Abstract
Cry11Aa of Bacillus thuringiensis subsp. israelensis is the most active toxin to Aedes aegypti in this strain. We previously reported that, in addition to a 65 kDa GPI (glycosylphosphatidylinositol)-anchored ALP (alkaline phosphatase), the toxin also binds a 250 kDa membrane protein. Since this protein is the same size as cadherin, which in lepidopteran insects is an important Cry toxin receptor, we developed an anti-AaeCad antibody. This antibody detects a 250 kDa protein in immunoblots of larval BBMVs (brush border membrane vesicles). The antibody inhibits Cry11Aa toxin binding to BBMVs and immunolocalizes the cadherin protein to apical membranes of distal and proximal caecae and posterior midgut epithelial cells. This localization is consistent with areas to which Cry11Aa toxin binds and causes pathogenicity. Therefore, the full-length Aedes cadherin cDNA was isolated from Aedes larvae and partial overlapping fragments that covered the entire protein were expressed in Escherichia coli. Using toxin overlay assays, we showed that one cadherin fragment, which contains CR7–11 (cadherin repeats 7–11), bound Cry11Aa and this binding was primarily through toxin domain II loops α8 and 2. Cadherin repeats CR8–11 but not CR7 bound Cry11Aa under non-denaturing conditions. Cry11Aa bound the cadherin fragment with high affinity with an apparent Kd of 16.7 nM. Finally we showed that this Cry11Aa-binding site could also be competed by Cry11Ba and Cry4Aa but not Cry4Ba. These results indicate that Aedes cadherin is possibly a receptor for Cry11A and, together with its ability to bind an ALP, suggest a similar mechanism of toxin action as previously proposed for lepidopteran insects.
Keywords: Bacillus thuringiensis, binding affinity, cadherin, Cry11Aa toxin, midgut, receptor
INTRODUCTION
Aedes aegypti is an important mosquito vector of dengue and yellow fever, diseases that are of increasing concern worldwide [1]. Chemical insecticides are primarily used for the control of these and other mosquito vectors. However, an increased incidence of insecticide resistance has been observed in many of these disease vectors. Consequently, formulations using Bacillus thuringiensis subsp. israelensis, a larvicide, are frequently used worldwide for the control of this insect vector [2].
B. thuringiensis produces insecticidal δ-endotoxin proteins (named Cry and Cyt toxins) during the sporulation phase. The subsp. israelensis produces four major insecticidal proteins (Cry4Aa, Cry4Ba, Cry10Aa and Cry11Aa) and three cytolytic proteins (Cyt1Aa, Cyt2Ba and Cyt1Ca) [3]. Among them, Cry11Aa is the most active toxin against Ae. aegypti. Upon ingestion by susceptible insect larvae, Cry11Aa crystals are first solubilized in the alkaline environment of larval midgut, and then the soluble protoxins are processed into 34 and 32 kDa fragments by gut proteases [4]. These two fragments form a heterodimer that retains insecticidal activity [5]. The activated fragments then bind specific receptors in microvilli of midgut epithelial cells, leading to membrane insertion and pore formation [2,6]. These latter processes are thought to lyse midgut cells, ultimately killing larval mosquitoes [2].
Several different receptors from lepidopteran and dipteran insects and nematodes have been reported to bind activated toxins. In case of lepidopteran-specific Cry1A toxins, four different protein receptors have been revealed: cadherin [7–9], a GPI (glycosylphosphatidylinositol)-anchored APN (aminopeptidase N) [10,11], a GPI-anchored ALP (alkaline phosphatase) [12] and a 270 kDa glycoconjugate [13]. Among these proteins, binding to a cadherin receptor is required for further toxin cleavage and activation that is critical for intoxication [2,6,14]. The same classes of protein receptors from dipteran insects have also been revealed. For example, an ALP from Ae. aegypti binds Cry11Aa [15], APNs from Anopheles quadrimaculatus and Anopheles gambiae bind Cry11Ba [16,17] and a cadherin-like protein from An. gambiae was identified as a receptor for Cry4Ba [18]. For the Cry11Aa toxin, a 250 kDa protein was shown to also bind this toxin in addition to the GPI-anchored ALP protein [15,19]. Consequently, it is likely that the 250 kDa protein may be a cadherin-like protein. In the present study we show that a cadherin protein indeed binds the Cry11Aa toxin.
The Cry1A toxin-binding domains in lepidopteran cadherin receptors have been mapped. For example, in the Manduca sexta cadherin protein, BtR1, three binding sites are known. The first, localized in CR7 (cadherin repeat 7), binds to domain II loop 2 of Cry1Ab toxin [20,21]. A second binding epitope in CR11 interacts with domain II loop α8 and loop 2 of Cry1Ab toxin [22]. A third Cry1Ab binding site is in CR12 [23], which has been shown in Heliothis virescens to bind domain II loop 3 of Cry1Ab and 1Ac toxins [24]. In the latter case, the toxin-binding region was narrowed further to residues 1422–1440 of CR12. In Bombyx mori, the CRs of BtR175 immediately adjacent to the membrane are involved in binding loops 2 and 3 of Cry1Aa [25]. In contrast, we know little about toxin-binding domains in dipteran cadherins.
To alleviate this knowledge gap, we identified a putative Cry11Aa toxin cadherin receptor cDNA from larval Ae. aegypti midgut. The toxin-binding domains in this cadherin were identified as well as toxin loop regions that are involved in interacting with the cadherin. This report shows that the cadherin receptor binds Cry11Aa toxin with high affinity. Finally, the spatial expression of Aedes cadherin was analysed in larval guts, and the expression correlates with areas which were previously shown be the sites of toxin binding and pathogenicity.
MATERIALS AND METHODS
Purification, activation and biotinylation of Cry11Aa toxin
Inclusions for Cry11Aa, Cry11Ba, Cry4Aa, Cry4Ba and loop α8 Cry11Aa mutant toxins were isolated from recombinant strains producing these toxins individually [26,27]. B. thuringiensis strains were grown in nutrient broth sporulation medium containing 12.5 μg/ml erythromycin at 30°C. Following cell autolysis, the spores and inclusions were harvested, washed three times with 1 M NaCl plus 10 mM EDTA, pH 8.0 and centrifuged. The resulting pellet was resuspended in 30 ml of the same buffer and purified by NaBr gradients as previously described [28].
The purified inclusions were solubilized in 50 mM Na2CO3, pH 10.5, except Cry11Aa mutant toxins were solubilized in 0.1 M NaOH, since they had lower solubility in the bicarbonate buffer, and then dialysed with 50 mM Na2CO3, pH 10.5. The solubilized toxins were activated by trypsin (1:20, w/w). The activated Cry11Aa toxin was purified by ion-exchange chromatography (Mono Q, FPLC), biotinylated (Amersham Biosciences kit) and then purified using a Sephadex G25 column.
Assembly, cloning and sequence analyses of the full-length Ae. aegypti cadherin cDNA
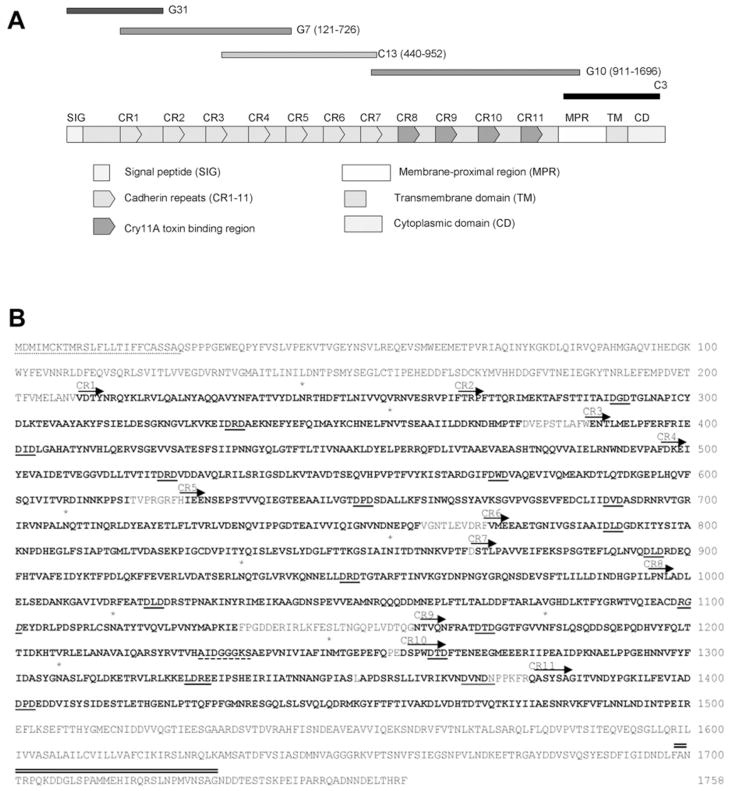
Sequence primers based on the Ae. aegypti genome (http://aaegypti.vectorbase.org) were designed to amplify five overlapping cDNA fragments (0.9 kb G31, 1.8 kb G7, 1.5 kb C13, 2.3 kb G10 and 1.0 kb C3, see Figure 3). These fragments covering the full-length cadherin cDNA were isolated from an Aedes midgut cDNA library or Aedes cDNA. Among them, the C3 fragment was obtained by 3′-RACE (3′-rapid amplification of cDNA ends). No 5′-RACE was performed, since the G31 fragment had stop codons before the predicted start codon. All fragments were cloned into the TA cloning vector, pCR2.1TOPO (Invitrogen) and fully sequenced at the Institute for Integrative Genome Biology (IIGB) at University of California, Riverside (UCR).
Figure 3. The amino acid sequence and structure features of the full-length Aedes cadherin protein.
(A) Five overlapping cadherin cDNA fragments, G31, G7, C13, G10 and C3 were amplified from an Aedes midgut cDNA library or whole Aedes cDNA samples and these fragments were assembled by five sequential steps of subcloning. This Aedes cDNA contains a complete ORF, which encodes a full-length Aedes cadherin protein. The protein contains a signal peptide (SIG), 11 cadherin repeats (CR1–11), a membrane-proximal region (MPR), a transmembrane domain (TM) and a cytoplasmic domain (CD). Toxin-binding regions were localized in CR8–11. (B) The putative N-terminal signal peptide is underlined with dots and the transmembrane domain is double underlined. The 11 cadherin repeat sequences are in bold. Putative calcium-binding sites are underlined and the integrin recognition sequence RGD (aa 1098–1101) is in italics and underlined. The ATP/GTP-binding site is dash-underlined and the predicted N-glycosylated residues (Asn) are labelled with an asterisk above the letter N.
The recombinant plasmid pCR2.1AaeCad was obtained by assembling the five overlapping fragments mentioned above. This resulting plasmid harboured the full-length cDNA encoding the Ae. aegypti cadherin ORF (open reading frame).
Sequence alignments and other sequence analyses were performed using NCBI blast programs and Lasergene (Dnastar). Signal peptide and the transmembrane domain were identified by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) and HMMTOP (http://www.enzim.hu/hmmtop/) respectively. The IRSEC Motifscan program was used to identify protein motifs in Aedes cadherin (http://myhits.isb-sib.ch/cgi-bin/motif_scan).
Expression of partial cadherin fragments and antibody preparation
To express partial cadherin proteins, the G7, C13 and G10 fragments (Table 1) were cloned into a suitable pQE30 series vector (Qiagen) to generate the plasmids pQE32G7, pQE32C13 and pQE30G10 respectively. Similarly, clones for each cadherin repeat, CR7 to CR11, were obtained by PCR using specific primers. The final constructs were transformed into the Escherichia coli M15(pREP4) strain. Protein expression was induced by addition of 1 mM IPTG (isopropyl β-D-thiogalactoside) and proteins were produced as inclusion bodies. The N-terminally His-tagged recombinant proteins were purified using Ni-NTA (Ni2+-nitrilotriacetate) resin (Qiagen) using urea and resolved by SDS/PAGE. The CR proteins were dialysed against PBS buffer and were kept at −80°C until needed.
Table 1.
Partial Aedes cadherin protein and synthetic peptide sequences
| Name | Sequence | Molecular mass (kDa) | Description |
|---|---|---|---|
| G7 | ITLVVEGD – –EAYETLFL | 68.5 | Amino acid sequence of residues 121–726 of cadherin, containing CR1–4 and partial CR5 |
| C13 | GTFTLTIV – –NELLDRDT | 57.4 | Amino acid sequence of residues 440–952 of cadherin, containing partial CR3 and CR4–7 |
| G10 | KTFPDLQK – -DFIGIDND | 87.7 | Amino acid sequence of residues 911–1696 of cadherin, containing partial CR7 and CR8–11 |
| CR7 | STLPAVVE – -INDHGPIL | 14.4 | Cadherin repeat 7, amino acid sequence of residues 869–994 of cadherin |
| CR8 | LPNLADLE – –NYMAPKIE | 15.6 | Cadherin repeat 8, amino acid sequence of residues 995–1133 of cadherin |
| CR9 | FPGDDERI – -MTGEPEFQ | 13.5 | Cadherin repeat 9, amino acid sequence of residues 1134–1255 of cadherin |
| CR10 | DSPWDTDF – -VNDNPPKFR | 13.5 | Cadherin repeat 10, amino acid sequence of residues 1258–1376 of cadherin |
| CR11 | QASYSAGI – -NDINTPEI | 14 | Caderin repeat 11, amino acid sequence of residues 1377–1500 of cadherin |
| Loop α-8 | GVSIPVNYNEWY | 1.4 | Amino acid sequence of residues 257–268 of Cry11Aa |
| Loop 1 | DIPARENIRGVH | 1.4 | Amino acid sequence of residues 298–309 of Cry11Aa |
| Loop 2 | FTQWFQSTLYG | 1.4 | Amino acid sequence of residues 386–396 of Cry11Aa |
| Loop 3 | LTYNRIEYDSPTTEN | 1.8 | Amino acid sequence of residues 447–461 of Cry11Aa |
The purified 6 × His-tagged G10 protein was separated by SDS/PAGE and the gels were stained and destained. Then, purified protein bands were excised from the gel, the bands were washed three times and used to immunize rabbits for antibody development according to standard protocols.
Alternatively, inclusion bodies were purified from bacterial cultures expressing cadherin fragments or repeats using a B-PER Bacterial Protein extraction reagent following the manufacturer’s instructions (Pierce). The inclusion bodies were dissolved in 0.1 M NaOH buffer for 1 h and then dialysed against 50 mM Na2CO3 (pH 10.5), and protein concentration was measured using the BCA (bicinchoninic acid) assay (Pierce). Total proteins extracted were analysed by SDS/PAGE and the percentage of protein consisting of cadherin fragments or repeats was measured by ImageJ software.
Immunolocalization of cadherin and Cry11A binding sites in Ae. aegypti larval guts
Ae. aegypti larval guts (4th instar) were dissected in PBS, transferred into 4% PFA (paraformaldehyde) and then fixed at room temperature (25°C) for 20 min (whole 4th instar larvae were fixed in 4%PFA overnight at 4°C). The tissues were then washed in PBST (PBS plus 0.1% Triton X-100) three times for 30 min, dehydrated in a 20, 40, 70 and 96% ethanol series in PBS for 30 min each and finally incubated in 100% ethanol for 1 h. The tissues were placed in ethanol/xylene mixtures (70/30, then 30/70) for 3 h per mixture and then in 100%xylene at room temperature overnight. Paraffin chips were added at 20–50% of total 100% xylene volume for 4–6 h at room temperature. The samples were then placed in 100% paraffin for 24 h at 55°C. The tissues were embedded first in paraffin blocks, then 8–10 μm thick sections were cut, placed on poly-L-lysine (Sigma) slides coated with 1% gelatin (Becton Dickson), and then the sections were dried for 1–2 days at 40°C.
For immunolocalization, the sections were washed with 100% xylene for 15 min twice, rehydrated in 100, 70, 40 and 20% ethanol for 5 min each and rinsed in deionized water and PBST. To detect sites of Cry11A binding, the sections were incubated with 100 nM Cry11A toxin and washed three times with PBST. Then the sections were incubated with Cry11A (1:1000) antibodies diluted in PBST/1% BSA at 4°C overnight and washed with PBST/0.1%BSA/2%goat serum for 20 min twice. Alternatively, to detect cadherin expression in tissues, the sections were incubated with anti-AaeCad (1:100) and then washed as described above. Finally, all sections were incubated for 1 h in the dark with secondary antibodies (Jackson Immunoresearch Labs). For cadherin and Cry11Aa detection, Cy3 (indocarbocyanine)-conjugated goat-anti-rabbit antibody diluted 1:1000 was used, and actin F was detected using Phalloidin-Alexa Fluor® 488 diluted 1:100. After washing twice in the same buffer, the sections were mounted in Shur/Mount medium (Electron Microscopy Science). The images were obtained using a laser-scanning confocal Zeiss Axioplan microscope (LSM Zeiss 510, at the Institute of Integrative Genome Biology, University of California Riverside, CA, U.S.A.) at × 40 and × 100 magnification. All images were imported into Adobe Photoshop for assembly and annotation.
Competition of Cry11Aa binding in isolated BBMVs (brush border membrane vesicles)
BBMVs were prepared from 4th instar Ae. aegypti larvae midguts as described previously [29]. The binding of Cry11Aa toxin to BBMVs was performed as described in [27]. Briefly, binding of 10 nM labelled toxin to 10 μg BBMV protein was performed in 100 μl binding buffer (PBS, 0.1% BSA and 0.1% Tween 20, pH 7.6). After 1 h at 25°C, the membrane pellet was separated by centrifugation for 10 min at 14000 g to remove unbound toxins and the pellet was then washed three times with binding buffer. Finally, the pellet was resuspended in 20 μl PBS, 4 μl 6 × Laemmli sample loading buffer (60% glycerol, 300 mM Tris/HCl, pH 6.8, 12 mM EDTA, 12% SDS, 864 mM 2-mercaptoethanol and 0.05% Bromophenol Blue). It was boiled for 5 min and then separated by SDS/PAGE and electro-transferred to nitrocellulose membranes (Immobilon, Amersham Biosciences). The membranes were incubated with streptavidin–peroxidase conjugate (1:1500 dilutions, Amersham Biosciences) for 1 h and then visualized using luminol (ECL™; enhanced chemiluminescence; Amersham Biosciences). For competition assays, the BBMVs were pre-incubated with different dilutions of the polyclonal anti-AaeCad or anti-Aedes NHE3 antibodies for 1 h at room temperature before incubation with biotinylated Cry11Aa. For competition with cadherin fragments, BBMVs were incubated in the presence or absence of CR9, CR10 or anti-AaeCad antiserum for 1 h at room temperature.
Western blotting of Ae. aegypti BBMV
An equal volume of 2 × sample loading buffer was added to 10 μg of Ae. aegypti BBMV protein and boiled and then loaded on to SDS/PAGE (9% gels) and electrotransferred to nitrocellulose membranes. After blocking, the membranes were incubated for 2 h with anti-AaeCad antibody (1:7500 dilution) followed by incubation with goat anti-rabbit HRP (horseradish peroxidase, 1:5000 dilution) secondary antibody (Sigma). Blots were developed by ECL™ (Amersham Biosciences).
Toxin overlay assay
Purified partial cadherin fragments (90 pmol) or cadherin repeats (30 pmol) were separated by SDS/PAGE (8% or 15% gels respectively), and then transferred on to PVDF membranes (Immobilon, Amersham Biosciences). The membranes were incubated with 3% BSA and then with 20 nM Cry11Aa toxins for 2 h. Unbound toxins were removed by washing the membrane four times with washing buffer (0.1% Tween 20 in PBS) for 15 min. The membranes were then incubated with rabbit anti-Cry11Aa antibody (1:2000 dilution) followed by a secondary goat anti-rabbit antibody conjugated to HRP (Amersham Biosciences) (1:5000 dilution). Finally, the membranes were exposed to luminol (ECL™, Amersham Biosciences). To determine the ability of synthetic peptides to compete with Cry11Aa toxin binding, different concentrations of the four loop peptides were mixed separately with biotinylated Cry11Aa toxins in washing buffer for 1 h at room temperature before incubating these mixtures with PVDF membranes containing the cadherin fragments or repeats.
Competitive ELISA
Dose-dependent binding curves were obtained using a modified ELISA [30]. In brief, 96-well plates were coated with 0.4 μg of Cry11Aa per well and then treated with blocking buffer (PBS, 0.1% Tween 20 and 0.5% gelatin) for 1 h at 37°C, washed and then 0.1–1000 nM G7, C13, G10 and cadherin repeats (CR7–11) protein solutions were transferred to the coated plates. After washing to remove unbound proteins, either anti-AaeCad antibody (1:2000) or anti-His6 antibody (1:5000) was added and incubated overnight at 4°C. Subsequently after additional washes, goat anti-rabbit antibody coupled to ALP (1:2000) or goat-anti-mouse antibody coupled to ALP (1:2000) was added to the wells and incubated for a further 2 h at 37°C. After further washing, ALP activity was revealed with freshly prepared substrate (3 mM nitrophenyl phosphate) and the absorbance read at 405 nm with a microplate reader (Molecular Devices). The G10 concentration that showed a linear range of binding was used for competitive ELISA below.
The kinetics of association of Cry11Aa toxin to G10 was measured by competitive ELISA [6]. G10 (8 nM) was equilibrated with increasing concentrations of Cry11Aa toxin (0.2 nM to 1000 nM) in 100 μl PBST for 1 h at room temperature. The mixtures were then transferred to plate wells previously coated with Cry11Aa toxin. The detection procedure was then continued as described above. Data were analysed by using Origin 6.1 (Origin Lab). The concentration corresponding to half maximal absorbance was considered the dissociation constant.
RESULTS
Expression of cadherin in larval Ae. aegypti guts
Previous work with lepidopteran insects showed that Cry1 toxins from B. thuringiensis bind with high affinity to cadherin proteins that are localized to epithelia of midgut cells [8,9,31,32]. Furthermore, these Cry toxins often bind the C-terminal end of cadherin protein [20,21,23,24].
Therefore, we screened all cadherin genes in the Aedes genome (http://www.vectorbase.org/index.php) and identified a protein with the highest homology to lepidopteran cadherins. Using sequences obtained from the Ae. aegypti genome, we amplified a portion of the C-terminus of Ae. aegypti cadherin that is most orthologous to lepidopteran cadherins. This partial cadherin fragment, named G10, was expressed as a His-tag protein, and the purified protein was used for the production of a rabbit anti-AaeCad antibody. To show that cadherin plays a role in binding of the Cry11Aa toxin to Ae. aegypti midgut epithelia, we performed a competition assay using BBMVs from this insect. Using biotinylated Cry11Aa, we showed the anti-AaeCad antibody competed readily with toxin binding (Figure 1A, lanes 2–4). In contrast, another antibody to the sodium-protein exchanger NHE3 that is also expressed in the midgut epithelia of this mosquito species did not compete in this assay (Figure 1A, lanes 5–6).
Figure 1. The Cry11Aa toxin binding is inhibited by an anti-AaeCad antibody and the cadherin protein is detected in Aedes BBMV.
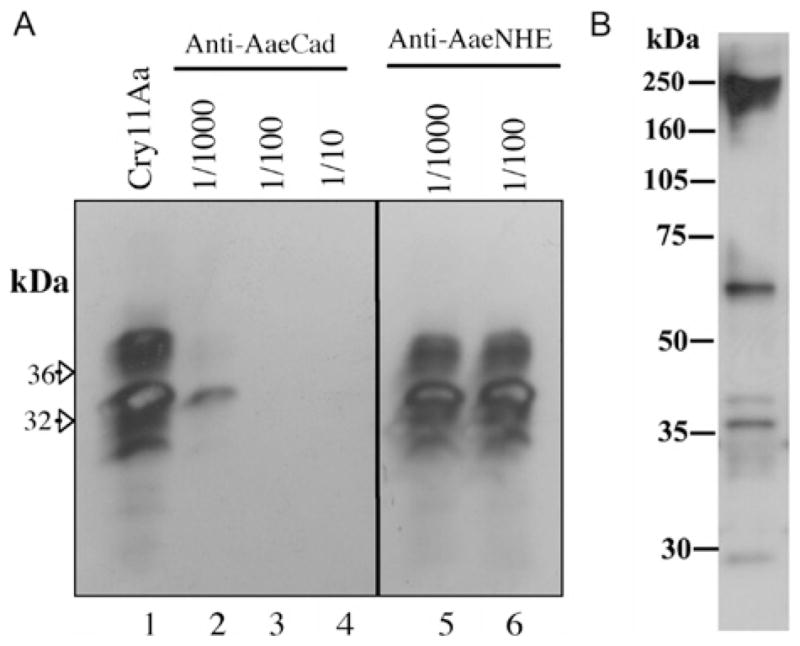
(A) Cry11Aa (5 nM) binding to BBMVs (10 μg) is competed with varying dilutions of anti-AaeCad antibody but not by an anti-NHE3 antibody that also detects a membrane-bound protein in Aedes midgut [55]. (B) Western blot of BBMV proteins (10 μg) analysed with the anti-AaeCad antibody diluted 1:7500.
The anti-AaeCad antibody detected a major band with a molecular size of 250 kDa and two less intense bands of 60 and 35 kDa in Ae. aegypti BBMVs (Figure 1B). In a degraded membrane preparation, there is a loss of the 250 kDa band, but the 60 and 35 kDa bands show an increase in intensity (results not shown). The 250 kDa protein thus represents the full-length cadherin, whereas the lower-molecular-mass bands detected may be degradation products or, alternatively, other cross-reacting proteins. In M. sexta, it has been shown that the homologous cadherin undergoes proteolytic degradation during larval development [33]. The larger molecular size of cadherin compared with the size predicted from the amino acid sequence may be due to post-translational modifications.
B. thuringiensis subsp. israelensis toxins disrupt the larval midgut [34] as does the Cry11Aa toxin [35]. Furthermore, the Cry4A, 4B and 11A toxins also bind to the apical brush border of caeca and posterior midgut cells [36]. Since cadherin is one of the putative receptors for these Cry toxins, we then determined if the distribution of cadherin in the larval gut of Ae. aegypti is similar to sites to which these Cry toxins have been shown to bind. As shown in Figure 2, the affinity-purified antibody shows intense immunofluorescence on the apical side of the distal and proximal caeca (Figures 2F and 2G respectively), and on the apical membrane of posterior midgut epithelial cells (Figures 2A, 2C and 2D). These are the same tissues that bind Cry toxins from B. thuringiensis subsp. israelensis and show subsequent pathological responses. No immunofluorescence was observed in cells of the anterior midgut (Figures 2B and 2E) or the hindgut (Figure 2A). No immunofluorescence was observed in control sections incubated with preimmune serum at the same dilution as the anti-AaeCad antibody (Figure 2H). In addition, the Cry11A toxin protein has an identical pattern of localization in Ae. aegypti larval tissues (Figures 2J, 2K and 2L). Cry11A toxin binds to the apical membrane of gastric caecae (Figure 2J) and the posterior midgut (Figures 2K and 2L), and no toxin binding was detected in the anterior midgut (Figure 2I).
Figure 2. Immunolocalization of cadherin and Cry11A toxin binding sites in larval gut of 4th instar Ae. aegypti larvae.
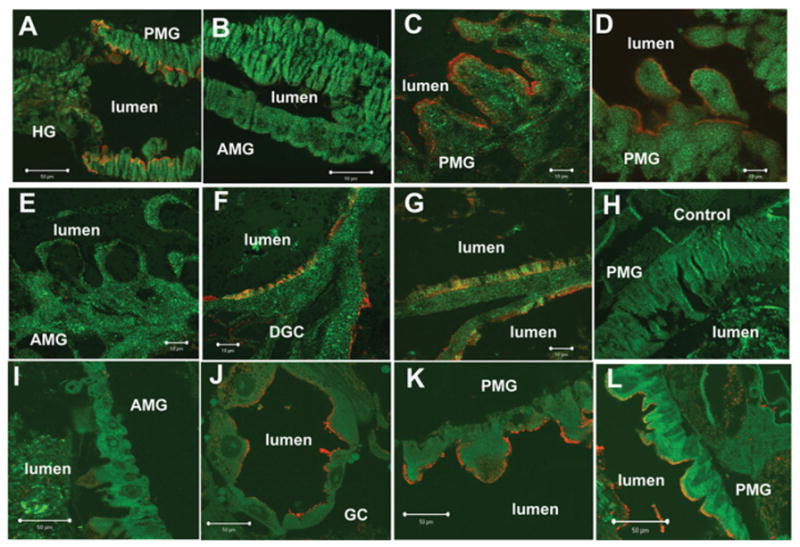
Whole larvae (C–H) and gut sections (A and B) were incubated with affinity purified anti-AaeCad antibody diluted 1:100. Whole larvae sections (I–L) were incubated with Cry11Aa (100 nM) and then with anti-Cry11A antibody diluted 1:1000. Cy3-linked secondary antibody (1:1000, red) was used to determine cadherin localization or Cry11A toxin-binding sites. The cell and tissue structures were visualized by phalloidin (Alexa Fluor® 488, 1:100, green). Red immunofluorescence shows cadherin localization on the apical side of the posterior midgut (A, C and D) but not in the apical membranes of anterior midgut (B and E) and hindgut (A) epithelial cells. Cadherin was also observed on the apical side of distal (F) and proximal (G) gastric caecae. No specific signal was observed in posterior midgut cells when tissues were probed with preimmune serum as a negative control (H). Cry11A toxin bound the apical membrane of epithelial cells in gastric caeca (J) and posterior midgut (K and L). No immunofluorescence was found in the apical membrane of anterior midgut (I). Scale bars, 50 μm (A, B, I–L); 100 μm (C–H). AMG, anterior midgut; DGC, distal gastric caecae; GC, gastric caecae; HG, hindgut; PGC, proximal gastric caecae; PMG, posterior midgut.
Isolation of full-length cadherin and its analysis
Since these initial results indicated an important role of the cadherin protein in binding to Ae. aegypti midgut epithelia, we then attempted to isolate the full-length cadherin. However, attempts to isolate the full-length clone using primers designed to known 5′ and 3′ ends were unsuccessful. Therefore, using primers designed to internal cadherin and vector sequences, the 5′ cDNA end (0.9 kb G31) was isolated from an Ae. aegypti larval midgut library made in pSPORT1. The 3′ end was amplified using RACE to give the 1.0 kb C3 fragment. Using sequences from these two cDNA ends and the G10 fragments, the rest of the Aedes cadherin cDNA was obtained as two fragments (Figure 3A).
These five overlapping partial fragments (Figure 3A) were assembled by five steps of sequential subcloning to generate the full-length Aedes cadherin cDNA of 5.97kb. This full-length cadherin transcript was expressed in an insect cell line that gave a protein of approx. 250 kDa (S. B. Lee, J. Chen. and S. S. Gill, unpublished work), which confirmed that this cDNA contains the complete ORF for Aedes cadherin. The cDNA encodes a protein of 1758 amino acids (Figure 3A). The cadherin protein is incorrectly annotated in the Aedes genome as two partial peptides (AAEL007478-PA, AAEL007488-PA), which lack the N-terminal end of the protein. The ORF is encoded by a gene of greater than 150 kb that contains 22 exons (http://www.vectorbase.org).
A comparison with other insect cadherins that are known Cry toxin receptors showed that the Aedes cadherin has the highest amino acid identity (47.2%) with An. gambiae cadherin [18], and shares 27–30.9% identity with lepidopteran cadherins [7,23,37–39].
The full-length Aedes cadherin contains a signal peptide (aa 1–25), 11 cadherin repeats (CR1–11), a membrane-proximal region (aa 1500–1598), a transmembrane domain (aa 1599–1630) and a cytoplasmic domain (aa 1631–1758) (Figure 3). Analysis of the sequence revealed 14 putative calcium-binding sites (one DXD in all cadherin repeats and the cytoplasmic domain respectively, one DXNDN and one LDRE exclusively in CR10; where X is any amino acid), nine glycosylation sites and one ATP/GTP-binding site (1228AIDGGGKS1253). An integrin recognition sequence RGD is located in CR8, but neither another integrin recognition sequence, LDV, nor cadherin cell adhesion recognition sequences, HAV, exists in Aedes cadherin (Figure 3B).
Aedes cadherin toxin-binding regions
Because the Cry1A toxin-binding sites on M. sexta and H. virescens cadherin have been localized to specific regions of the protein [20–24], we also determined the Cry11Aa toxin-binding sites on the cadherin. Three partial fragments covering all the cadherin repeats were cloned, expressed and purified by Ni-NTA agarose and then analysed with Cry11Aa toxin overlay assays. As shown in Figure 4(A), only G10, which contains CR7–11, bound Cry11Aa toxin strongly, whereas G7 and C13, containing CR1–5 and CR3–7 respectively, rarely bound Cry11Aa toxin. In ELISA we obtained the same results, with only the G10 peptide and not the G7 and C13 peptides binding Cry11Aa (Supplementary Figure S1 at http://www.BiochemJ.org/bj/424/bj4240191add.htm). To determine further the toxin-binding regions on G10, each of the cadherin repeats in G10 (CR7–11, Table 1) was amplified, cloned, expressed in E. coli separately (Figure 4B) and used for analysing toxin binding. Using non-denaturing conditions and ELISA, we showed that Cry11A binds CR8–11, but not CR7 (Figure 4C). Interestingly, in toxin overlay assays only CR9 and CR10 bound Cry11Aa toxin (Supplementary Figure S2 at http://www.BiochemJ.org/bj/424/bj4240191add.htm). Furthermore, CR10 can compete with Cry11Aa binding to BBMVs, when used as a competitor in a BBMV-binding competition assay (Table 2).
Figure 4. In toxin overlay assays and ELISA, the Cry11Aa toxin binds cadherin fragments and cadherin repeats.
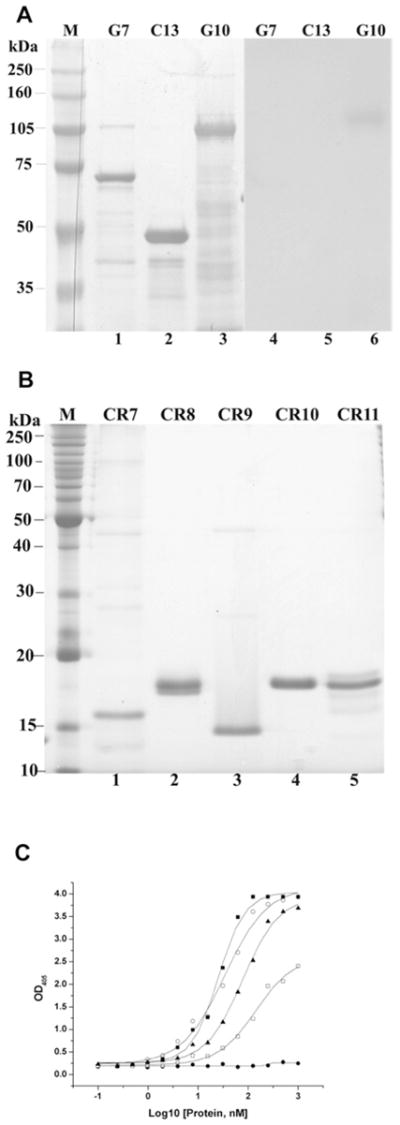
(A) Partial cDNA fragments, G7, C13 and G10 (90 pmol each) were separated by SDS/PAGE (8 % gel) and stained using Coomassie Blue (lanes 1–3). These fragments were electotransferred to a PVDF membrane, which was incubated with 20 nM Cry11Aa toxin. Unbound toxin was removed by washing, the membrane was incubated with anti-Cry11Aa antiserum (1:2000) and then visualized by luminol (lanes 5–7). (B) The purified cadherin repeats CR7–11 (30 pmol each) were resolved by SDS/PAGE (15 % gel) and stained using Coomassie Blue (lanes 1–5). (C) The cadherin repeats, CR8 (○), CR9 (■), CR10 (□) and CR11 (▲) show dose-dependent binding to immobilized Cry11Aa (0.4 μg), but CR7 (●) does not bind Cry11Aa. OD405, A 405.
Table 2.
Cadherin repeats can compete with Cry11Aa toxin binding to BBMV
| Competitor | Excess amount of protein | Toxin binding to BBMVs (%) |
|---|---|---|
| CR9 | 100-fold | 102 |
| 250-fold | 101 | |
| CR10 | 100-fold | 83 |
| 1000-fold | 29 |
Identification of Cry11Aa loop regions that interact with Aedes cadherin
The loop regions of many Cry toxins are known sites through which these toxins bind their receptors. These domains have been determined through a combination of mutations of the loop regions [40,41] or use of loop peptides in competition assays [22,27,42]. To determine which of the predicted Cry11 loop regions are involved in binding to Aedes cadherin, synthetic peptides corresponding to the four Cry11Aa loop regions, loop α-8, loop 1, loop 2 and loop 3 were synthesized (Table 1). These four peptides together with Cry11Aa were then used as competitors to biotinylated Cry11Aa in toxin overlay assays. Both loop α-8 and α-2 could compete with Cry11Aa binding to G10, CR9 and CR10 (Table 3). Loop 1 and loop 3 peptides showed little competition with toxin binding to these fragments (Table 3). As a positive control, excess unlabelled Cry11Aa also competed with the binding of biotinylated Cry11Aa toxin to G10 (Table 3).
Table 3.
Loop region peptides compete with Cry11Aa toxin binding to cadherin fragments and repeats
| Partial cadherin fragments | Toxin binding to peptide fragment in the presence of competitors (%)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Non-labelled toxin
|
Loop α-8
|
Loop 1
|
Loop 2
|
Loop 3
|
||||||
| 1:100 | 1:1000 | 1:100 | 1:1000 | 1:100 | 1:1000 | 1:100 | 1:1000 | 1:100 | 1:1000 | |
| G10 | 71 ± 3 | 40 ± 10 | 65 ± 4 | 54 ± 6 | 87 ± 3 | 90 ± 2 | 71 ± 3 | 70 ± 19 | 93 ± 8 | 101 ± 18 |
| CR9 | - | - | 73 ± 35 | 37 ± 28 | 102 ± 14 | 99 ± 8 | 34 ± 11 | 39 ± 4 | 104 ± 14 | 105 ± 2 |
| CR10 | - | - | 73 ± 13 | 65 ± 14 | 103 ± 13 | 108 ± 15 | 62 ± 8 | 54 ± 21 | 94 ± 16 | 108 ± 13 |
Determination of binding affinity between Cry11Aa toxin and G10
To measure Cry11Aa affinity to the cadherin fragment G10, a competitive ELISA was performed. As shown in Figure 5(A) (insert), the G10 protein binds Cry11Aa toxin in a dose-dependent manner. Moreover, a H. virescens cadherin fragment, PP9, that binds the Cry1A toxins, did not bind the Cry11Aa toxin. The G10 concentration that binds Cry11Aa in the linear range was used for the subsequent competitive ELISA. In this competitive ELISA, increasing Cry11Aa concentrations (0–1000 nM) displaced the G10 protein from binding immobilized Cry11Aa (Figure 5A). The apparent dissociation constant (Kd) for G10 binding to Cry11Aa toxin was 16.8 nM. These affinity levels are similar to the 20–30 nM binding affinity of Cry11Aa toxin to BBMVs prepared from the midguts of larval Culex pipiens (S.M. Dai and S.S. Gill, unpublished work).
Figure 5. Cry11Aa toxin binds the G10 cadherin fragment with high affinity.

(A) The cadherin fragment G10 shows dose-dependent binding (insert, ●) to immobilized Cry11Aa (0.4 μg), but a cadherin fragment from the Heliothis virescens cadherin [24] (insert, ■) does not bind Cry11Aa. Bound G10 was determined with an anti-AaeCad antiserum followed by incubation with secondary antibody and colour detection. The binding affinity, 16.7 nM, of Cry11Aa toxin to the cadherin fragment was determined in a competition assay using 80 nM G10 with increasing concentrations of Cry11Aa toxin (0–1000 nM). OD405, A 405. (B) The Cry11Aa loop α8 mutant V262A (○) retains its ability to bind the G10 fragment and competes with Cry11Aa binding, whereas the mutant E266A (▲) does not bind the Aedes cadherin fragment. Wild-type Cry11Aa binding is shown as ■ in (A, main graph) and (B). Maximal binding was normalized to the maximal absorbance obtained in the absence of G10. Error bars indicate standard deviation obtained using three separate experiments.
Mutagenesis in loop α8 (257GVSIPVNYNEWY268) of Cry11A toxin was performed previously, and the mutants V262A and E266A were shown to be less toxic to Ae. aegypti larva and were also less efficient in competing with Cry11A toxin binding to BBMVs [27]. In the present study, V262A showed similar binding characteristics to the G10 cadherin fragment as wild-type Cry11A, but in contrast the E266A mutant showed no binding to this cadherin fragment (Figure 5B).
B. thuringiensis subsp. israelensis produces two other mosquitocidal toxins, Cry4A and Cry4B, in addition to Cry11Aa. The Cry4A toxin can compete with the Cry11Aa toxin binding site at more than 100-fold concentration of the toxin, whereas Cry4B shows no competition at the concentrations tested. In contrast, Cry11Ba, a toxin isolated from B. thuringiensis subsp. jegathesan, competes well with the Cry11Aa-toxin binding site on the G10 cadherin fragment (Figure 6).
Figure 6. Mosquitocidal Cry11Ba and Cry4Aa toxins, but not Cry4Ba, compete with the Cry11Aa-binding site on the Aedes cadherin fragment.
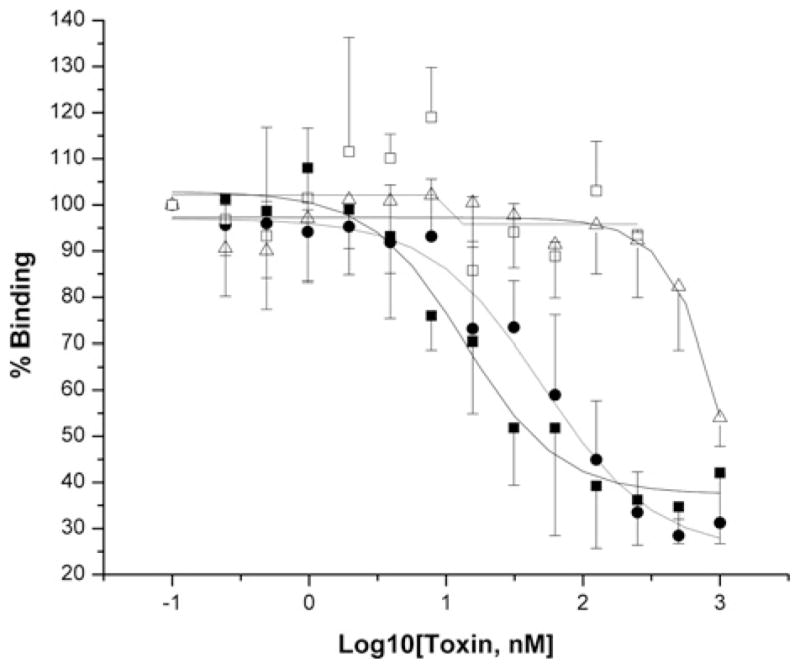
Cry11Ba (●) and Cry4Aa (△) show displacement of Cry11Aa (■) binding to G10, whereas Cry4Ba (□) does not compete. Binding assays were performed as described in the legend to Figure 5 and in the Materials and methods section. Error bars indicate standard deviation obtained using three separate experiments.
DISCUSSION
We previously showed in the disease vector Ae. aegypti that not only did a 65 kDa ALP protein act as a receptor protein, but that an unidentified 250 kDa membrane protein also bound the Cry11Aa toxin [27]. Here we show the 250 kDa protein is a cadherin, which binds Cry11Aa. Therefore, just as in lepidopteran insects, a single toxin can bind both a cadherin and a GPI-anchored protein. The Cry11Aa toxin binds Aedes cadherin with high affinity. The binding affinity of the G10 cadherin fragment is slightly lower than that reported for Cry1A toxin binding to M. sexta and H. virescens cadherin fragments [23,24,43].
In a toxin overlay assay that is traditionally used to map toxin-binding domains, the cadherin toxin-binding domain was mapped to CR9 and CR10 of Aedes cadherin (Supplementary Figure S1A). However, in ELISA experiments, Cry11A binds CR8–11, but not CR7 (Figure 4C). Since the latter experiments were done using non-denaturing conditions, the results suggest that CR11, which is the most proximal cadherin repeat to the cell membrane, and CR8–10, could both be involved in Cry11Aa toxicity. Indeed, CR10 inhibits Cry11Aa toxin binding to larval gut BBMVs. In An. gambiae, the Cry4Ba toxin also showed limited binding to the CR11-MPED peptide [18]. In contrast, the last cadherin repeat (CR12) is detected to specifically bind Cry1 toxins in lepidopteran insects [23,24]. It is important to note that the CR12 peptide of M. sexta was detected to bind Cry1A toxin under native conditions but not in denatured conditions [23,44]. It is likely that the CR11s of mosquito cadherins are critical for toxicity. In An. gambiae, the CR11-MPED peptide synergizes the toxicity of Cry4Ba to larvae, suggesting that this peptide fragment contains a Cry4Ba toxin-binding site [18].
Three Cry1A toxin-binding regions have been mapped in the cadherin of lepidopteran insects. These include CR7, CR11 and CR12 [20–24,44] which interact with loops 2, α8 and 3 of Cry1A toxins respectively. In the present study, we show that CR9 and CR10 bind the Cry11Aa toxin through domain II loops α8 and 2. A Cry11Aa mutant in loop α8, E266A, lost all ability to bind the G10 fragment that contains CR9 and CR10. These results are consistent with assays that show loop α8 peptides can compete with Cry11Aa binding to BBMVs [27]. Another mutant in this loop, V262A, retains a similar binding affinity with the G10 cadherin fragment as the wild-type Cry11Aa toxin. The lower toxicity of this mutant [27] could be caused by its lower solubility in larval midgut because it has lower solubility in carbonate buffer (results not shown) or potentially by decreased binding to other receptors, such as ALP, since loop α8 also plays a role in binding to the latter [27]. In addition, Cry11Aa binds the Cyt1A protein, which functions as a surrogate receptor by means of domain II loops α8 and β4 [30]. These results suggest that the exposed loop α8 of Cry11Aa toxin is an important epitope, including its ability to bind cadherin.
Fernandez et al. [27] showed that in addition to loop α8, loop 3 also can compete with Cry11Aa toxin binding to BBMV. Since this loop region binds native CR12 of lepidopteran insects, it is conceivable that Cry11Aa loop 3 would bind to native Aedes cadherin CR11. In any case, multiple toxin-binding sites on the cadherin receptors are probably essential for triggering the toxin conformation change, which facilitates the cleavage of helix α1 that promotes subsequent toxin oligomerization [2,14,45].
A number of proteins, including ALP, APNs and cadherins have been identified as potential receptors of mosquitocidal Cry proteins. This diversity in toxin-binding proteins is not unusual considering the diversity of Cry toxins in the mosquitocidal strain B. thuringiensis subsp. israelensis [3] and the different mosquito species that have been studied. In An. quadrimaculatus and An. gambiae, the highly active Cry11Ba binds an APN [16,17] and the Cry4Ba toxin targets a cadherin-like protein from An. gambiae [18]. In Ae. aegypti, the Cry11Aa toxin binds an ALP from Ae. aegypti [15], the cadherin reported in the present study and also two different aminopeptidases [46].
This diversity in the toxin-binding proteins is also observed in lepidopteran insects [2]. In this case, the Cry1A toxins are thought to first bind a cadherin protein with high affinity [9] followed by cleavage of helix α-1 by an unknown protease [14]. The proteolysed toxin undergoes oligomerization through coil-coiled helices [47] and the oligomer subsequently binds to a GPI-anchor protein, such as an ALP or an APN, thereby facilitating insertion into lipid rafts in cell membranes [2,28]. The model was validated recently where Cry1A toxins that lack helix α-1 were shown to kill lepidopteran larvae that are resistant because of mutated cadherins [19].
Since a similar set of toxin-binding proteins have been identified in mosquitoes, the mechanism of action of mosquitocidal Cry toxins is probably very comparable with that observed with lepidopteran-active toxin. Consequently, as a model we propose that the Cry11Aa toxin initially targets the cadherin identified here through CR9, CR10 and/or CR11 and subsequently, after proteolysis and oligomerization, binds to the ALP previously identified [15,27]. Both the cadherin and ALP proteins are expressed in the apical side of distal and proximal caeca and apical regions of posterior midgut of Ae. aegypti larvae. Of note, these are the same tissues which bind Cry toxins from B. thuringiensis subsp. israelensis [36] and show subsequent toxin pathogenicity [34]. The targeting of both caecae and posterior midgut but not the anterior midgut is not surprising, since the cells in these two regions have very similar repertoires of proteins and the pH in this region of the gut is closer to neutral. In contrast, the anterior midgut has an alkaline environment and is structurally and functionally distinct [48].
There are, however, some important differences in Cry toxin action in mosquitoes. For example, the Cry11Ba, a toxin related to Cry11Aa, binds an APN with high affinity in two anopheline mosquitoes [16,17]. These observations suggest that Cry11Ba could bind this APN directly without prior binding to cadherin and oligomerization. If these initial observations are further validated in mosquitoes, this evidence would suggest there is additional complexity to the mechanism of action of lepidopteran toxins that has hitherto not been appreciated.
Mosquitoes exposed to only the Cry11Aa toxin can develop resistance after selection for 16–20 generations with higher levels of resistance observed after further selection. Resistance development was slower when this toxin was used in combination with the Cry4A and Cry4B toxins [49]. The mosquitoes also developed resistance when only the Cry4A and Cry4B toxins were used [49]. The rapid development of resistance suggests it is likely that changes in the cadherin gene can lead to the Cry11Aa resistance observed in Culex. This resistance would be similar to the high levels of Cry1A resistance observed in lepidopteran larvae that is associated with mutations or deletions in cadherin [7,37,50].
Both the Cry11Aa and Cry11B toxins show high levels of toxicity to both Aedes and Culex species [51,52]. The ability of the Cry11B toxin to compete with Cry11Aa in binding assays suggests that both toxins use the same binding site on the G10 cadherin fragment and also are probably in the midgut of mosquitoes. Indeed, in Cry11Aa-resistant Culex mosquitoes, the Cry11B protein shows low level cross-resistance [53,54]. The Cry11Aa and Cry11B toxins also show cross-resistance to Culex mosquitoes resistant to Cry4A and Cry4B [54]. It is not surprising, therefore, that Cry4A shows some competition with the Cry11Aa toxin-binding site. However, the low level of competition observed suggests that in Aedes the Cry4A toxin probably binds to another binding site on cadherin or to another protein altogether.
Several protein receptors and their interaction with specific toxins have been reported in lepidopteran insects and more recently in mosquitoes. These investigations provide a deeper understanding of toxin-action mechanisms and for the improvement of B. thuringiensis toxin use in insect pest control programmes. For example, the M. sexta Bt-R1 region most proximal to the cell membrane (CR12-MPED) can synergize Cry1A toxicity against lepidopteran pest insects [43] and modified Cry1A toxins lacking helix α1 can kill cadherin-silenced M. sexta and toxin-resistant Pectinophora gossypiella that have cadherin deletion mutations, which may be useful against pests resistant to standard toxins [19]. It is expected that the CR toxin-binding domains in Aedes cadherin could also function as a synergist against dipteran insects, as recently reported for An. gambiae cadherin [18]. Additionally, knowledge on the interaction between Aedes caderin and Cry11Aa toxin together with the molecular mechanism of the Cyt1A synergism with Cry11Aa toxin [30] could lead to dipteran-specific toxin modification and improvement.
Supplementary Material
Acknowledgments
FUNDING
Research was funded in part through grants from the National Institutes of Health [grant number 1R01 AI066014], Dirección General de Asuntos del Personal Académico/Universidad Nacional Autónoma de México [grant numbers IN218608 and IN210208-N] and CONACyT (Consejo Nacional de Ciencia y Tecnología) [grant number U48631-Q].
Abbreviations used
- ALP
alkaline phosphatase
- APN
aminopeptidase N
- BBMV
brush border membrane vesicle
- CR
cadherin repeat
- Cy3
indocarbo-cyanine
- GPI
glycosylphosphatidylinositol
- HRP
horseradish peroxidase
- Ni-NTA
Ni2+-nitrilotriacetate
- ORF
open reading frame
- PBST
PBS plus 0.1 % Triton X-100
- PFA
paraformaldehyde
- RACE
rapid amplification of cDNA ends
Footnotes
AUTHOR CONTRIBUTIONS
Jianwu Chen, Alejandra Bravo, Mario Soberon and Sarjeet Gill designed and performed the research and jointly wrote and edited the manuscript. Karlygash Aimanova performed the immunohistochemical experiments and Luisa Fernandez performed the BBMV competition assay, and both contributed towards the manuscript for their respective sections.
References
- 1.Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 2.Bravo A, Gill SS, Soberon M. Bacillus thuringiensis: mechanisms and use. In: Gilbert LI, Kostas I, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 6. Elsevier; 2005. pp. 175–205. [Google Scholar]
- 3.Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MT, Harris D, Zaritsky A, Parkhill J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2002;68:5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai SM, Gill SS. In vitro and in vivo proteolysis of the Bacillus thuringiensis subsp. israelensis CryIVD protein by Culex quinquefasciatus larval midgut proteases. Insect Biochem Mol Biol. 1993;23:273–283. doi: 10.1016/0965-1748(93)90008-g. [DOI] [PubMed] [Google Scholar]
- 5.Yamagiwa M, Sakagawa K, Sakai H. Functional analysis of two processed fragments of Bacillus thuringiensis Cry11A toxin. Biosci Biotechnol Biochem. 2004;68:523–528. doi: 10.1271/bbb.68.523. [DOI] [PubMed] [Google Scholar]
- 6.Bravo A, Gomez I, Conde J, Munoz-Garay C, Sanchez J, Miranda R, Zhuang M, Gill SS, Soberon M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophys Acta. 2004;1667:38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Gahan LJ, Gould F, Heckle DG. Identification of a gene associated with Bt resistance in Heliothis virescens. Science. 2001;293:857–860. doi: 10.1126/science.1060949. [DOI] [PubMed] [Google Scholar]
- 8.Nagamatsu Y, Toda S, Yamaguchi F, Ogo M, Kogure M, Nakamura M, Shibata Y, Katsumoto T. Identification of Bombyx mori midgut receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin. Biosci Biotechnol Biochem. 1998;62:718–726. doi: 10.1271/bbb.62.718. [DOI] [PubMed] [Google Scholar]
- 9.Vadlamudi RK, Ji TH, Bulla LA., Jr A specific binding protein from Manduca sexta for the insecticidal toxin of Bacillus thuringiensis subsp. berliner. J Biol Chem. 1993;268:12334–12340. [PubMed] [Google Scholar]
- 10.Gill SS, Cowles EA, Francis V. Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J Biol Chem. 1995;270:27277–27282. doi: 10.1074/jbc.270.45.27277. [DOI] [PubMed] [Google Scholar]
- 11.Knight PJ, Knowles BH, Ellar DJ. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J Biol Chem. 1995;270:17765–17770. doi: 10.1074/jbc.270.30.17765. [DOI] [PubMed] [Google Scholar]
- 12.Jurat-Fuentes JL, Adang MJ. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur J Biochem. 2004;271:3127–3135. doi: 10.1111/j.1432-1033.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- 13.Valaitis AP, Jenkins JL, Lee MK, Dean DH, Garner KJ. Isolation and partial characterization of gypsy moth BTR-270, an anionic brush border membrane glycoconjugate that binds Bacillus thuringiensis Cry1A toxins with high affinity. Arch Insect Biochem Physiol. 2001;46:186–200. doi: 10.1002/arch.1028. [DOI] [PubMed] [Google Scholar]
- 14.Gomez I, Sanchez J, Miranda R, Bravo A, Soberon M. Cadherin-like receptor binding facilitates proteolytic cleavage of helix alpha-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002;513:242–246. doi: 10.1016/s0014-5793(02)02321-9. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez LE, Aimanova KG, Gill SS, Bravo A, Soberon M. A GPI-anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem J. 2006;394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdullah MA, Valaitis AP, Dean DH. Identification of a Bacillus thuringiensis Cry11Ba toxin-binding aminopeptidase from the mosquito, Anopheles quadrimaculatus. BMC Biochem. 2006;7:16. doi: 10.1186/1471-2091-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R, Hua G, Andacht TM, Adang MJ. A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae. Biochemistry. 2008;47:11263–11272. doi: 10.1021/bi801181g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua G, Zhang R, Abdullah MA, Adang MJ. Anopheles gambiae cadherin AgCad1 binds the Cry4Ba toxin of Bacillus thuringiensis israelensis and a fragment of AgCad1 synergizes toxicity. Biochemistry. 2008;47:5101–5110. doi: 10.1021/bi7023578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soberon M, Pardo-Lopez L, Lopez I, Gomez I, Tabashnik BE, Bravo A. Engineering modified Bt toxins to counter insect resistance. Science. 2007;318:1640–1642. doi: 10.1126/science.1146453. [DOI] [PubMed] [Google Scholar]
- 20.Gomez I, Miranda-Rios J, Rudino-Pinera E, Oltean DI, Gill SS, Bravo A, Soberon M. Hydropathic complementarity determines interaction of epitope 869HITDTNNK876 in Manduca sexta Bt-R1 receptor with loop 2 of domain II of Bacillus thuringiensis Cry1A toxins. J Biol Chem. 2002;277:30137–30143. doi: 10.1074/jbc.M203121200. [DOI] [PubMed] [Google Scholar]
- 21.Gomez I, Oltean DI, Gill S, Bravo A, Soberon M. Mapping the epitope in cadherin-like receptors involved in Bacillus thuringiensis Cry1A toxins interaction using phage display. J Biol Chem. 2001;276:28906–28912. doi: 10.1074/jbc.M103007200. [DOI] [PubMed] [Google Scholar]
- 22.Gomez I, Dean DH, Bravo A, Soberon M. Molecular basis for Bacillus thuringiensis Cry1Ab toxin specificity: two structural determinants in the Manduca sexta Bt-R1 receptor interact with loops α-8 and 2 in domain II of Cy1Ab toxin. Biochemistry. 2003;42:10482–10489. doi: 10.1021/bi034440p. [DOI] [PubMed] [Google Scholar]
- 23.Hua G, Jurat-Fuentes JL, Adang MJ. Bt-R1a extracellular cadherin repeat 12 mediates Bacillus thuringiensis Cry1Ab binding and cytotoxicity. J Biol Chem. 2004;279:28051–28056. doi: 10.1074/jbc.M400237200. [DOI] [PubMed] [Google Scholar]
- 24.Xie R, Zhuang M, Ross LS, Gomez I, Oltean DI, Bravo A, Soberon M, Gill SS. Single amino acid mutations in the cadherin receptor from Heliothis virescens affect its toxin binding ability to Cry1A toxins. J Biol Chem. 2005;280:8416–8425. doi: 10.1074/jbc.M408403200. [DOI] [PubMed] [Google Scholar]
- 25.Nagamatsu Y, Koike T, Sasaki K, Yoshimoto A, Furukawa Y. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal CryIAa toxin. FEBS Lett. 1999;460:385–390. doi: 10.1016/s0014-5793(99)01327-7. [DOI] [PubMed] [Google Scholar]
- 26.Delecluse A, Bourgouin C, Klier A, Rapoport G. Specificity of action on mosquito larvae of Bacillus thuringiensis israelensis toxins encoded by two different genes. Mol Gen Genet. 1988;214:42–47. doi: 10.1007/BF00340177. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez LE, Perez C, Segovia L, Rodriguez MH, Gill SS, Bravo A, Soberon M. Cry11Aa toxin from Bacillus thuringiensis binds its receptor in Aedes aegypti mosquito larvae through loop α-8 of domain II. FEBS Lett. 2005;579:3508–3514. doi: 10.1016/j.febslet.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 28.Zhuang M, Oltean DI, Gomez I, Pullikuth AK, Soberon M, Bravo A, Gill SS. Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation. J Biol Chem. 2002;277:13863–13872. doi: 10.1074/jbc.M110057200. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen-Leroux C, Charles JF. Binding of Bacillus sphaericus binary toxin to a specific receptor on midgut brush-border membranes from mosquito larvae. Eur J Biochem. 1992;210:585–590. doi: 10.1111/j.1432-1033.1992.tb17458.x. [DOI] [PubMed] [Google Scholar]
- 30.Perez C, Fernandez LE, Sun J, Folch JL, Gill SS, Soberon M, Bravo A. Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci USA. 2005;102:18303–18308. doi: 10.1073/pnas.0505494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aimanova KG, Zhuang M, Gill SS. Expression of Cry1Ac cadherin receptors in insect midgut and cell lines. J Invertebr Pathol. 2006;92:178–187. doi: 10.1016/j.jip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Brown MR, Hua G, Adang MJ. Comparison of the localization of Bacillus thuringiensis Cry1A δ-endotoxins and their binding proteins in larval midgut of tobacco hornworm, Manduca sexta. Cell Tissue Res. 2005;321:123–129. doi: 10.1007/s00441-005-1124-6. [DOI] [PubMed] [Google Scholar]
- 33.Candas M, Francis BR, Griko NB, Midboe EG, Bulla LA., Jr Proteolytic cleavage of the developmentally important cadherin BT-R1 in the midgut epithelium of Manduca sexta. Biochemistry. 2002;41:13717–13724. doi: 10.1021/bi026323k. [DOI] [PubMed] [Google Scholar]
- 34.Charles JF, de Barjac H. Action of crystals of Bacillus thuringiensis var. israelensis on the midgut of Aedes aegypti L larvae, studied by electron microscopy. Ann Microbiol (Paris) 1983;134A:197–218. [PubMed] [Google Scholar]
- 35.Zalunin IA, Chaika S, Dronina MA, Revina LP. Cytopathological effect of Bacillus thuringiensis israelensis endotoxins on the intestines of Aedes aegypti mosquito larvae. Parazitologiia. 2002;36:337–344. [PubMed] [Google Scholar]
- 36.Ravoahangimalala O, Charles JF. In vitro binding of Bacillus thuringiensis var. israelensis individual toxins to midgut cells of Anopheles gambiae larvae (Diptera: Culicidae) FEBS Lett. 1995;362:111–115. doi: 10.1016/0014-5793(95)00220-4. [DOI] [PubMed] [Google Scholar]
- 37.Morin S, Biggs RW, Sisterson MS, Shriver L, Ellers-Kirk C, Higginson D, Holley D, Gahan LJ, Heckel DG, Carriere Y, et al. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc Natl Acad Sci USA. 2003;100:5004–5009. doi: 10.1073/pnas.0831036100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagamatsu Y, Toda S, Koike T, Miyoshi Y, Shigematsu S, Kogure M. Cloning, sequencing, and expression of the Bombyx mori receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin. Biosci Biotechnol Biochem. 1998;62:727–734. doi: 10.1271/bbb.62.727. [DOI] [PubMed] [Google Scholar]
- 39.Vadlamudi RK, Weber E, Ji I, Ji TH, Bulla LA., Jr Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J Biol Chem. 1995;270:5490–5494. doi: 10.1074/jbc.270.10.5490. [DOI] [PubMed] [Google Scholar]
- 40.Rajamohan F, Cotrill JA, Gould F, Dean DH. Role of domain II, loop 2 residues of Bacillus thuringiensis CryIAb delta-endotoxin in reversible and irreversible binding to Manduca sexta and Heliothis virescens. J Biol Chem. 1996;271:2390–2396. doi: 10.1074/jbc.271.5.2390. [DOI] [PubMed] [Google Scholar]
- 41.Rajamohan F, Hussain SR, Cotrill JA, Gould F, Dean DH. Mutations at domain II, loop 3, of Bacillus thuringiensis CryIAa and CryIAb delta-endotoxins suggest loop 3 is involved in initial binding to lepidopteran midguts. J Biol Chem. 1996;271:25220–25226. doi: 10.1074/jbc.271.41.25220. [DOI] [PubMed] [Google Scholar]
- 42.Likitvivatanavong S, Aimanova K, Gill SS. Loop residues of the receptor binding domain of Bacillus thuringiensis Cry11Ba toxin are important for mosquitocidal activity. FEBS Lett. 2009;585:2021–2030. doi: 10.1016/j.febslet.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Hua G, Jurat-Fuentes JL, Abdullah MA, Adang MJ. Synergism of Bacillus thuringiensis toxins by a fragment of a toxin-binding cadherin. Proc Natl Acad Sci USA. 2007;104:13901–13906. doi: 10.1073/pnas.0706011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorsch JA, Candas M, Griko NB, Maaty WS, Midboe EG, Vadlamudi RK, Bulla LA. Cry1A toxins of Bacillus thuringiensis bind specifically to a region adjacent to the membrane-proximal extracellular domain of BT-R1 in Manduca sexta: involvement of a cadherin in the entomopathogenicity of Bacillus thuringiensis. Insect Biochem Mol Biol. 2002;32:1025–1036. doi: 10.1016/s0965-1748(02)00040-1. [DOI] [PubMed] [Google Scholar]
- 45.Soberon M, Fernandez LE, Perez C, Gill SS, Bravo A. Mode of action of mosquitocidal Bacillus thuringiensis toxins. Toxicon. 2007;49:597–600. doi: 10.1016/j.toxicon.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Aimanova KG, Pan S, Gill SS. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem Mol Biol. 2009 doi: 10.1016/j.ibmb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jimenez-Juarez N, Munoz-Garay C, Gomez I, Gill SS, Soberon M, Bravo A. The pre-pore from Bacillus thuringiensis Cry1Ab toxin is necessary to induce insect death in Manduca sexta. Peptides. 2008;29:318–323. doi: 10.1016/j.peptides.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onken H, Moffett DF. Revisiting the cellular mechanisms of strong luminal alkalinization in the anterior midgut of larval mosquitoes. J Exp Biol. 2009;212:373–377. doi: 10.1242/jeb.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Georghiou GP, Wirth MC. Influence of exposure to single versus multiple toxins of Bacillus thuringiensis subsp. israelensis on development of resistance in the mosquito Culex quinquefasciatus (Diptera: Culicidae) Appl Environ Microbiol. 1997;63:1095–1101. doi: 10.1128/aem.63.3.1095-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Chen H, Wu S, Yang Y, Xu X, Wu Y. Identification and molecular detection of a deletion mutation responsible for a truncated cadherin of Helicoverpa armigera. Insect Biochem Mol Biol. 2006;36:735–740. doi: 10.1016/j.ibmb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Chang C, Yu YM, Dai SM, Law SK, Gill SS. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delecluse A, Rosso ML, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microbiol. 1995;61:4230–4235. doi: 10.1128/aem.61.12.4230-4235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheong H, Dhesi RK, Gill SS. Marginal cross-resistance to mosquitocidal Bacillus thuringiensis strains in Cry11A-resistant larvae: presence of Cry11A-like toxins in these strains. FEMS Microbiol Lett. 1997;153:419–424. doi: 10.1111/j.1574-6968.1997.tb12605.x. [DOI] [PubMed] [Google Scholar]
- 54.Wirth MC, Delecluse A, Federici BA, Walton WE. Variable cross-resistance to Cry11B from Bacillus thuringiensis subsp. jegathesan in Culex quinquefasciatus (Diptera: Culicidae) resistant to single or multiple toxins of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 1998;64:4174–4179. doi: 10.1128/aem.64.11.4174-4179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pullikuth AK, Aimanova K, Kang’ethe W, Sanders HR, Gill SS. Molecular characterization of sodium/proton exchanger 3 (NHE3) from the yellow fever vector, Aedes aegypti. J Exp Biol. 2006;209:3529–3544. doi: 10.1242/jeb.02419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.