Abstract
Monogenic nephrotic syndrome (NS) is responsible for only a small percentage of cases of NS, however information from the studies of this unique cohort have dramatically improved our understanding of disease pathogenesis. The use of genetic testing in the management of patients with NS poses a unique challenge to clinicians with regard to whom to test and how to utilize the information obtained from testing in the clinical setting. In our view, there are presently not enough data to justify routine testing of all patients with NS; however, testing is warranted in patients with congenital nephrotic syndrome (CNS; onset at 0-3 months), infantile NS (onset at 3-12 months), a family history of NS, and those in whom NS is associated with other congenital malformations. The family should be given complete and unbiased information on the potential benefits and risks associated with therapy, including the reported outcomes of treatment in patients with similar mutations. Based on the data available in the literature, there is probably no indication for intensive immunosuppressive treatment in monogenic NS if complete or partial remission has not been achieved within 6 weeks of starting treatment. We advocate routine genetic testing of family members of individuals with genetic forms of NS prior to living-related kidney transplantation. There is a strong need for prospective multi-center studies to more completely determine the burden of disease due to monogenic NS, as well as randomized controlled trials to clarify the presence or absence of clinical responses of monogenic NS to available therapies.
INTRODUCTION
Nephrotic syndrome (NS) is a common condition seen in nephrology clinics in both developed and developing countries, although the prevalence in the general population is estimated to be only 16/100,000 children.1 Most pediatric cases in the Western Hemisphere are steroid-sensitive nephrotic syndrome (SSNS). Conversely, about 20% of cases are steroid-resistant (SRNS).1-2 The SRNS variants are often due to focal segmental glomerulosclerosis (FSGS) and are often characterized by rapid progression to end-stage kidney disease (ESKD), requiring dialysis and kidney transplantation within two to ten years of diagnosis. The pathogenesis of NS has not been clearly delineated. However, a landmark study by the Tryggvason group in 1998 identified mutations in nephrin (NPHS1), an essential component of the podocyte slit diaphragm, as a cause of congenital NS (CNS).3 This finding introduced the importance of the podocyte in the pathogenesis of NS, and has led to the proposal that most cases of NS are due to podocyte defects, and should therefore be classified as “podocytopathies”.4 Since the finding of nephrin mutations, there have been several studies reporting mutations in other genes in both pediatric and adult onset NS.5-21 Notably, most of the genes identified to date localized to the podocyte or the slit diaphragm, thereby confirming the importance of podocyte dysfunction in the pathogenesis of NS. A rare exception is LAMB2, whose gene product is enriched in the glomerular basement membrane.8. Tables 1 and 2 catalogue the genes identified to date in both autosomal recessive (AR) and autosomal dominant (AD) NS as well as the phenotype associated with each of these genes. Moreover, the most prevalent AR genes and AD genes are listed in Box 1. Despite these exciting findings, there are no large-scale population studies to delineate the disease burden due to monogenic NS. Nevertheless, extrapolation from selected adult case series seems to suggest that <10% of all cases of SRNS/FSGS, and <2% of all cases of NS, are probably due to single gene defects22. This raises the question of the clinical utility, relevance, and cost-effectiveness of large scale genetic testing in patients presenting with NS. There is therefore a need for the development of clear guidelines for genetic testing for both clinicians and researchers, as well as continued dialogue on the challenges posed by judicious clinical utilization of the information from both positive and negative genetic tests. This review articulates our collective opinion on the clinical indications for genetic testing in NS and the clinical utility of genetic testing in monogenic NS based on the evidence available in the literature to date. This article will focus on single gene defects, while complex inheritance issues are beyond the scope of this review.
Table 1.
Etiology of autosomal recessive nephrotic syndrome
| Genes | Locus | Phenotype |
|---|---|---|
| NPHS1/nephrin | 19q13.1 | Congenital nephrotic syndrome |
| FSGS | ||
| NPHS2/Podocin | 1q25-q31 | FSGS |
| NPHS3/PLCE1 | 10q23-q24 | DMS |
| FSGS | ||
| SMARCAL1 | 2q34-q36 | Syndromic immune complex nephritis (Schimke immune-osseous dysplasia) |
| LAMB2 | 3p21 | Syndromic DMS (Pierson syndrome) |
| FSGS | ||
| SCARB2 | 4q21.1 | Syndromic nephrotic syndrome |
| Nephrotic syndrome with C1q deposits | ||
| Progressive myoclonic epilepsy | ||
| COQ6 | 14q24 | Syndromic nephrotic syndrome |
| FSGS/DMS | ||
| Sensorineural deafness | ||
| Seizure disorder | ||
| Ataxia | ||
| PTPRO/GLEPP1 | 12p12 | FSGS/MCD |
| MYO1E | 15q21 | FSGS |
| ITGA3 | 17q21 | Syndromic nephrotic syndrome |
| Congenital nephrotic syndrome | ||
| Interstitial lung disease | ||
| Skin fragility | ||
FSGS= Focal segmental glomerulosclerosis, DMS= Diffuse mesangial sclerosis, MCD= Minimal change disease.
Table 2.
Etiology of autosomal dominant nephrotic syndrome
| Genes | Locus | Phenotype |
|---|---|---|
| CD2AP # | 6p12.3 | FSGS |
| WT1 + | 11p13 | Syndromic nephrotic syndrome |
| Denys Drash syndrome | ||
| Frasier syndrome | ||
| DMS | ||
| FSGS | ||
| α-actinin 4 | 19q13 | FSGS |
| TRPC6 | 11q21-q22 | FSGS |
| LMX1B | 9q34.1 | Nail patella syndrome |
| INF2 | 14q32 | FSGS |
| Syndromic FSGS (Charcot Marie tooth disease | ||
| ARHGAP24 | 4q20 | FSGS |
AR disease have been reported
Most mutations in WT1 are de-novo or germ line mutations
GENETICS OF NEPHROTIC SYNDROME
Monogenic NS may be inherited as an autosomal recessive (AR) or autosomal dominant (AD) condition, and may be either isolated or part of a multi-systemic inherited disorder. The major causes of AR NS are mutations in nephrin (NPHS1), podocin (NPHS2), and phospholipase C epsilon 1 (PLCE1) (Box 1). Indeed, two large cohort studies of children with hereditary NS have reported that 18-26% of all genetic causes of NS were due to mutations in NPHS223-24. In similar studies limited to NS in the first year of life, Hinkes et al. reported that mutations in NPHS1 and NPHS2 were responsible for 60% of all cases of infantile NS25. In another study restricted to children with the pathologic findings of diffuse mesangial sclerosis (DMS) alone, mutations in PLCE1 were found to be responsible for >25% of all cases of NS26. As in many other genetic conditions, most AR forms of NS typically have their onset in childhood, are highly penetrant, and are more likely to be associated with other malformations. In contrast, the major genetic causes of AD forms of NS include mutations in inverted formin 2 (INF2), transient receptor potential cation channel protein, type C6 (TRPC6) and actinin-alpha 4 (ACTN4) (Box 1). In three different studies, it was reported that mutations in INF2 are probably responsible for 16% of all cases of AD NS/FSGS in a predominantly adult population15, 27-28. Unlike AR disease, AD forms of NS typically have their onset during adulthood, are incompletely penetrant, and may even be asymptomatic. More recently, due to the availability of high throughput genomic techniques, genetic risk factors and complex inheritance due to gene/gene or gene/environment interactions are now being reported29-30. For example, variants in MYH9 and APOL1 were recently reported as risk factors for NS with pathological changes of FSGS in African American patients29-30. Presently there are no population-based data to clarify the overall disease burden due to single-gene defect forms of NS. However, the overall consensus is that they probably represent only a tiny fraction of all cases of NS, while the large majority of cases remain idiopathic.
The prevalence of single gene defects also varies with age, and sometimes with ethnicity. In studies that are limited to familial cases, up to 80% of cases in the first 3 months of life (CNS) and 50% in the first year of life, are due to mutations in one of four genes, specifically NPHS1, NPHS2, WT1 and LAMB225. In our collective experience, even among cases of obvious familial NS the cause is still unknown in about 80% of cases. Thus a negative result with candidate gene screening does not exclude hereditary disease. The primary aims of genetic testing are to establish a diagnosis in symptomatic individuals, and to also predict risk in family members. Thus there is a clear need for the development of standardized criteria for genetic testing in both children and adults who present with NS.
IS ROUTINE GENETIC TESTING FOR NEPHROTIC SYNDROME JUSTIFIED?
The clinician should consider the use of genetic testing similarly to any other diagnostic test. Before ordering a test, three pertinent questions should be addressed: 1) Is the result of this test likely to aid in diagnosis?, 2) Is the test result likely to alter the management of the patient or better inform a discussion of likely outcomes and prognosis?, and 3) Is the test result from the proband likely to produce data on risk to other family members? We believe the answers to at least one of these three questions should be “yes” before ordering genetic testing in NS. As discussed in the previous section, <2% of currently available genetic tests for NS are likely to identify a disease-causing mutation, raising the important question of cost versus benefit. Additionally, a negative test does not exclude monogenic NS, as hereditary NS and FSGS has been found to be profoundly heterogeneous, and multiple genes are almost certainly yet to be found. Based on these concepts, we do not recommend universal genetic testing at the present time, but fully support testing in subsets of NS patients in whom there would likely be actionable findings.
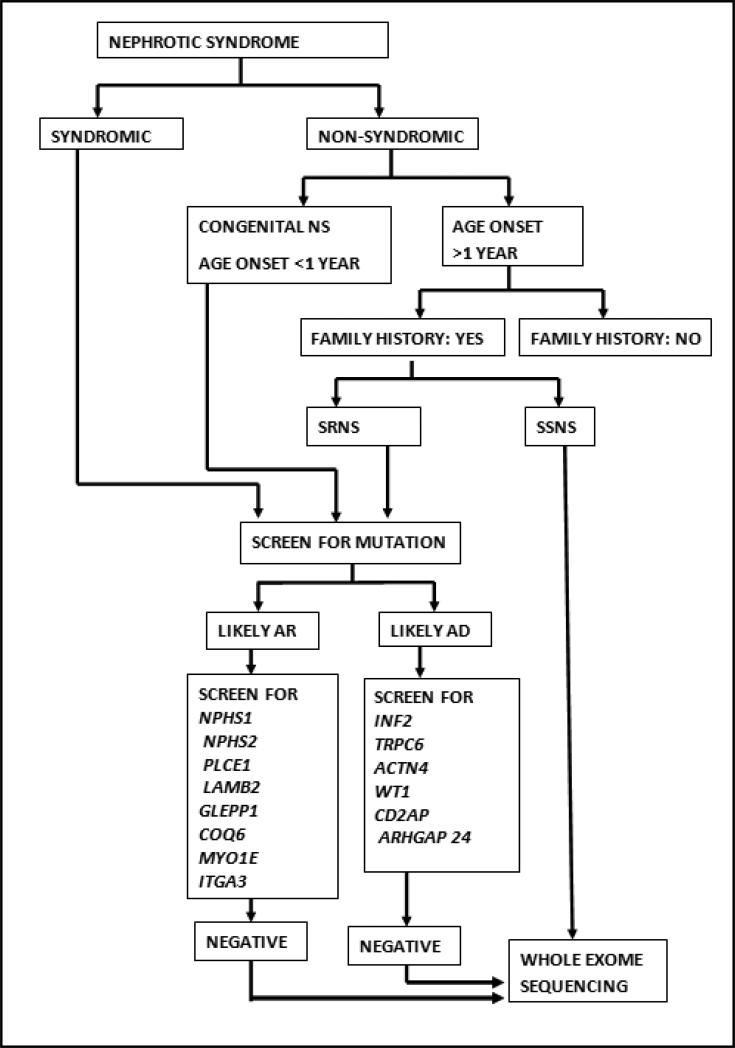
In a recent study of pediatric and adult onset NS in a Spanish cohort, Santin et al. found that a family history of NS and early onset disease were the most important risk factors for finding pathogenic mutations in known NS genes31. Thus, based on the epidemiologic data presented in the previous section and our personal experience, we recommend genetic testing for the following categories of patients: 1) All children presenting with congenital NS (NS with onset in the first 3 months of life), 2) All children presenting with infantile NS (NS onset from 3-12 months of life) 3) All patients in whom NS is part of other multiple malformations/syndromes, and 4) All patients with a family history of NS or chronic kidney disease (Box 1). Specifically, we propose the use of the algorithm shown in Figure 1 to determine which NS patients should receive genetic testing, and which specific tests should be ordered. While genetic testing will add to the overall cost of the management of a patient with NS, it is still less expensive than the monetary and emotional costs of chronic kidney disease. The advantages of genetic testing should not be viewed from only the point of view of therapeutic intervention. Other important advantages include better information for families on the likely clinical course of disease; better information to enhance the selection of appropriate transplant donors, and the ability to provide genetic counseling for other family members.
Figure 1.
Algorithm for genetic testing in nephrotic syndrome
METHODS OF TESTING
The availability of rapid and cost-effective genetic tests should not be seen as a replacement for detailed and meticulous clinical evaluation of patients by clinicians. Instead, it should be seen as a challenge to obtain a thoughtful family history and to construct pedigrees within the limited time available during consultation. Data generated from this type of clinical encounter will help in streamlining the choice of tests for the patient. In the U.S., the methods employed by most commercial laboratories (Clinical Laboratory Improvement Amendments: CLIA-certified laboratories), is the use of exon primers to identify mutations in panels of candidate genes for specific disorders. This is based on the understanding that mutations in single gene defects have invariably been in the coding regions of the gene of interest. The advantage of this is that the return of the results to patients is relatively expeditious and interpretation of positive results does not require robust bioinformatics support. However, interpretation of negative results is more problematic, since the patient may have either a mutation in non-coding regions of candidate genes or mutations in novel genes. The rapid technological advances in “next generation sequencing” will likely decrease the need for this candidate gene approach and may replace it as the test of choice in the near future. With this method, either the whole-genome (i.e. the total genome), or the coding regions of all the genes in the human genome (i.e. the whole-exome) are sequenced. The major advantage of this method is that the yield of positive results is likely to be higher because the test is not limited to known candidate genes, and mutations may be identified in previously unrecognized novel genes. Discovery of novel genes will not only aid in diagnosis, but will also have the potential to improve our understanding of the molecular mechanisms of disease in NS as well as provide potential molecular drug targets for therapy. At present, the use of whole-exome sequencing in genetic diagnosis is limited to research laboratories; the enormous amount of data generated with this method requires complex and well-organized bioinformatics support for analysis. In addition, whole-exome sequencing may identify deleterious variants that are immaterial to the disease being investigated. Most individuals have multiple ‘deleterious’ mutations in their genome that do not appear to cause morbidity or mortality32. Such technical advances also highlight the urgent clinical need for the development of accurate, ethical and concise approaches (such as clinical care guidelines) for communicating incidental but potentially actionable findings to affected families during the confined timeframes of clinic visits.
GENETIC TESTING AND CLINICAL UTILITIES
Availability of genetic testing
One of the most important early challenges for clinicians caring for patients with NS is to determine what genetic tests for NS are currently available. Fortunately, there is an outstanding free resource (www.genetests.org) which provides updated information about the availability of genetic testing for a vast array of genetic diseases, including NS. Sponsored by the National Center for Biotechnology Information (NCBI – http://www.ncbi.nlm.nih.gov/), the website enables users to search for information using relevant criteria, including the disease, gene, protein, laboratory performing the test, or laboratory director.
Insurance coverage for genetic testing
Insurance coverage for genetic testing in NS remains another important clinical challenge. Not all insurers currently reimburse for genetic testing in NS, since this is not yet considered the “standard of care”. Insurers may not reimburse “research” or “experimental” patient costs, or compensate for results from non-CLIA-approved research laboratories. Unfortunately, the lack of insurance coverage makes genetic testing for NS financially prohibitive for many families world-wide.
Treatment decisions in patients with positive genetic tests
There is undisputable evidence from published studies that the majority of cases of hereditary and familial NS due to single gene defects are steroid-resistant. The largest series so far are two studies by the Hildebrandt group that showed that virtually all cases of NS due to NPHS2 mutations were steroid-resistant, and that mutations in NPHS2 were rare in a cohort of children with steroid-sensitive NS23, 33. Furthermore, another study showed steroid-resistance in all patients with monogenic NS (irrespective of the genetic mutation) and only a partial response in a small percentage of patients34. However, there are anecdotal reports in the literature that individuals with NS due to genetic mutations may be responsive to immunomodulatory or chemotherapeutic agents12, 35-36. Moreover, some agents now used for the treatment of NS may also affect the podocyte actin cytoskeleton in addition to their immunosuppressive properties37-38. A recent study provides evidence that in addition to the effect on the immune system, the anti-proteinuric effect of cyclosporine may result from blocking the calcineurin-mediated dephosphorylation of synaptopodin, leading to stabilization of the actin cytoskeleton which is often disrupted in most cases of familial NS/FSGS37. As there is a direct relationship between the clinical response to therapy and the risk of progression to end-stage kidney disease (ESKD), and considering the devastating impact of ESKD on a growing child or adult, the decision on the treatment of individuals with monogenic NS needs to be pragmatic. In our opinion, confirmation of a positive genetic test should be followed by a detailed unbiased discussion with the family. They should be advised that the chances of a clinical response to therapy in monogenic NS are low compared with non-genetic diseases, and that significant side effects are often associated with the currently available therapies. However, they should also be informed that response to therapy, even if it is only a partial response, may prevent or slow the progression to ESKD requiring dialysis and transplantation. Having provided these key details to the family, the physician should support the family in the decision-making process. For clinicians, the knowledge that the patient has a genetic mutation should help in modifying the intensity and duration of immunosuppression, considering the systemic and renal toxicity of most of these agents. Irrespective of the results of genetic testing, the use of angiotensin-converting enzyme inhibitors (ACEI) and lipid lowering agents that are known to slow the rate of progression of glomerular diseases should also be included as part of the standard supportive care for all patients with SRNS, including those with genetic forms of NS39-43
Genetic testing and kidney transplantation
It has been shown in several studies that individuals with NS/FSGS due to single gene defects have a reduced risk of recurrence of FSGS in their kidney allograft23, 44-46. In a recent survey of children with FSGS who received at least one kidney transplant, Jungraithmayr et al reported a recurrence rate of 36%46. Of note, 11 of these patients had FSGS due to NPHS2 mutations and none had recurrence of disease in their kidney allograft.
The reason for the low rate of recurrence of disease in genetic FSGS suggests that this is a kidney-specific disease unlike idiopathic FSGS that frequently recurs after transplantation. It should also be noted that the risk of recurrence is different for different genes, and that exposure to new antigens from the donated kidney may lead to the development of de novo immune complex disease47. Indeed, a small percentage of individuals with NS due to nephrin mutations have been known to develop heavy proteinuria post-transplant, and to have circulating anti-nephrin antibodies47.
Together, these data support the use of pre-transplant genetic testing in patients with NS. Such testing will provide clinicians with information that may be helpful in predicting the post- transplant course, and in guiding post-transplant management.
A much more controversial issue is the utility of genetic testing in kidney donors. We do not recommend routine genetic testing for single gene defects in kidney donors because of the low incidence of these mutations in the general population. However, there is a need for careful thought prior to embarking on living-related donor (LRD) kidney transplantation from family members of patients with known single gene defects. The risk obviously is different between AD and AR conditions. Theoretically, in AR disease, an unaffected individual with a heterozygous change would not be expected to develop disease, although the risk of developing kidney disease due to unknown modifier genes and/or environmental factors is not clear. The situation is even more problematic for AD disease, where a mutation may be incompletely penetrant, and affected individuals may develop disease later in life or even remain asymptomatic throughout their life. Our group previously reported on donors who developed FSGS and ESKD years after donating kidneys to affected siblings/family members with familial FSGS48. In one of the families reported, we identified the disease-causing mutation in this family and found that the recipient, the donor and other affected members of the family carried the same mutation, confirming that this was the cause of the disease in the family (unpublished observation). These observations emphasize the importance of obtaining a complete family history prior to LRD donation. Such findings lend support to the use of genetic testing in living-related donors of individuals with genetic forms of NS. We do not support the use of LRD donation from families with known hereditary disease and unknown gene defect. If the gene defect is known, LRD should only be accepted with CLIA-certified testing of donors to ensure that they do not have the mutation.
Genetic testing in clinical trials
Historically, genetic testing for NS has not been performed as a standard investigation in clinical trials due to the fact that the tests were not routinely available. However with widespread availability of some tests now, recent NS clinical trials have begun to incorporate genetic testing at entry into their protocols, typically for NPHS2 (Podocin) and WT1 (Wilms Tumor1). Although therapeutic selection during NS trials has not yet been driven by such genetic results, it is highly likely that in the near future such genetic results will begin to be used to directly compare treatment responses for various selected homozygous and heterozygous mutations. We believe that due to the paucity of data regarding treatment responses in monogenic NS/FSGS, apart from congenital NSwhere there is a well-established management algorithm, patients with monogenic NS should not be excluded from clinical trials. However, there should be more strict criteria for withdrawal of the individuals with monogenic NS once they are enrolled, in order to mitigate the potential systemic toxicity of trial drugs/agents.
SUMMARY
In conclusion, recent advances in molecular genetics and genomic science have opened the door to dramatically improve our understanding of the molecular basis of NS, and enable precise diagnosis of disease. However, these advances also pose significant challenges for the clinical use of this information, both for clinicians and for their patients and families. Despite these challenges, the advances in genetic diagnosis have created an unprecedented opportunity to develop a “personalized” approach to provide improved care for both children and adults with NS.
BOX 1: Major nephrotic syndrome genes and indications for genetic testing.
Autosomal recessive genes
■ NPHS2 (Podocin)
■ NPHS1 (Nephrin)
■ PLCE1 (Phospholipase C epsilon 1)
Autosomal dominant genes
■ INF2 (Inverted formin 2)
■ TRPC6 (transient receptor potential cation channel C6)
■ ACTN4 (Actinin alpha 4)
■ WT1 (Wilms tumor 1)
Indications for genetic testing
■ Congenital nephrotic syndrome (onset first three months of life)
■ Infantile nephrotic syndrome (onset 3-12 months of life)
■ Family history of nephrotic syndrome
■ Multiple extra-renal manifestations
Footnotes
COMPETING INTERESTS STATEMENT
MP Winn is a Consultant to Athena diagnostics.
REFERENCES
- 1.McKinney PA, Feltbower RG, Brocklebank JT, Fitzpatrick MM. Time trends and ethnic patterns of childhood nephrotic syndrome in Yorkshire, UK. Pediatr Nephrol. 2001;16:1040–1044. doi: 10.1007/s004670100021. [DOI] [PubMed] [Google Scholar]
- 2.International Study of Kidney Disease in Children The primary nephrotic syndrome in children. Identification of patients with minimal change nephrotic syndrome from initial response to prednisone. J Pediatr. 1981;98:561–564. doi: 10.1016/s0022-3476(81)80760-3. [DOI] [PubMed] [Google Scholar]
- 3.Kestila M, et al. Positionally cloned gene for a novel glomerular protein-nephrin- is mutated in congenital nephrotic syndrome. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 4.Wiggins RC. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int. 2007;71:1205–1214. doi: 10.1038/sj.ki.5002222. [DOI] [PubMed] [Google Scholar]
- 5.Shih NY, et al. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 286:312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- 6.Dreyer SD, et al. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet. 1998;19:47–50. doi: 10.1038/ng0598-47. [DOI] [PubMed] [Google Scholar]
- 7.Boute N, et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 8.Zenker M, et al. Human laminin beta2 deficiency causes congenital nephrosis with mesangial sclerosis and distinct eye abnormalities. Hum Mol Genet. 2004;13:2625–32. doi: 10.1093/hmg/ddh284. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan JM, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 10.Boerkoel CF, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 11.Winn MP, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308:1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 12.Hinkes B, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 13.Niaudet P, Gubler MC. WT1 and glomerular diseases. Pediatr Nephrol. 2006;21:1653–1660. doi: 10.1007/s00467-006-0208-1. [DOI] [PubMed] [Google Scholar]
- 14.Berkovic SF, et al. Array-based gene discovery with three unrelated subjects shows SCARB2/LIMP-2 deficiency causes myoclonus epilepsy and glomerulosclerosis. Am J Hum Genet. 2008;82:673–684. doi: 10.1016/j.ajhg.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown EJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer O, et al. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- 17.Heeringa SF, et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akilesh S, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest. 2011;121:4127–4137. doi: 10.1172/JCI46458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mele C, et al. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med. 2011;365:295–306. doi: 10.1056/NEJMoa1101273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozaltin F, et al. Disruption of PTPRO causes childhood-onset nephrotic syndrome. Am J Hum Genet. 2011;89:139–147. doi: 10.1016/j.ajhg.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Has C, et al. Integrin α3 mutations with kidney, lung, and skin disease. N Engl J Med. 2012;366:1508–1514. doi: 10.1056/NEJMoa1110813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Büscher AK, et al. Mutations in podocyte genes are a rare cause of primary FSGS associated with ESRD in adult patients. Clin Nephrol. 2012;78:47–53. doi: 10.5414/cn107320. [DOI] [PubMed] [Google Scholar]
- 23.Ruf RG, et al. Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol. 2004;15:722–732. doi: 10.1097/01.asn.0000113552.59155.72. [DOI] [PubMed] [Google Scholar]
- 24.Hinkes B, et al. Specific podocin mutations correlate with age of onset in steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2008;19:365–371. doi: 10.1681/ASN.2007040452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinkes BG, et al. Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics. 2007;119:e907–919. doi: 10.1542/peds.2006-2164. [DOI] [PubMed] [Google Scholar]
- 26.Gbadegesin R, et al. Mutations in PLCE1 are a major cause of isolated diffuse mesangial sclerosis (IDMS). Nephrol Dial Transplant. 2008;23:1291–1297. doi: 10.1093/ndt/gfm759. [DOI] [PubMed] [Google Scholar]
- 27.Gbadegesin RA, et al. Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis. Kidney Int. 2012;81:94–99. doi: 10.1038/ki.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyer O, et al. Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis. J Am Soc Nephrol. 2011;22:239–245. doi: 10.1681/ASN.2010050518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopp JB, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santín S, et al. Clinical utility of genetic testing in children and adults with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2011;6:1139–1148. doi: 10.2215/CJN.05260610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gbadegesin R, et al. Mutational analysis of NPHS2 and WT1 in frequently relapsing and steroid-dependent nephrotic syndrome. Pediatr Nephrol. 2007;22:509–513. doi: 10.1007/s00467-006-0377-y. [DOI] [PubMed] [Google Scholar]
- 34.Büscher AK, et al. Immunosuppression and renal outcome in congenital and pediatric steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2010;5:2075–2084. doi: 10.2215/CJN.01190210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitamura A, et al. A familial childhood-onset relapsing nephrotic syndrome. Kidney Int. 2007;71:946–951. doi: 10.1038/sj.ki.5002110. [DOI] [PubMed] [Google Scholar]
- 36.Wasilewska AM, et al. Effect of cyclosporin A on proteinuria in the course of glomerulopathy associated with WT1 mutations. Eur J Pediatr. 2011;170:389–391. doi: 10.1007/s00431-010-1278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ransom RF, Lam NG, Hallett MA, Atkinson SJ, Smoyer WE. Glucocorticoids protect and enhance recovery of cultured murine podocytes via actin filament stabilization. Kidney Int. 2005;68:2473–2483. doi: 10.1111/j.1523-1755.2005.00723.x. [DOI] [PubMed] [Google Scholar]
- 38.Faul C, et al. The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat Med. 2008;14:931–938. doi: 10.1038/nm.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman JE, Watson AR. Hyperlipidaemia, diet and simvastatin therapy in steroid-resistant nephrotic syndrome of childhood. Pediatr Nephrol. 1996;10:171–174. doi: 10.1007/BF00862065. [DOI] [PubMed] [Google Scholar]
- 40.Sanjad SA, et al. Management of hyperlipidemia in children with refractory nephrotic syndrome: the effect of statin therapy. J Pediatr. 1997;130:470–474. doi: 10.1016/s0022-3476(97)70213-0. [DOI] [PubMed] [Google Scholar]
- 41.Valdivielso P, et al. Atorvastatin in dyslipidaemia of the nephrotic syndrome. Nephrology (Carlton) 2003;8:61–64. doi: 10.1046/j.1440-1797.2003.00143.x. [DOI] [PubMed] [Google Scholar]
- 42.Ellis D, et al. Long-term antiproteinuric and renoprotective efficacy and safety of losartan in children with proteinuria. J Pediatr. 2003;143:89–97. doi: 10.1016/S0022-3476(03)00279-8. [DOI] [PubMed] [Google Scholar]
- 43.Prescott WA, Jr, et al. The potential role of HMG-CoA reductase inhibitors in pediatric nephrotic syndrome. Ann Pharmacother. 2004;38:2105–2114. doi: 10.1345/aph.1D587. [DOI] [PubMed] [Google Scholar]
- 44.Conlon PJ, et al. Spectrum of disease in familial focal and segmental glomerulosclerosis. Kidney Int. 1999;56:1863–1871. doi: 10.1046/j.1523-1755.1999.00727.x. [DOI] [PubMed] [Google Scholar]
- 45.Weber S, et al. NPHS2 mutation analysis shows genetic heterogeneity of steroid-resistant nephrotic syndrome and low post-transplant recurrence. Kidney Int. 2004;66:571–579. doi: 10.1111/j.1523-1755.2004.00776.x. [DOI] [PubMed] [Google Scholar]
- 46.Jungraithmayr TC, et al. Screening for NPHS2 mutations may help predict FSGS recurrence after transplantation. J Am Soc Nephrol. 2011;22:579–585. doi: 10.1681/ASN.2010010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuusniemi AM, et al. Plasma exchange and retransplantation in recurrent nephrosis of patients with congenital nephrotic syndrome of the Finnish type (NPHS1). Transplantation. 2007;83:1316–1323. doi: 10.1097/01.tp.0000262569.27890.64. [DOI] [PubMed] [Google Scholar]
- 49.Winn MP, et al. Focal segmental glomerulosclerosis: a need for caution in live-related renaltransplantation. Am J Kidney Dis. 1999;33:970–974. doi: 10.1016/s0272-6386(99)70435-x. [DOI] [PubMed] [Google Scholar]