believed that most cases of cholera are not reported due to poor surveillance systems. Fatality rates are 5% of total cases in Africa and less than 1% elsewhere (10).
Cholera exists as a seasonal disease in Nigeria, occurring annually mostly during rainy seasons. In most of the affected areas, cholera looks endemic since the cases occur round the year but with different magnitudes. It may occur in southern part of the state and later shift to the northern part. The disease is more pronounced in the border areas of the affected states, and the reason is attributed to migration of the population and personal hygiene of the affected people.
In Nigeria, the Federal Ministry of Health reported 37,289 cases and 1,434 deaths between January and October 2010 (11). Reported cases almost tripled compared to those of 2009, and 80% were women and children.
In 2010, cholera outbreak had been more widespread and deadly than in almost 20 years (11). The cholera epidemic was first reported at Taraba state, and it quickly spread to 17 more states, including some states in the southern part of Nigeria.
In a separate report by Federal Ministry of Health, it was shown that, in 2011 alone (till 18 November 2011), a total of 22,797 cases of cholera with 728 deaths and case-fatality rate (CFR) 3.2% has been recorded (12). This is, however, not as high as the recorded cholera cases in 2010. Hence, WHO described the 2010 cholera outbreak as the worst in Nigeria since 1991 when 7,654 people died of cholera out of 59,478 cases (13).
The toxigenic strains of V. cholerae produce the strains of V. cholerae carrying the cholera toxin gene (ctx) and have the potential to produce cholera toxin (CT). These toxigenic strains are responsible for cholera epidemics. Water is recognized as the most important vehicle for cholera transmission (2).
In Africa and, indeed, Nigeria which is one of the most-hit countries, the cause of cholera has been attributed to heavy seasonal rainfall and poor sanitation.
During the 2010 cholera outbreak in Nigeria, Nigerian Institute of Medical Research Emergency Response Team (NIMRERT) was deployed in some affected states, specifically Borno, Bauchi and Gombe states with the aim of isolating and characterizing the circulating causative agents during outbreaks, using serologic characterization, biotyping, and PCR analysis of the ctxA, wbeO1, and wbfO139 genes.
Ethical approval was obtained from the Institutional Review Board (NIMR-IRB) of Nigerian Institute of Medical Research before the start of the project.
MATERIALS AND METHODS
Sample collection
Rectal swabs were collected from the in-patients in the various treatment centres and/or camps visited and then inoculated as specified below. Three rectal swabs were collected from each participant or legal representative who consented during the outbreak, with the aid of the sterile swab sticks.
In total, 201 samples were collected from the three states visited—Borno, Gombe, and Bauchi—comprising 171 rectal swabs, 20 water samples (i.e. six from well, three from borehole, four from sachet, two from stream, one from tap, and four disinfectant solutions), 5 stool samples, 5 vomitus samples (from some patients among the 171). One of the swabs was used in inoculating the Cary-Blair transport medium, the other streaked directly on the thiosulphate-citrate bile salts-sucrose agar (TCBS), and the third smeared directly on slide for Gram-staining. Other samples, such as vomitus, water, sewage, and disinfectants (sodium hypochlorite) were collected and inoculated in alkaline peptone water and incubated for further processing. The water samples were collected randomly from the drinking-water sources of the participants, such as well, borehole, sachet, stream, and tap while the vomitus samples were collected from some of the in-patients. The disinfectants used in the treatment sites were also sampled to rule out contamination while the sewage samples were randomly selected based on information given by the participants in the questionnaire.
The samples were further processed on arrival at the laboratory.
It is pertinent to mention that the samples were collected from new cases without antibiotics and some prolonged admission cases that had been on antibiotics.
Laboratory diagnosis
Isolation and confirmation of V. cholerae strains
Samples transported in Cary-Blair medium were separately inoculated into alkaline peptone water (pH 8.6) for growth enrichment at 37 oC for 24 h. Each bacterial culture was then subcultured onto TCBS agar at 37 0C for 24 h. Suspected Vibrio cholerae colonies were picked and subcultured for purity in a new TCBS plate called the purity plate. Each pure isolate of V. cholerae was serotyped for Ogawa and Inaba classification by slide agglutination, using serotype-specific antisera (Biorad, USA). Serogrouping of the isolates, based on the use of serogroup O1 and O139-specific antisera, was done using the slide-agglutination technique. Isolates other than suspected V. cholerae were also identified after preparing purity plates on MacConkey agar, using standard methods.
Characterization of the isolated strains
The isolated organisms were identified by growth of yellow colonies on TCBS agar and production of purple colour on reaction with oxidase reagent. The isolates were tested for lactose and glucose fermentation, using Kligers Iron agar (KIA), motility, and indole and urease production (MIU). Methyl Red test and String test were also performed.
Storage of strains
Each isolate was stored in a cryovial containing glycerol (20%) tryptone soy broth at −20 0C.
Serologic characterization
Suspected colonies from TCBS agar were serogrouped with O1 and O139-specific antisera by slide agglutination. These were then serotyped with Ogawa and Inaba antisera (BioRad, USA).
Antibiotics susceptibility testing
The V. cholerae isolates were subcultured onto Mueller Hinton agar and their susceptibility patterns determined following procedure as described by CLSI (14). The antibiotics used were: augmentin, ofloxacin, gentamicin, nalidixic acid, nitrofurantoin, cotrimoxazole, amoxicillin, tetracycline, and doxycycline.
Genomic DNA preparation
Genomic DNA of each isolate was prepared by overnight boiling of broth culture and purification of the isolated DNA.
Amplification of the cytotoxin (ctxA), wbeO1 and wbfO139 genes
The ctxA gene VCT1 [5’-CTCAGACGGGATTTGTTAGGCACG-3’], sense strand and VCT2 [5’-TCTATCTCTGTAGCCCCTATTACG-3’], antisense strand, the wbeO1 gene : O1 wbeF [5’- GTTTCACTGAACAGATGGG-3’], O1 wbeR [5’-GGTCATCTGTAAGTACAAC-3’] and wbfO139 gene O139F2 [5’-AGCCTCTTTATTACGGGTGG-3’], sense strand and O139R2 [5’-GTCAAACCCGATCGTAAAGG-3’] from each isolate were amplified by PCR with 50-200 ng of DNA mixed with PCR buffer (20 mM Tris-HCl, pH 8.3; 50 mM KCl) containing 200 micromole each of dNTP, 1.5 mM MgCl2, 30 picomole each of forward and reverse primer, and 1 unit of Taq polymerase. The reaction was conducted in a thermocycler (Eppendorf Mastercycler pro, Hamburg), with an initial denaturation at 95 0C for 5 min. This was followed by 35 rounds of amplification defined by denaturation at 95 0C for 1 min, primer annealing done at 60 0C for 1 min for the ctxA gene and the wbeO1 gene while the annealing temperature for the wbfO139 gene was 55 °C for 1 min. The primer extension for all three genes was at 72 °C for 1 min. A final extension step at 72 °C for 5 min was done, and the PCR product was kept at 4 °C prior to analysis.
Agarose gel electrophoresis
The PCR products were electrophoresed horizontally on 1.5% agarose gel that was prestained with ethidium bromide at 100 volt for 45 min, and 100 bp ladder (Promega, USA) was used as DNA molecular weight standard. The amplicon-sizes were visualized under UV light for ctxA gene, wbeO1, and wbf O139 genes to be 302 bp, 192 bp, and 449 bp respectively. Photo documentation was done with the aid of a photo-documentation system by CLINX Science Instruments, China.
RESULTS
In total, 201 samples were collected from the three states visited—Borno, Gombe, and Bauchi—and the distribution is shown in Table 1.
Table 1.
Type and sources of samples collected according to state
| State | Rectal swab | Stool | Vomitus | Water sample | Disinfectant solution (Hypochlorite) | Total | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Borehole | Sachet | Well | Stream | Tap | ||||||
| Bauchi | 100 | 1 | 3 | 3 | 4 | 1 | 112 | |||
| Borno | 48 | 2 | 1 | - | 2 | - | - | - | 4 | 57 |
| Gombe | 23 | 2 | 1 | - | 2 | 2 | 1 | 1 | 0 | 32 |
| Total | 171 | 5 | 5 | 3 | 4 | 6 | 2 | 1 | 4 | 201 |
Ninety-six isolates of V. cholerae were characterized, and all were of Ogawa serotype. All the vomitus samples grew V. cholerae O1 Ogawa; the organism was also isolated from all the water samples and the disinfectant solution collected, except in the sachet water from Gombe state (Table 2).
Table 2.
Distribution of isolated organisms according to state
| State | V. cholerae O1 | V. cholerae non-O1 | Inaba | Ogawa | Classical biotype | El Tor biotype | V. mimicus | V. parahaemolyticus | Proteus spp. | P. aeruginosa | K. pneumonia |
| Bauchi | 48 | 32 | Nil | 48 | 48 | Nil | 20 | 6 | 8 | 4 | Nil |
| Borno | 35 | 20 | Nil | 35 | 35 | Nil | 10 | 10 | 12 | 6 | 4 |
| Gombe | 13 | 2 | Nil | 13 | 13 | Nil | 4 | 4 | 3 | 2 | Nil |
| Total | 96 | 54 | Nil | 96 | 96 | Nil | 34 | 20 | 23 | 12 | 4 |
Fifty-four of the isolated organisms were of the Vibrio species other than V.cholerae, and those species were identified as V. mimicus and V. parahaemolyticus. Other organisms isolated were Proteus species, Pseudomonas aeruginosa, and Klebsiella species (Table 2).
Generally, all the isolated Vibriocholerae strains were sensitive to ofloxacin, gentamicin in all the states and moderately sensitive to tetracycline and doxycycline in Gombe state. Most of these were also moderately sensitive to nitrofurantoin and nalidixic acid, though not a drug of choice. The organisms were resistant to augmentin, cotrimoxazole, and amoxicillin (Table 3).
Table 3.
Antibiotics susceptibility patterns of the isolated Vibrio cholerae strains
| Antibiotics | Bauchi | Borno | Gombe | Remarks | |||
| Sensitive (%) | Resistant (%) | Sensitive (%) | Resistant (%) | Sensitive (%) | Resistant (%) | ||
| Augmentin | 18 | 82 | 8 | 92 | 32 | 68 | Resistant |
| Ofloxacin | 92 | 8 | 97 | 3 | 79 | 21 | Sensitive |
| Gentamicin | 89 | 11 | 92 | 8 | 63 | 37 | Sensitive |
| Nalidixic acid | 60 | 40 | 82 | 11 | 21 | 79 | Moderate |
| Nitrofurantoin | 78 | 22 | 85 | 15 | 68 | 32 | Moderate |
| Cotrimoxazole | 17 | 83 | 5 | 95 | 0 | 100 | Resistant |
| Amoxicillin | 0 | 100 | 0 | 100 | 26 | 74 | Resistant |
| Tetracycline | 89 | 11 | 93 | 7 | 58 | 42 | Sensitive |
| Doxycycline | 91 | 9 | 98 | 2 | 68 | 32 | Sensitive |
Sixty-nine (72%) of the V. cholerae strains were ctxA-positive, and none of the non-O1 strains tested was positive for wbfO139 gene.
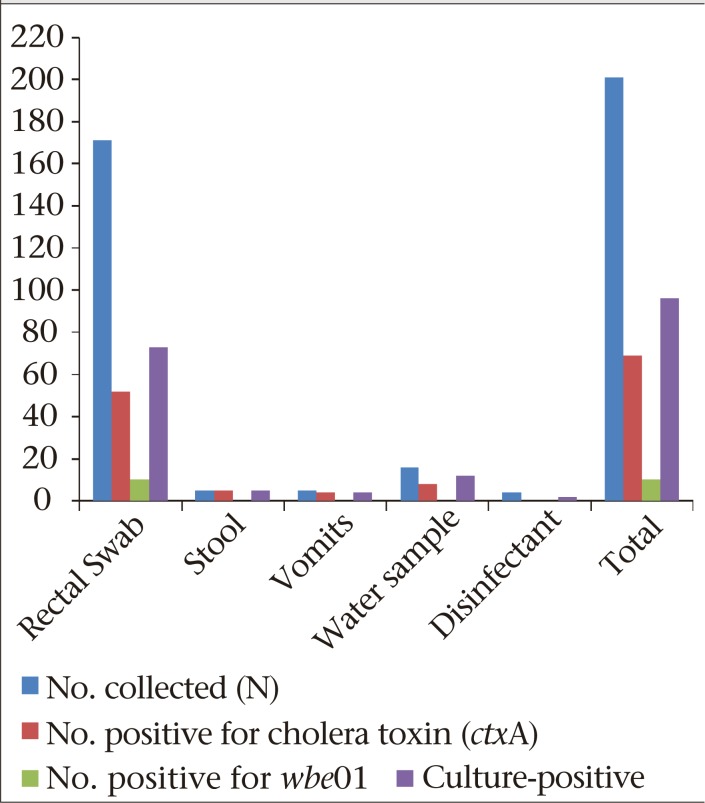
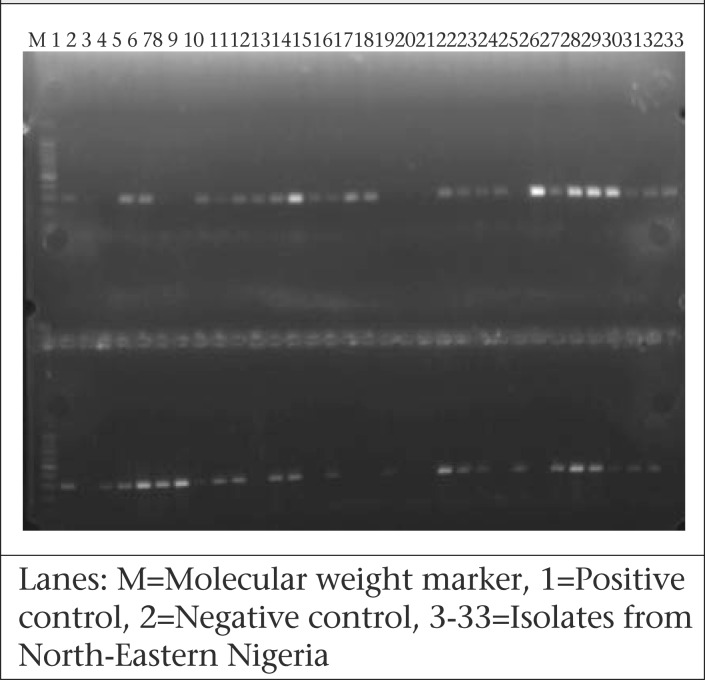
Figure 1 shows the frequency of distribution of ctxA gene and wbeO1 amongst culture-positive samples while Figure 2 shows agarose gel electrophoresis of ctxA gene amongst V. cholerae isolates from North-Eastern Nigeria. Figure 3 shows one of the sources of water used by the patients that was contaminated with cholera germ.
Figure 1.
Cholera toxin (ctxA) and wbeO1 from culture-positive samples
Figure 2.
Representative agarose gel electrophoresis of V. cholerae O1 strains positive for ctxA gene, isolated during the 2010 cholera outbreak in North-Eastern Nigeria
Figure 3.
Photo of a well-water source for water vendors in the study area
DISCUSSION
The results from this study showed that the V. cholerae strains isolated from the 2010 outbreak in North-Eastern Nigeria were all of Ogawa serotypes and Classical biotype. This is interesting because, in the previous outbreaks in Nigeria, it had always been Ogawa serotype (15,16); however, not many reports have been published on the Classical biotype. A few reported so far have shown the El Tor biotype to predominate and affirming it as the seventh pandemic of cholera (17). In a report by Opaj et al. (17), the isolates were obtained from North-Central Nigeria, and these were of Inaba serotype, although some non-diarrhoeal stool was tested in this study, and these were not outbreak strains. Authors concluded, however, that V. cholerae was endemic in their environment. In an earlier report from the same environment in 1991, the Ogawa serotype was reported to be prevalent probably because these were outbreak strains (16).
From a recent work (18), two V. cholerae isolates obtained from a previous outbreak in Nigeria in 2008 (region not stated) were confirmed as Ogawa serotype and biotype El Tor, using PCR analysis of biotyping genes tcpA. Our study and previous studies showed that the V. cholerae O1 strains circulating in Nigeria included both Inaba and Ogawa, although there might be a possibility that Inaba could result from endemicity while the outbreak strains could be of Ogawa serotype. A previous study has reported that the switching of serotypes resulted from a change in the genetic make-up of the wbtTgene, which determined the specificity of Ogawa (19).
Although the Classical biotype has been reported to be extinct, it is still present in Nigeria. There seems to be a shift between the Classical and El Tor biotype as reported in some studies where El Tor strains that produced cholera toxin of the Classical type were isolated and were, therefore, designated El Tor variants (20).
Outbreaks of cholera epidemic are usually characterized by endotoxin-producing V. cholerae and possibly antibiotics-resistant type. In the 2010 outbreak in Nigeria, as also observed in other outbreaks in Nigeria (1991, 1992, and 1996), the V. cholerae O1 were generally sensitive to ofloxacin, gentamicin, and moderately sensitive to nalidixic acid in all states, except Gombe state where resistance to tetracycline and doxycycline was as high as 42% and 32% respectively.
On the other hand, the 2010 outbreak strains of Haiti were resistant to nalidixic acid, though not the drug of choice (21). Both 2010 outbreak strains from Nigeria and Haiti were, however, resistant to cotrimoxazole, and the Nigerian strains were moderately sensitive to tetracycline.
In a previous study by Jos in Nigeria (17), all V. cholerae isolates were resistant to ampicillin, penicillin, chloramphenicol, and cloxacillin while these were sensitive to tetracycline (the drug of choice), ofloxacin, and erythromycin. Generally, from this study and previous ones, the isolates were sensitive to tetracycline, the drug of choice.
According to a recent report by Bhattacharya et al. (22), tetracycline-resistant V. cholerae O1 have been reported in a number of studies on major epidemics from Latin America, Tanzania, Bangladesh, and India. Resistance rates of as high as 76% were reported after a 5-month extensive use of the drug for treatment and prophylaxis.
Drug-resistant V. cholerae strains are, therefore, a global health concern since infections resulting from these could be more severe and difficult to treat. In addition, there could be higher case-fatality rates, prolonged hospitalizations, increased healthcare costs, and probably more secondary infections. An outbreak in Guinea-Bissau led to increased case-fatality rates from 1% to 5.3% after the outbreak strain acquired multidrug resistance (23).
It is, therefore, paramount that antimicrobials are given to patients suffering from cholera only when they have severe dehydration or other conditions that truly warrant their use. This could help reduce spread of antimicrobial drug resistance.
In addition, continuous surveillance of antimicrobial drug susceptibility patterns of V. cholerae should be done to lessen illness and death from the disease.
The current investigation went further than previous studies in Nigeria to confirm the production of cholera toxin by these strains, and it was found that 72% of the isolated strains had the ctxA gene responsible for the production of cholera toxin. However, this study has not done in-depth molecular studies to look into cholera toxin gene B (ctxB)-sequencing as well as screening for clonal analysis using the pulsed-field gel electrophoresis. This aspect of the work would form the next stage in the study, in which we hope to answer some of these questions concerning our outbreak strains.
None of the non-O1 V. cholerae strains was V. cholerae O139 as there was no amplification with the wbfO139 gene. This goes to confirm that there is currently no V. cholerae O139 in Nigeria, and our strains are not similar to those from Asia that have reported cases of V. cholerae O139 (24,25). This was also buttressed by an in-depth study compared to the Haiti outbreak strains (18).
Conclusions
It was observed in this study that the same serotype of V. cholerae has been circulating during outbreaks in Nigeria, and these were of the O1 Classical biotype and Ogawa serotype. More than two-thirds of these carried the enterotoxin (ctxA) gene while no O139 was isolated in the study. Nevertheless, the current drug of choice proved effective but concerted efforts should be made to unravel the factors escalating the outbreak while other preventive channels, such as health education and awareness, vaccine, and clean water and sanitation, are other plausible measures.
REFERENCES
- 1.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–14. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Cholera, 2000. Wkly Epidemiol Rec. 2001;76:233–40. [PubMed] [Google Scholar]
- 3.Kaper JB, Morris JG, Jr, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith DC, Kelly-Hope LA, Miller MA. Review of reported cholera outbreaks worldwide, 1995-2005. Am J Trop Med Hyg. 2006;75:973–7. [PubMed] [Google Scholar]
- 5.Chen C-H, Shimada T, Elhadi N, Radu S, Nishibuchi M. Phenotypic and genotypic characteristics and epidemiological significance of ctx+ strains of Vibrio cholerae isolated from seafood in Malaysia. Appl Environ Microbiol. 2004;70:1964–72. doi: 10.1128/AEM.70.4.1964-1972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alam M, Nusrin S, Islam A, Bhuiyan NA, Rahim N, Delgado G. Cholera between 1991 and 1997 in Mexico was associated with infection by classical, El, Tor, and El Tor variants of Vibrio cholerae. J Clin Microbiol. 2010;48:3666–74. doi: 10.1128/JCM.00866-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bompangue Nkoko D, Giraudoux P, Plisnier PD, Tinda AM, Piarroux M, Sudre B, et al. Dynamics of cholera outbreaks in Great Lakes region of Africa, 1978-2008. Emerg Infect Dis. 2011;17:2026–34. doi: 10.3201/eid1711.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alam M, Hasan NA, Sultana M, Nair GB, Sadique A, Faruque ASG. Diagnostic limitations to accurate diagnosis of cholera. J Clin Microbiol. 2010;48:3918–22. doi: 10.1128/JCM.00616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Global Task Force on Cholera Control (2010). Weekly Epidemiological Record: cholera articles (http://www.who.int/cholera/statistics/en, accessed on 12 January 2010).
- 10.World Health Organization. Cholera in 1997. Wkly Epidemiol Rec. 1998;73:201–8. [PubMed] [Google Scholar]
- 11.Nigerian Guardian Newspaper (2010) Cholera news: cholera cases now 37, 289, 1434 deaths. (http://ngrguardiannews.com/2010/10/28/20959, accessed on 28 October 2010).
- 12.Nigeria. Federal Ministry of Health Weekly Epidemiology Report. 2011;1(26):1–8. [Google Scholar]
- 13.World Health Organization. Cholera vaccines: WHO position paper. Wkly Epidemiol Rec. 2010;85:117–28. [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute . Quality manual. 3rd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2003. pp. 69–79. [Google Scholar]
- 15.Adesiyun AA, Adekeye JO, Umoh JU, Nadarajah M. Studies on well water and possible health risks in Katsina, Nigeria. J Hyg (Lond) 1983;90:199–205. doi: 10.1017/s0022172400028862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oyediran ABO. Disease epidemics. Med Today. 1991;19:30. [Google Scholar]
- 17.Opajobi SA, Kandakai-Olukemi YT, Mawak JD, Olukemi MA, Bello CSS. Vibrio cholerae 01 infections in Jos, Nigeria. Afr J Clin Exper Microbiol 20045:260–264.
- 18.Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, et al. Characterization of toxigenic Vibrio cholerae from Haiti, 2010-2011. Emerg Infect Dis. 2011;17:2122–9. doi: 10.3201/eid1711.110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tapchaisri P, Na-Ubol M, Tiyasuttipan W, Chaiyaroj SC, Yamasaki S, Wongsaroj T. Molecular typing of Vibrio cholerae O1 isolates from Thailand by pulsed-field gel electrophoresis. J Health Popul Nutr. 2008;26:79–87. [PMC free article] [PubMed] [Google Scholar]
- 20.Ang GY, Yu CY, Balqis K, Elina HT, Azura H, Hani MH. Molecular evidence of cholera outbreak caused by a toxigenic Vibrio cholerae O1 El Tor variant strain in Kelantan, Malaysia. J Clin Microbiol. 2010;48:3963–9. doi: 10.1128/JCM.01086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sjölund-Karlsson M, Reimer A, Folster JP, Walker M, Dahourou GA, Batra DG. Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg Infect Dis. 2011;17:2151–4. doi: 10.3201/eid1711.110720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharya K, Kanungo S, Sur D, Lal Sarkar B, Manna B, Lopez AL, et al. Tetracycline-resistant Vibrio cholerae O1, Kolkata, India. Emerg Infect Dis. 2011;17:568–9. doi: 10.3201/eid1703.101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalsgaard A, Forslund A, Petersen A, Brown DJ, Dias F, Monteiro S. Class 1 integron-borne, multiple-antibiotic resistance encoded by a 150-kilobase conjugative plasmid in epidemic vibrio cholerae O1 strains isolated in Guinea-Bissau. J Clin Microbiol. 2000;38:3774–9. doi: 10.1128/jcm.38.10.3774-3779.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair GB, Ramamurthy T, Bhattacharya SK, Mukhopadhyay AK, Garg S, Bhattacharya MK. Spread of Vibrio cholerae O139 Bengal in India. J Infect Dis. 1994;169:1029–34. doi: 10.1093/infdis/169.5.1029. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, et al. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20:201–7. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]