Abstract
The present study aims to investigate coelomocytes, immune mediators cells in the echinoderm Holothuria tubulosa, as an unusual source of antimicrobial and antibiofilm agents. The activity of the 5kDa peptide fraction of the cytosol from H. tubulosa coelomocytes (5-HCC) was tested against a reference group of Gram-negative and Gram-positive human pathogens. Minimal inhibitory concentrations (MICs) ranging from 125 to 500 mg/ml were determined against tested strains. The observed biological activity of 5-HCC could be due to two novel peptides, identified by capillary RP-HPLC/nESI-MS/MS, which present the common chemical-physical characteristics of antimicrobial peptides. Such peptides were chemically synthesized and their antimicrobial activity was tested. The synthetic peptides showed broad-spectrum activity at 12.5 mg/ml against the majority of the tested Gram-positive and Gram-negative strains, and they were also able to inhibit biofilm formation in a significant percentage at a concentration of 3.1 mg/ml against staphylococcal and Pseudomonas aeruginosa strains.
The immune mediators in H. tubulosa are a source of novel antimicrobial peptides for the development of new agents against biofilm bacterial communities that are often intrinsically resistant to conventional antibiotics.
Keywords: Biofilm, Staphylococci, Antimicrobial peptides (AMP), Innate immunity
Introduction
The survival and fitness of echinoderms in a marine environment, a habitat heavily populated by micro-organisms, suggest that they have developed a potent defensive mechanism that is part of an innate immune system (Tincu and Taylor 2004). The absence of fouling on many of these species indicates an effective defensive strategy to prevent the adhesion of bacteria, the first organisms that allow the formation of biofilm structures and fouling. With the aim of discovering new antimicrobial molecules, the echinoderm immune system was investigated as an unusual source of novel antimicrobial peptides (AMPs) with antibiofilm activity.
AMPs are small molecular weight proteins with broad spectrum antimicrobial activity against bacteria, viruses, and fungi. These evolutionarily conserved peptides are widely distributed in living organisms. Molecules with microbiocidal properties are found since in ameboid protozoa (Leippe 1999), prokaryotes (Pag and Sahl 2002), and plants (Garcia-Olmedo et al. 1998). Usually AMPs are positively charged and have both a hydrophobic and hydrophilic side that enables the molecule to be soluble in aqueous environments yet also enter lipid-rich membranes and kill target cells by using diverse mechanisms.
By focusing on the AMPs of echinoderms, many peptides have been discovered. In the coelomocytes (the immune mediators in echinoderms) of the sea star Asterias rubens a number of potential AMPs that are fragments of actin, histone HA2 and filamin were identified (Maltseva et al. 2004). In a recent study, the smallest peptide fragment of a beta-thymosin detected in the coelomocytes of Mediterranean sea-urchin Paracentrotus lividus showed the common chemical-physical characteristics of AMPs (Schillaci et al. 2012). In addition, two cystein-rich AMPs, named centrocyns, have been identified in the green sea urchin Strongylocentrotus droebachiensis (Li et al. 2010).
Recently Haug et al. (2002) described an antimicrobial peptide in the supernatant of the egg homogenate of Cucumaria frondosa with a high antibacterial activity, especially against Gram-positive bacteria. In Cucumaria echinata a peptide of 20 amino acids was described, corresponding to the sequence from residue 332 of hemolytic lectin CEL-III, which exhibited strong antibacterial activity toward Gram-positive bacteria. The antibacterial activity of this peptide was correlated with its membrane-permeabilizing activity that perturbs bacterial cell membranes, leading to enhancement of their permeability (Hatakeyama et al. 2004).
Over the last years, the study on natural AMPs has attracted considerable interest as a new class of antimicrobial drugs with antibiofilm activity (Batoni et al. 2011; Herrmann et al. 2010).
The growth of bacteria as biofilms is responsible for biomaterial and device-associated infections (Donlan 2001). Furthermore it is an important virulence factor of pathogens, like Staphylococcus aureus and Pseudomonas aeruginosa, in the development of the chronic form of their infections in human beings, such as native valve endocarditis, burn or wound infections, cystic fibrosis-associated infections, otolaryngological infections, etc. (Post et al. 2007). Biofilms show multi-factorial resistance to conventional antibiotics. Currently no effective therapies that target microbial biofilms exist; the prompt removal of the contaminated device or surgical intervention remain the most effective means to treat biofilm-associated infections (Brady et al. 2008). Therefore novel anti-biofilm agents, treatments and strategies are needed (Projan and Youngman 2002).
With the aim of searching for new antimicrobial agents as alternatives to conventional antibiotics, we focused on the coelomocytes of the echinoderm Holothuria tubulosa (sea-cucumber). The antimicrobial activity of a 5kDa peptide fraction from coelomocyte cytosol of H. tubulosa (5-HCC) was evaluated against a group of important reference strains of well-known human pathogens. Nine peptides were detected in the content of 5-HCC, and two of them were chemically synthesized and tested for their antimicrobial and anti-biofilm activities.
Materials and methods
Animals and bleeding procedure
H. tubulosa (sea cucumber) were collected from the Gulf of Palermo (Mongerbino 38°06’58” nord, 13°30’26” est) at a depth of 10 m near a prairie of Posidonia oceanica and maintained until their use in a refrigerated aquarium at 15°C and fed with commercial invertebrate food (Algamac 2000 Bio-Marine CA USA). The coelomic fluid (CF) was collected by cutting the anterior-dorsal part of the animals with a scalpel and draining the fluid into a plastic cup containing an isosmotic anticoagulant solution (0.5 M NaCl, 20 mM Tris–HCl, 30 mM EDTA; pH 7.4) (ISO-EDTA). Coelomocytes were separated from plasma by centrifugation at 900 g for 10 min at 4°C, washed twice in ISO-EDTA and suspended in the same medium at 5 × 106 cells/ml in ISO–EDTA. Total cell counts were determined using an improved Neubauer haemocytometer and dead cells were evaluated by using the eosin-y exclusion test (0.5% in ISO-EDTA).
Preparation of coelomocyte lysate supernatant (CLS)
Acid-soluble protein extracts were prepared according to a previously described method (Mercado et al. 2005) with some slight modifications. Coelomocytes (1 × 107) were suspended in a solution of 10% acetic acid in ISO without EDTA, sonicated (Sonifier Branson, model B-15 Danbury, CT USA) for 1 min at 0°C (1 pulse per second, 70% duty cycle) and centrifuged at 27,000 × g for 30 min at 4°C to remove any precipitate. After centrifugation, the supernatants were filtered (0.45 μM, Millex™, Millipore Corp.) and freeze dried to be later re-dissolved in H2O.
A 5 kDa peptide fraction of the cytosol from coelomocytes (5-HCC) was obtained by using a filter with a membrane with a nominal size of 5 kDa (Ultrafree-0.5 PBCC Centrifugal filter Unit, Amicon Millipore, MA, USA).
Identification of 5kDa peptide fraction (5-HCC) components
Capillary RP-HPLC/nESI-MS/MS analysis
HPLC-grade water and CH3CN were provided by Carlo Erba (Milan, Italy). Capillary RP-HPLC/nESI-MS/MS analysis of the in-gel enzymatic digests was performed using an Ultimate 3000 LC system combined with an autosampler and a 1:100 flow splitter (Dionex Corporation, Sunnyvale, CA, USA), coupled on-line with a linear ion trap nano-electrospray mass spectrometer (LTQ, Thermo Fischer Scientific, San Jose, CA). Ionization was performed with liquid junction using an uncoated capillary probe (20 mm i.d.; New Objective, Woburn, MA, USA). 2 mg of 5-HCC was freeze dried and re-dissolved in 0.1% aqueous formic acid (FA) (1 mg/ml). 10 μl of this solution was directly loaded onto a C18 μ-precolumn cartridge (0.3 μm × 5 mm, 100 Å, 5 μm, PepMap, Dionex) equilibrated with 0.1% aqueous formic acid (FA) at a flow rate of 20 μl/min for 4 min. Then, the solution was switched onto a reversed-phase C18 column (0.18 × 150 mm, 300 Å, 5 μm, BioBasic, ThermoFisher Scientific) and peptides were separated by elution at room temperature with a linear gradient of solvent B (ACN + 0.5% FA) in A (H2O + 0.5% FA) from 20% to 55% in 40 min at a flow rate of 1.5 μl/min. The nano-ESI source operated under the following conditions: capillary temperature 220°C, source voltage 2 kV, capillary voltage 45 V. Repetitive mass spectra were acquired in positive ion mode in the m/z range 450–2000. Characterization of peptide ions was performed by the data-dependent method as follows: 1) full MS scan (mass-to-charge ratio 450–2000); 2) zoom scan of the three most intense ions (isolation width: 2 Da) and 3) MS/MS of these three ions (activation Q 0.250; activation time 30 ms; normalized collision energy 27 a.u.). Mass calibration was made using a standard mixture of caffeine (Mr 194.1 Da), MRFA peptide (Mr 423.6 Da) and Ultramark (Mr 1621 Da). Data acquisition was performed using the Excalibur v. 1 · 4 software (ThermoFisher Scientific).
Database search and peptide identification via MASCOT Engine
MS/MS data were used to perform peptide and protein identifications by searching in the NCBI non-redundant protein sequence database of Echinodermata (60697 entries, updated June 2011) using the MOWSE algorithm as implemented in the MS search engine MASCOT server (Matrix Science, London, UK, version 2.3). MASCOT was set up as follows: i) assuming no enzyme as cleavage specificity; ii) with a fragment ion mass tolerance of ±0.6 Da and a parent ion mass (PIM) tolerance of ± 1.6 Da; iii) specifying oxidation of methionine, transformation of N-terminal glutamine and N-terminal glutamic acid residue in the pyroglutamic acid form as variable modifications.
In MASCOT searches, proteins scores (as MudPIT score) were derived from ions scores as a non-probabilistic basis for ranking protein hits, disregarding all individual peptides with scores below 20. Only proteins that met the following criteria were accepted as unambiguously identified: 1) MASCOT score >76 (probability-based MOWSE score: -10*log(P), where P is the probability that the observed match is a random event; score > 76 indicate identity or extensive homology (P < 0 · 05)).
Because all protein hits derived from the MASCOT search did not satisfy the above reported criteria, all the MS/MS spectra were also subjected to manual interpretation assisted by the PepNovo software (http://proteomics.ucsd.edu/Software/PepNovo.html) in order to obtain peptide sequence information.
Synthetic peptides
Two of the nine identified peptides, peptides H1 and H2, were purchased from CASLO, Lyngby, Denmark. The peptides were synthesized using Fmoc solid phase technology and the peptide content and purity were determined by high performance liquid chromatography (HPLC) and mass spectrometry (MS) analysis.
Antimicrobial assays
Microbial strains
The strains Staphylococcus aureus ATCC 25923, Staphylococcus aureus ATCC 29213, Staphylococcus aureus ATCC 6538, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 15442 were used in the assay. The good biofilm producer Staphylococcus epidermidis ATCC 35984 was also included in the analysis.
Minimum inhibitory concentrations (MIC)
MICs of 5-HCC and of chemically synthesized peptides were determined by a micro-method as previously described (Schillaci et al. 2008; Schillaci et al. 2005). Tryptic Soy Broth (TSB, Sigma) containing 2% glucose was used as medium.
Biofilm capability evaluation (Safranin method)
The staphylococcal strains and P. aeruginosa ATCC15442 were tested for their ability to form biofilms. Briefly, bacteria were grown in TSB containing 2% glucose overnight at 37°C in a shaking bath and then diluted 1:200 to a suspension with optical density (OD) of about 0.040 at 570 nm (Pitts et al. 2003). Polystyrene 24-well tissue culture plates were filled with 1 ml of diluted suspension and incubated for 24 hours at 37°C. The wells were then washed three times with 1 ml of sterile phosphate-buffered saline (PBS) and stained with 1 ml of safranin 0.1% v/v for 1 min. The excess stain was removed by placing the plates under running tap water. Plates were dried overnight in an inverted position at 37°C. Safranin-stained adherent bacteria in each well were re-dissolved to homogeneity in 1 ml of 30% v/v glacial acetic acid, and the OD was read at 492 nm. Each assay was performed in triplicate and repeated at least twice.
Interference with biofilm formation assay
The procedure described above was used to evaluate the activity of synthesized H1 and H2 in the prevention of biofilm formation. Polystyrene 24-well tissue culture plates were filled with 1 ml of diluted bacterial suspension, obtained and diluted as previously described, and sub-MIC concentrations (6.2 and 3.1 mg/ml) of peptides 7 and 8 were directly added to the bacterial suspension at time zero and incubated at 37°C for 24 hours. The wells were then washed and stained with safranin as per the biofilm forming assay. By comparing the average optical density (O.D.) of the growth control wells with the sample wells, the following formula was used to calculate the percentages of inhibition for each concentration of the sample:
| (1) |
Hemolytic assay
The assay was performed as described by (Arizza et al. 2013). In brief, freshly collected rabbit erythrocytes (RE) with heparin, kindly provided by the “Zooprophylaxis Institute of Sicily” (Palermo, Italy), were washed (400 g for 10 min at 4°C) to remove the buffy coat, and the erythrocytes obtained were washed three times with phosphate-buffered saline (PBS: 6 mM KH2PO4; 30 mM Na2HPO4; 0.11 M NaCl; pH 7.4) and suspended in 10 ml PBS to obtain an 80 × 106 cells/ml suspension. Aliquots of 200 μl of RE suspension were mixed with 200 μl of H1 and H2 concentrations (1.5, 3.2, 6.2 and 50 mg/ml) prepared in PBS, two-fold sequentially diluted v/v with a PBS. After a 1 h incubation at 37°C, the reaction mixture was centrifuged at 800 g for 15 min at 4°C to remove debris and residual erythrocytes. The O.D. of hemoglobin release was measured spectrophotometrically at a 541 nm wavelength. Spontaneous hemoglobin release (0%) was estimated incubating the RE with PBS while the complete hemolysis (100%) was assessed incubating the erythrocytes in a solution of 0.1% Triton-X 100 in distilled water and the hemolysis percentage was calculated according to the following equation:
| (2) |
Statistical analysis
Each experiment was performed in triplicate. The values were the mean of three assays ± SD. Significance was determined by using the Student's t-test and differences were considered significant at P < 0.05.
Results
Conformational analysis of 5kDa peptide fraction (5-HCC) components
With the aim of detecting the presence of novel antimicrobial peptides and determining their sequence, the 5-HCC fraction was subjected to capillary RP-HPLC-HPLC/nESI-MS/MS. The MS/MS data obtained were used to query the protein database of Echinodermata and to perform de novo peptide sequencing.
With this approach, nine principal peptides with molecular masses ranging from 805.5 to 2215.7 Da were identified. Their sequences are reported in Table 1.
Table 1.
Chemical-physical characteristics and amino acid sequences of peptides detected in 5-HCC content
| Peptides | ESI-MS calculated MH + Da | Total net charge (pH 7) | Total hydrophobic ratio (%) | pI | Sequences, and hydrophobic amino acids on the same face (underlined) |
|---|---|---|---|---|---|
| #1 |
805.5 |
-1.8 |
28.57 |
4.47 |
FTDESHA |
| #2 |
919.2 |
-0.6 |
50.00 |
6.48 |
LSELLHHA |
| #3 |
1003.4 |
-0.6 |
44.44 |
6.48 |
GVLSELLHH |
| #4 |
1017.4 |
-0.6 |
55.56 |
6.48 |
VLSELLHHA |
| #5 |
1074.4 |
-0.6 |
50.00 |
6.48 |
GVLSELLHHA |
| #6 |
1367.6 |
-0.4 |
41.67 |
6.77 |
HLGHHALDDLLK |
| #7 |
1389.5 |
+0.9 |
41.67 |
7.56 |
HLGHHALDHLLK |
| #8 |
1547.6 |
+0.9 |
42.86 |
7.56 |
ASHLGHHALDHLLK |
| #9 | 2111.3 | -3.1 | 27.78 | 4.23 | EVKPNLTEKIEDLSQQMD |
By using an antimicrobial peptide database (Wang and Wang 2004), we observed that only two of the nine peptides, Holothuroidin 1 (H1) and Holothuroidin 2 (H2), identified as peptide #7 and #8, whose molecular weights are respectively 1389.5 and 1547.6 Da, were likely to be novel AMPs, due to their chemical-physical characteristics and similarity with already defined AMPs. They had a sequence of 12 and 14 amino acids (a.a.), respectively, and the H2 differed only for the first two a. a. (A1 and S2), which were not present in H1. They are cationic peptides mainly enriched by residues such as lysine hystidine and arginine, with a net charge of +0.9, calculated by using the protein calculator v3.3. (http://www.scripps.edu/~cdputnam/protcalc.html) and a deduced pI of 7.5 calculated by using a web tool (http://isoelectric.ovh.org/) (Table 1).
Both H1 and H2 peptides showed a similarity ≥ 35% respectively with protonectins, a peptide present in the venom of the neotropical social wasp Agelaia pallipes pallipes, with a potent antimicrobial action against both Gram-positive and Gram-negative bacteria (Mendes et al. 2004) (Table 2) and signiferins, a naturally occurring cationic peptide produced by an Australian frog, Crinia signifera, that showed a wide spectrum of activity against Gram-positive and -negative bacteria including B. cereus, E. faecalis, L. lactis, L. innocua, M. luteus, S. aureus, S. epidermidis and S. uberis (Maselli et al. 2004) (Table 3). The other identified peptides (#1-#6 and #9) in H. tubulosa are negatively charged and have very little likelihood of being AMPs.
Table 2.
Sequence comparation of Holothuroidin 1 with other AMPs
| Peptides | Sequence | Similarity (%) | Species |
|---|---|---|---|
| Holothurioidin 1 |
H LGHHALDHL LK |
|
Holothuria tubulosa |
| Protonectin |
I LG + TIL + GL LKGL |
42.85 |
Agelaia pallipes pallipes (Mendes et al. 2004) |
| Polybia-CP |
I LG + TIL + GL LKSL |
42.85 |
Polybia paulista (Souza et al. 2005) |
| Macropin 1 |
G FG + MAL + KL LKKVL |
40.00 |
Macropis fulvipes (Slaninova et al. 2012) |
| Signiferin 2.2 |
IIGH LIKTALGFLGL+ |
37.50 |
Crinia signifera (Maselli et al. 2004) |
| Temporin-1DRa | HFLG++TLVNLAKKIL | 37.50 | Rana aurora draytonii (Conlon et al. 2007) |
Table 3.
Sequence comparison of Holothuroidin 2 with other AMPs
| Peptides | Sequence | Similarity (%) | Species |
|---|---|---|---|
| Holothurioidin 2 |
ASHLGHHA LDHL LK |
|
Holothuria tubulosa |
| Frenatin 3 |
GLMSVLG + HAVGNVLGGL FKS |
38.09 |
Litoria infrafrenata (Raftery et al. 1996) |
| Signiferin 2.2 |
IIGHLIKTA LGFLGL+ |
37.5 |
Crinia signifera (Maselli et al. 2004) |
| Signiferin 2.1 |
IIGHLIKTA LGMLGL+ |
37.5 |
Crinia signifera (Maselli et al. 2004) |
| Temporin 1Ja | ILPLVG+N LLND+LL+ | 37.5 | Rana japonica (Isaacson et al. 2002) |
A Schiffer-Edmundson helical wheel projection (Schiffer and Edmundson 1967) of H1 and H2, performed using Helical Wheel Projection (rzlab.ucr.edu/script/wheel/wheel.cgi), indicated that the α-helixes of both peptides had considerable amphipathic character, with the polar and mainly cationic residues D8, H1, H4, H5, H9 and K12 segregated on one polar face and the hydrophobic or non-polar residues A6, L2, L7, L10, L11 on the opposite apolar face for H1 (Figure 1A), and D10, H3, H6, H7, H11, and K14 segregated on the polar face and the hydrophobic or non-polar residues A1, A8, G5, L4, L9, L12, L13, S2 placed on the opposite apolar face for H2 Figure 1B). Their mean hydrophobic moments and hydrophobic arc size range were respectively 5.28 and 193.6° for H1 and 5.08 and 173.3° for H2 (Figure 1).
Figure 1.
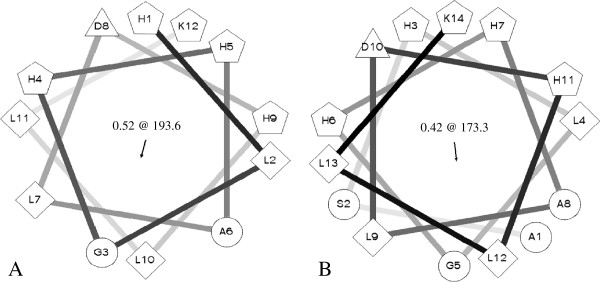
Edmundson wheel projection of (A) Holothuroidin 1 and (B) Holothuroidin 2 calculated by the Helical Wheel Projection web tool. Residues are schematically represented by geometric figures. Hydrophilic residues = circles, hydrophobic residues = diamonds, potentially negatively charged = triangles, and potentially positively charged = pentagons. In the centre mean relative hydrophobic moment and hydrophobic arc (@) are indicated.
The use of DeepView software (Swiss Institute of Bioinformatics - http://spdbv.vital-it.ch/) made it possible to resolve the secondary alpha-elical structure, which for both peptides was shown to be a periodic distribution of hydrophobic and hydrophilic residues, residues 4 – 10 for H1 and residues 2 – 12 for H2, in which the hydrophobic face is opposed to the hydrophilic one (Figure 2).
Figure 2.
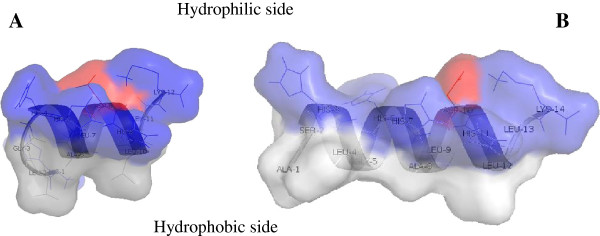
A ribbon representation of Holoturoidin 1 (A) and Holoturoidin 2 (B). The amphipathic nature of the peptide is shown in this representation with the hydrophilic side above and the hydrophobic side below the polypeptide backbone. The potential surface is superimposed. Color code: acidic residues in red, basic residues in blue, and hydrophobic residues in white.
Antimicrobial activity of 5-HCC
The antimicrobial activity of the 5kDa peptide fraction from coelomocyte cytosol (5-HCC) of H. tubulosa expressed as minimum inhibitory concentrations (MICs) against microbial reference strains are listed in Table 4. The 5-HCC resulted active against all tested reference microbial strains with MIC values ranging from 125 to 500 mg/ml.
Table 4.
Antibacterial activity of 5HCC assayed by MIC
|
Bacterial strains |
MIC |
|---|---|
| Values in vitro (mg/ml) | |
|
S. aureus ATCC 29213 |
500 |
|
S. aureus ATCC 25923 |
250 |
|
E. faecalis ATCC 29212 |
500 |
| P. aeruginosa ATCC 15442 | 500 |
Antimicrobial activity of synthetic peptides
The synthetic peptides H1 and H2 were screened for their antimicrobial activity against the same group used for the evaluation of biological activity of 5-HCC. The H2 resulted active at 12.5 mg/ml against planktonic Gram-positive (staphylococcal reference strains and E. faecalis ATCC 29212) and against P. aeruginosa ATCC 15442. A similar activity at the same concentration was observed for H1 but it resulted not active at the maximum tested concentration against S. aureus ATCC 29213, and E. faecalis ATCC 29212. See Table 5.
Table 5.
Antibacterial activity of synthetic Holothuroidin 1 and Holothuroidin 2 assayed by MIC
|
Bacterial strains |
MIC (mg/ml) |
|
|---|---|---|
| Holothuroidin 1 | Holothuroidin 2 | |
|
S. aureus ATCC 29213 |
>12.5 |
12.5 |
|
S. aureus ATCC 25923 |
12.5 |
12.5 |
|
S. aureus ATCC 6538 |
12.5 |
12.5 |
|
S. epidermidis ATCC 35984 |
12.5 |
12.5 |
|
E. faecalis ATCC 29212 |
>12.5 |
12.5 |
| P. aeruginosa ATCC 15442 | 12.5 | 12.5 |
Interference with biofilm formation
The impact of H1 and H2 on the biofilm formation of two staphylococcal reference strains, S. aureus ATCC 25923, and S. epidermidis ATCC 35984, were evaluated at concentrations of 3.2 and 1.5 mg/ml (well below the MIC values obtained against the planktonic forms) by staining with safranin adherent bacteria. The H1 inhibited biofilm formation was 51.8% for S. aureus ATCC 25923 and 68.5% for S. epidermidis ATCC 35984 at 3.2 mg/ml; the inhibition was 37.9% for S. aureus and 58.2% for S. epidermidis at the lower concentration of 1.5 mg/ml. The same concentrations were used to analyze the effect of H2 on biofilm formation. Higher inhibitions of 57.7% and 73.8% at 3.2 mg/ml and 40.5% and 59.7% at 1.5 mg/ml were observed on S. aureus and S. epidermidis biofilms respectively.
H1 and H2 also interfere with the biofilm formation of P. aeruginosa ATCC 15442 at the sub-MIC concentrations of 6.2 and 3.1 mg/ml. Inhibition percentages of 69.9% and 62.7% for H1 and 64.3% and 43.8% for H2 were observed (Figure 3).
Figure 3.
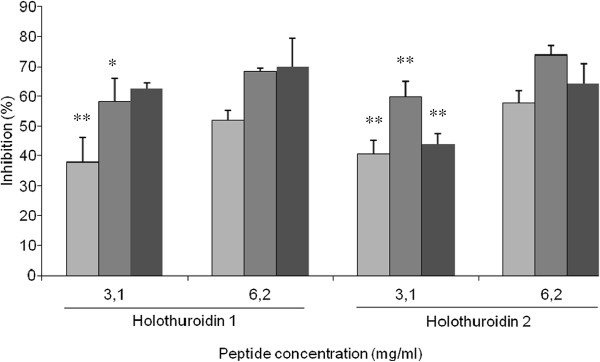
Inhibition activity of Holothuroidin 1 and 2 on S. aureus ATCC 25923, S. epidermidis ATCC 35984 (light gray square symbol) and P. aeruginosa ATCC 15442 (dark gray square symbol) biofilms. The data are expressed as the mean of three different experiments ± standard deviation. * = p < 0.05; ** = p < 0.01.
Hemolytic assay
The hemolytic activity of antimicrobial peptides against mammalian erythrocytes is often used as to measure their cytotoxicity. The hemolytic experiment performed to evaluate the toxicity of the peptides H1 and H2 did not show a measurable toxic effect against RBC (~ 1%) at MIC concentrations (Figure 4). A slight hemolytic activity, about 12.5%, was evident only at the highest concentration (50 mg/ml) (Figure 4).
Figure 4.
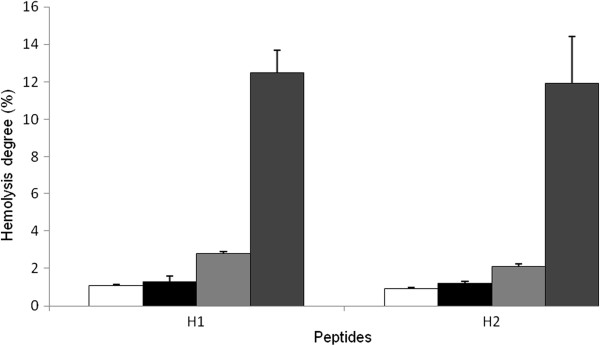
Hemolytic activity of H1 and H2 peptides from Holothuria tubulosa hemocytes against rabbit blood cells at different concentrations: white square symbol = 1.5 mg/ml; Black square symbol = 3.2; gray square symbol = 6.2; dark gray square symbol = 50 mg/ml. Data are the mean value of three separate experiments and expressed as percentage of hemolysis ± SD.
Discussion
Antimicrobial peptides (AMPs) represent the first line of defense against pathogens in several organisms (Hancock and Lehrer 1998). The majority of AMPs, constituted by a short stretch of amino acids residues (10–100 maximum), present the following common features: broad antimicrobial spectrum, small sizes, a net positive charge due to an excess number of cationic residues, an over-representation of one or two amino acids (Hughes and Fenical 2010).
Recently, a variety of peptides with antimicrobial properties have been isolated from echinoderms (Andersson et al. 1983; Bryan et al. 1994; Ridzwan et al. 1995). Antimicrobial activity has also been reported in gonads of the asteroid Marthasterias glacialis (Stabili and Pagliara 1994). We have already demonstrated that echinoderms such as Paracentrotus lividus contain, in the low molecular weight fraction (<5kDa) of acid precipitate from their coelomocytes, peptides with antimicrobial activity against staphylococci antibiofilm (Schillaci et al. 2010; Schillaci et al. 2012).
In this study, the fraction of low molecular weight peptide (<5kDa) from coelomocytes of the H. tubulosa (5-HCC) showed broad antimicrobial activity against many Gram-positive and Gram-negative pathogens tested. The use of capillary RP-HPLC/nESI-MS/MS allowed the characterization in the amino acid sequence of nine principal peptides in the 5-HCC content. Only two peptides, #7 and #8, called respectively Holothuroidin 1 and Holothuroidin 2, of the total nine peptides found in 5-HCC, are likely to be AMPs (Wang and Wang 2004).
Structurally they share the characteristics of many antimicrobial peptides: they are cationic peptides and possess a significant proportion (~ 30%) of hydrophobic residues (Hancock and Lehrer 1998; Zasloff 2002). They may form, as secondary structures, an α-helical, which is the most common structure of AMPs in nature and which is essential, and often sufficient, for antimicrobial activity (Mor and Nicolas 1994; Skerlavaj et al. 1996; Storici et al. 1994; Tossi et al. 1994). The helixes of two peptides present an asymmetrical arrangement of the hydrophobic residues, specifically: four hydrophobic residues on the same face and a total hydrophobic ratio of 41.67% for H1, and for H2, a hydrophobic face opposite to the hydrophilic one and a total hydrophobic ratio of 42.86%. This characteristic conformation of the peptides, in the course of microbial membrane invasion, allows the non-polar face of their α-helical structure to interact with the membrane lipid core while at the same time permitting its hydrophilic face to engage in electrostatic interactions with the membrane lipid headgroup region (Phoenix et al. 2002).
Both peptides are not long enough (when in an alpha-helical conformation) to traverse the lipid bilayer of a bacterial cell. Indeed, the `barrel stave' mechanism requires a minimum of 20 residues (Bechinger 1997; Epand et al. 1995; Shai 1995), so probably some aggregation of Holothuroidin molecules on the membrane surface may be needed before the bacterial cell wall can be breached, e.g. via the so-called “carpet” mechanisms for the disruption of the bacterial membrane (Epand et al. 1995; Shai 1995).
In our study we observed that synthetic H1 and H2 did not show, in PBS, hemolytic activity, probably because the high ionic strength of PBS allows negatively charged sialic acid present on the erythrocyte membrane to neutralize the peptides. On the basis of this evidence, they could be classified as peptide antibiotics (Saberwal and Nagaraj 1995). The two peptides showed a modest broad-spectrum activity against planktonic strains (MIC equal to 12.5 mg/ml). However, they showed a good inhibition of biofilm formation in vitro at lower MIC concentrations when evaluated against the planktonic forms on two staphylococcal reference strains and P. aeruginosa ATCC 15442. Such additional antimicrobial effect could be due to different mechanisms of action during the biofilm formation: a) an interference with the initial adhesion of microbial cells to the surface could be due to modifications of the microbial membranes, b) the killing of the early bacterial colonizers, or c) the inhibition of Quorum Sensing (QS), i.e. the intercellular communication system involved in biofilm formation (Batoni et al. 2011).
Whatever the supposed mechanism of action, the prevention of biofilm formation – rather than its elimination – is an interesting strategy to contrast the growth, as a sessile community, of many pathogens.
The extensive clinical use of classical antibiotics has led to the growing emergence of many medically relevant resistant strains of bacteria biofilm (Patel 2005). Therefore, the development of a new class of antibiotics has become critical. The cationic antimicrobial peptides could represent such a new class of antibiotics (Andreu and Rivas 1998; Hancock 1997; Sitaram and Nagaraj 2002). The development of resistance to membrane active peptides whose sole target is the cytoplasmic membrane is not expected because this would require substantial changes in the lipid composition of cell membranes of microorganisms (Hancock 2001).
Novel AMPs derived from coelomocytes of H. tubulosa are attractive candidates for developing useful antimicrobial agents against surface adherent staphylococci and P. aeruginosa of clinical interest.
The anti-adhesion properties of the peptides H1 and H2 could be useful to contrast staphylococcal and P.aeruginosa biofilms involved in food spoilage and biofilm formation of food transmitted pathogens. Moreover, the tested peptides are a good starting point to design new synthetic derivatives with modified chemical-physical properties, with the aim to improve their antimicrobial activity against pathogens and their pharmaceutical potential (Brogden and Brogden 2011; Huang et al. 2010).
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Domenico Schillaci, Email: domenico.schillaci@unipa.it.
Maria Grazia Cusimano, Email: mgcusimano@libero.it.
Vincenzo Cunsolo, Email: vcunsolo@unict.it.
Rosaria Saletti, Email: rsaletti@unict.it.
Debora Russo, Email: debora.russo@unipa.it.
Mirella Vazzana, Email: mirella.vazzana@unipa.it.
Maria Vitale, Email: maria.vitale@izzsicilia.it.
Vincenzo Arizza, Email: vincenzo.arizza@unipa.it.
Acknowledgements
Financial support from Fondi di Ateneo 2007, Università degli Studi di Palermo (Italy), is gratefully acknowledged.
References
- Andersson L, Lidgren G, Bohlin L, Magni L, Ogren S, Afzelius L. Studies of Swedish marine organisms. I. Screening of biological activity. Acta Pharm Suec. 1983;3(6):401–414. [PubMed] [Google Scholar]
- Andreu D, Rivas L. Animal antimicrobial peptides: an overview. Biopolymers. 1998;3(6):415–433. doi: 10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Arizza V, Vazzana M, Schillaci D, Russo D, Giaramita FT, Parrinello N. Gender differences in the immune system activities of sea urchin Paracentrotus lividus. Comparative biochemistry and physiology Part A. Molecular & integrative physiology. 2013;3(3):447–455. doi: 10.1016/j.cbpa.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Batoni G, Maisetta G, Brancatisano FL, Esin S, Campa M. Use of antimicrobial peptides against microbial biofilms: advantages and limits. Curr Med Chem. 2011;3(2):256–279. doi: 10.2174/092986711794088399. [DOI] [PubMed] [Google Scholar]
- Bechinger B. Structure and functions of channel-forming peptides: magainins, cecropins, melittin and alamethicin. J Membrane Biol. 1997;3(3):197–211. doi: 10.1007/s002329900201. [DOI] [PubMed] [Google Scholar]
- Brady RA, Leid JG, Calhoun JH, Costerton JW, Shirtliff ME. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol Med Microbiol. 2008;3(1):13–22. doi: 10.1111/j.1574-695X.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- Brogden NK, Brogden KA. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int J Antimicrob Ag. 2011;3(3):217–225. doi: 10.1016/j.ijantimicag.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan PJ, McClintock JB, Watts SA, Marion KR, Hopkins TS. In: Echinoderms Through Time. David B, Guille A, Feral J-P, Roux M, editor. Balkema, Rotterdam; 1994. Antimicrobial activity of ethanolic extracts of echinoderms from the northern Gulf of Mexico; pp. 17–23. [Google Scholar]
- Conlon JM, Coquet L, Leprince J, Jouenne T, Vaudry H, Kolodziejek J, Nowotny N, Bevier CR, Moler PE. Peptidomic analysis of skin secretions from Rana heckscheri and Rana okaloosae provides insight into phylogenetic relationships among frogs of the Aquarana species group. Regul Pept. 2007;3(2-3):87–93. doi: 10.1016/j.regpep.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Donlan RM. Biofilms and device-associated infections. Emerg Infect Dis. 2001;3(2):277–281. doi: 10.3201/eid0702.700277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM, Shai YC, Segrest JP, Anantharamaiah GM. Mechanisms for the modulation of membrane bilayer properties by amphipathic helical peptides. Biopolymers. 1995;3(5):319–338. doi: 10.1002/bip.360370504. [DOI] [PubMed] [Google Scholar]
- Garcia-Olmedo F, Molina A, Alamillo JM, Rodriguez-Palenzuela P. Plant defense peptides. Biopolymers. 1998;3(6):479–491. doi: 10.1002/(Sici)1097-0282(1998)47:6<479::Aid-Bip6>3.0.Co;2-K. [DOI] [PubMed] [Google Scholar]
- Hancock REW. Peptide antibiotics. Lancet. 1997;3(9049):418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- Hancock RE. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;3(3):156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;3(2):82–88. doi: 10.1016/S0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- Hatakeyama T, Suenaga T, Eto S, Niidome T, Aoyagi H. Antibacterial activity of peptides derived from the C-terminal region of a hemolytic lectin, CEL-III, from the marine invertebrate Cucumaria echinata. J Biochem. 2004;3(1):65–70. doi: 10.1093/jb/mvh007. [DOI] [PubMed] [Google Scholar]
- Haug T, Kjuul AK, Styrvold OB, Sandsdalen E, Olsen OM, Stensvag K. Antibacterial activity in Strongylocentrotus droebachiensis (Echinoidea), Cucumaria frondosa (Holothuroidea), and Asterias rubens (Asteroidea) J Invertebr Pathol. 2002;3(2):94–102. doi: 10.1016/S0022-2011(02)00153-2. [DOI] [PubMed] [Google Scholar]
- Herrmann G, Yang L, Wu H, Song Z, Wang H, Hoiby N, Ulrich M, Molin S, Riethmuller J, Doring G. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J Infect Dis. 2010;3(10):1585–1592. doi: 10.1086/656788. [DOI] [PubMed] [Google Scholar]
- Huang YB, Huang JF, Chen YX. Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell. 2010;3(2):143–152. doi: 10.1007/s13238-010-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CC, Fenical W. Antibacterials from the sea. Chemistry (Weinheim an der Bergstrasse, Germany) 2010;3(42):12512–12525. doi: 10.1002/chem.201001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Soto A, Iwamuro S, Knoop FC, Conlon JM. Antimicrobial peptides with atypical structural features from the skin of the Japanese brown frog Rana japonica. Peptides. 2002;3(3):419–25. doi: 10.1016/S0196-9781(01)00634-9. [DOI] [PubMed] [Google Scholar]
- Leippe M. Antimicrobial and cytolytic polypeptides of amoeboid protozoa--effector molecules of primitive phagocytes. Dev Comp Immunol. 1999;3(4–5):267–279. doi: 10.1016/S0145-305X(99)00010-5. [DOI] [PubMed] [Google Scholar]
- Li C, Haug T, Moe MK, Styrvold OB, Stensvag K. Centrocins: isolation and characterization of novel dimeric antimicrobial peptides from the green sea urchin, Strongylocentrotus droebachiensis. Dev Comp Immunol. 2010;3(9):959–968. doi: 10.1016/j.dci.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Maltseva AL, Aleshina GM, Kokryakov VN, Krasnodembsky EG, Ovchinnikova TV. New antimicrobial peptides from coelomocytes of sea star Asterias rubens. Biologiya. 2004;3:8. [Google Scholar]
- Maselli VM, Brinkworth CS, Bowie JH, Tyler MJ. Host-defence skin peptides of the Australian Common Froglet Crinia signifera: sequence determination using positive and negative ion electrospray mass spectra. Rapid Commun Mass Sp. 2004;3(18):2155–2161. doi: 10.1002/Rcm.1602. [DOI] [PubMed] [Google Scholar]
- Mendes MA, de Souza BM, Marques MR, Palma MS. Structural and biological characterization of two novel peptides from the venom of the neotropical social wasp Agelaia pallipes pallipes. Toxicon. 2004;3(1):67–74. doi: 10.1016/j.toxicon.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Mercado L, Schmitt P, Marshall SH, Arenas G. Gill tissues of the mussel Mytilus edulis chilensis: A new source for antimicrobial peptides. Electron J Biotechn. 2005;3(3):284–290. doi: 10.2225/vol8-issue3-fulltext-2. [DOI] [Google Scholar]
- Mor A, Nicolas P. The Nh2-terminal alpha-helical domain 1–18 of Dermaseptin is responsible for antimicrobial activity. J Biol Chem. 1994;3(3):1934–1939. [PubMed] [Google Scholar]
- Pag U, Sahl HG. In: Peptide Antibiotics: Discovery, Mode of Actions, and Applications. Dutton CJ, Haxell MA, McArthur HAI, Wax RG, editor. Dekker, M, New York; 2002. Lanthionine-containing bacterial peptides; pp. 47–80. [Google Scholar]
- Patel R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res. 2005. pp. 41–47. [DOI] [PubMed]
- Phoenix DA, Harris F, Daman OA, Wallace J. The prediction of amphiphilic alpha-helices. Curr Protein Pept Sc. 2002;3(2):201–221. doi: 10.2174/1389203024605368. [DOI] [PubMed] [Google Scholar]
- Pitts B, Hamilton MA, Zelver N, Stewart PS. A microtiter-plate screening method for biofilm disinfection and removal. J Microbiol Methods. 2003;3(2):269–276. doi: 10.1016/S0167-7012(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Post JC, Hiller NL, Nistico L, Stoodley P, Ehrlich GD. The role of biofilms in otolaryngologic infections: update 2007. Curr Opin Otolaryngol Head Neck Surg. 2007;3(5):347–351. doi: 10.1097/MOO.0b013e3282b97327. [DOI] [PubMed] [Google Scholar]
- Projan SJ, Youngman PJ. Antimicrobials: new solutions badly needed. Curr Opin Microbiol. 2002;3(5):463–465. doi: 10.1016/S1369-5274(02)00364-8. [DOI] [PubMed] [Google Scholar]
- Raftery MJ, Waugh RJ, Bowie JH, Wallace JC, Tyler MJ. The structures of the frenatin peptides from the skin secretion of the giant tree frog Litoria infrafrenata. J Pept Sci. 1996;3(2):117–24. doi: 10.1002/psc.60. [DOI] [PubMed] [Google Scholar]
- Ridzwan BH, Kaswandi MA, Azman Y, Fuad M. Screening for antibacterial agents in three species of sea cucumbers from coastal areas of Sabah. Gen Pharmacol. 1995;3(7):1539–1543. doi: 10.1016/0306-3623(95)00041-0. [DOI] [PubMed] [Google Scholar]
- Saberwal G, Nagaraj R. Cell-Lytic and antibacterial peptides that act by perturbing the barrier function of membranes - facets of their conformational features, structure-function correlations and membrane-perturbing abilities. Bba-Biomembranes. 1995;3(1):159–159. doi: 10.1016/0304-4157(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Schiffer M, Edmundson AB. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967;3(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillaci D, Petruso S, Sciortino V. 3,4,5,3 ',5 '-Pentabromo-2-(2 '-hydroxybenzoyl)pyrrole: a potential lead compound as anti-Gram-positive and anti-biofilm agent. Int J Antimicrob Ag. 2005;3(4):338–340. doi: 10.1016/j.ijantimicag.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Schillaci D, Arizza V, Dayton T, Camarda L, Di Stefano V. In vitro anti-biofilm activity of Boswellia spp. oleogum resin essential oils. Lett Appl Microbiol. 2008;3(5):433–438. doi: 10.1111/j.1472-765X.2008.02469.x. [DOI] [PubMed] [Google Scholar]
- Schillaci D, Arizza V, Parrinello N, Di Stefano V, Fanara S, Muccilli V, Cunsolo V, Haagensen JJ, Molin S. Antimicrobial and antistaphylococcal biofilm activity from the sea urchin Paracentrotus lividus. J Appl Microbiol. 2010;3(1):17–24. doi: 10.1111/j.1365-2672.2009.04394.x. [DOI] [PubMed] [Google Scholar]
- Schillaci D, Vitale M, Cusimano MG, Arizza V. Fragments of beta-thymosin from the sea urchin Paracentrotus lividus as potential antimicrobial peptides against staphylococcal biofilms. Ann N Y Acad Sci. 2012;3:79–85. doi: 10.1111/j.1749-6632.2012.06652.x. [DOI] [PubMed] [Google Scholar]
- Shai Y. Molecular recognition between membrane-spanning polypeptides. Trends Biochem Sci. 1995;3(11):460–464. doi: 10.1016/S0968-0004(00)89101-X. [DOI] [PubMed] [Google Scholar]
- Sitaram N, Nagaraj R. The therapeutic potential of host-defense antimicrobial peptides. Curr Drug Targets. 2002;3(3):259–267. doi: 10.2174/1389450023347786. [DOI] [PubMed] [Google Scholar]
- Skerlavaj B, Gennaro R, Bagella L, Merluzzi L, Risso A, Zanetti M. Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J Biol Chem. 1996;3(45):28375–28381. doi: 10.1074/jbc.271.45.28375. [DOI] [PubMed] [Google Scholar]
- Slaninová J, Mlsová V, Kroupová H, Alán L, Tůmová T, Monincová L, Borovičková L, Fučík V, Ceřovský V. Toxicity study of antimicrobial peptides from wild bee venom and their analogs toward mammalian normal and cancer cells. Peptides. 2012;3(1):18–26. doi: 10.1016/j.peptides.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Souza BM, Mendes MA, Santos LD, Marques MR, César LM, Almeida RN, Pagnocca FC, Konno K, Palma MS. Structural and functional characterization of two novel peptide toxins isolated from the venom of the social wasp Polybia paulista. Peptides. 2005;3(11):2157–64. doi: 10.1016/j.peptides.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Stabili L, Pagliara P. Antibacterial protection in Marthasterias glacialis eggs: characterization of lysozyme-like activity. Comp Biochem Physiol B Biochem Mol Biol. 1994;3(4):709–713. doi: 10.1016/0305-0491(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Storici P, Scocchi M, Tossi A, Gennaro R, Zanetti M. Chemical synthesis and biological-activity of a novel antibacterial peptide deduced from a pig myeloid cDNA. FEBS Lett. 1994;3(3):303–307. doi: 10.1016/0014-5793(94)80214-9. [DOI] [PubMed] [Google Scholar]
- Tincu JA, Taylor SW. Antimicrobial peptides from marine invertebrates. Antimicrob Agents Chemother. 2004;3(10):3645–3654. doi: 10.1128/AAC.48.10.3645-3654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi A, Scocchi M, Skerlavaj B, Gennaro R. Identification and characterization of a primary antibacterial domain in Cap18, a lipopolysaccharide-binding protein from rabbit leukocytes. FEBS Lett. 1994;3(1–2):108–112. doi: 10.1016/0014-5793(94)80395-1. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang G. APD: the antimicrobial peptide database. Nucleic Acids Res. 2004;3(Database issue):D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;3(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]


