Abstract
The tryptophan synthase α2β2 bi-enzyme complex catalyzes the last two steps in the synthesis of L-tryptophan (L-Trp). The α-subunit catalyzes cleavage of 3-indole-D-glycerol 3’-phosphate (IGP) to give indole and D-glyceraldehyde 3’-phosphate (G3P). Indole is then transferred (channeled) via an interconnecting 25 Å-long tunnel, from the α-subunit to the (β-subunit where it reacts with L-Ser in a pyridoxal 5’-phosphate-dependent reaction to give L-Trp and a water molecule. The efficient utilization of IGP and L-Ser by tryptophan synthase to synthesize L-Trp utilizes a system of allosteric interactions that (1) function to switch the α-site on and off at different stages of the β-subunit catalytic cycle, and (2) prevent the escape of the channeled intermediate, indole, from the confines of the α- and β-catalytic sites and the interconnecting tunnel. This review discusses in detail the chemical origins of the allosteric interactions responsible both for switching the α-site on and off, and for triggering the conformational changes between open and closed states which prevent the escape of indole from the bienzyme complex.
Introduction
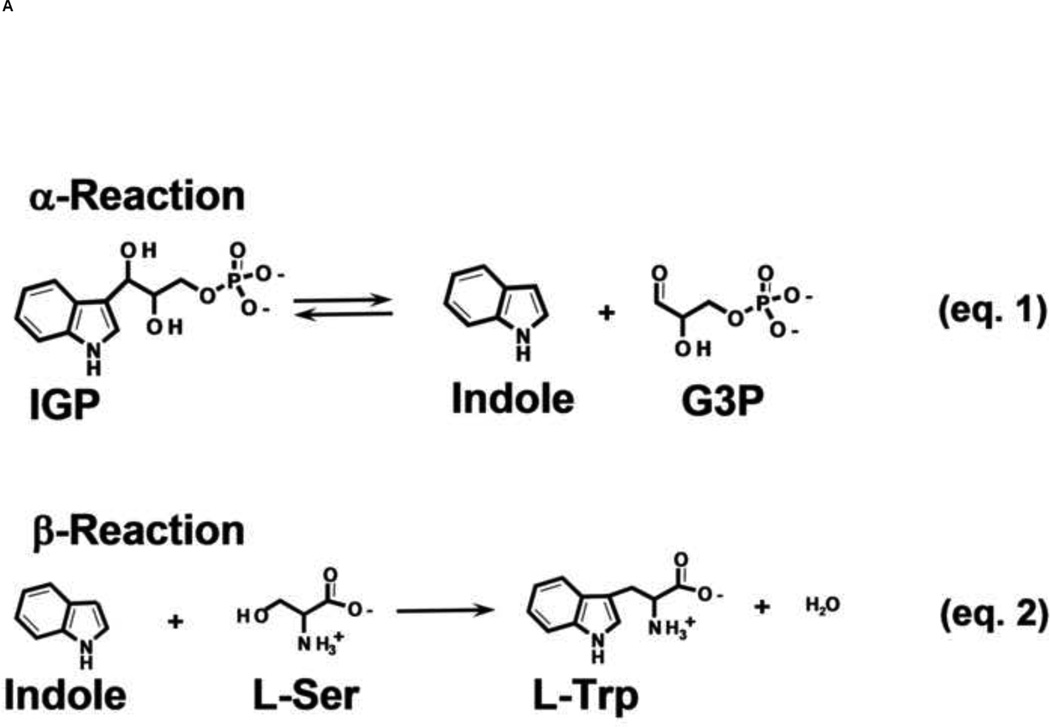
The acquisition of the amino acid, L-tryptophan (L-Trp), is critically important for the survival of all living organisms. Higher organisms obtain L-Trp through their diet. Bacteria, yeasts, molds and plants evolved to synthesize L-Trp using an elaborate set of protein catalysts encoded by the Tryptophan Operon (Trp Operon) [1, 2]. Expression of the Trp Operon proteins is subject to tight regulation, reflecting both the importance of L-Trp to the cell and the scarcity of the precursor metabolites. In gram negative bacteria, the α- and β-subunits of the tryptophan synthase bienzyme complex are the last two proteins encoded by the Trp Operon. These two enzymes carry out the last two steps in the biosynthesis of L-Trp (Figure 1A, eqs. 1 and 2) [2, 3].
Figure 1.
(A) Equations 1 and 2, the α- and β-reactions, respectively, catalyzed by tryptophan synthase [2]
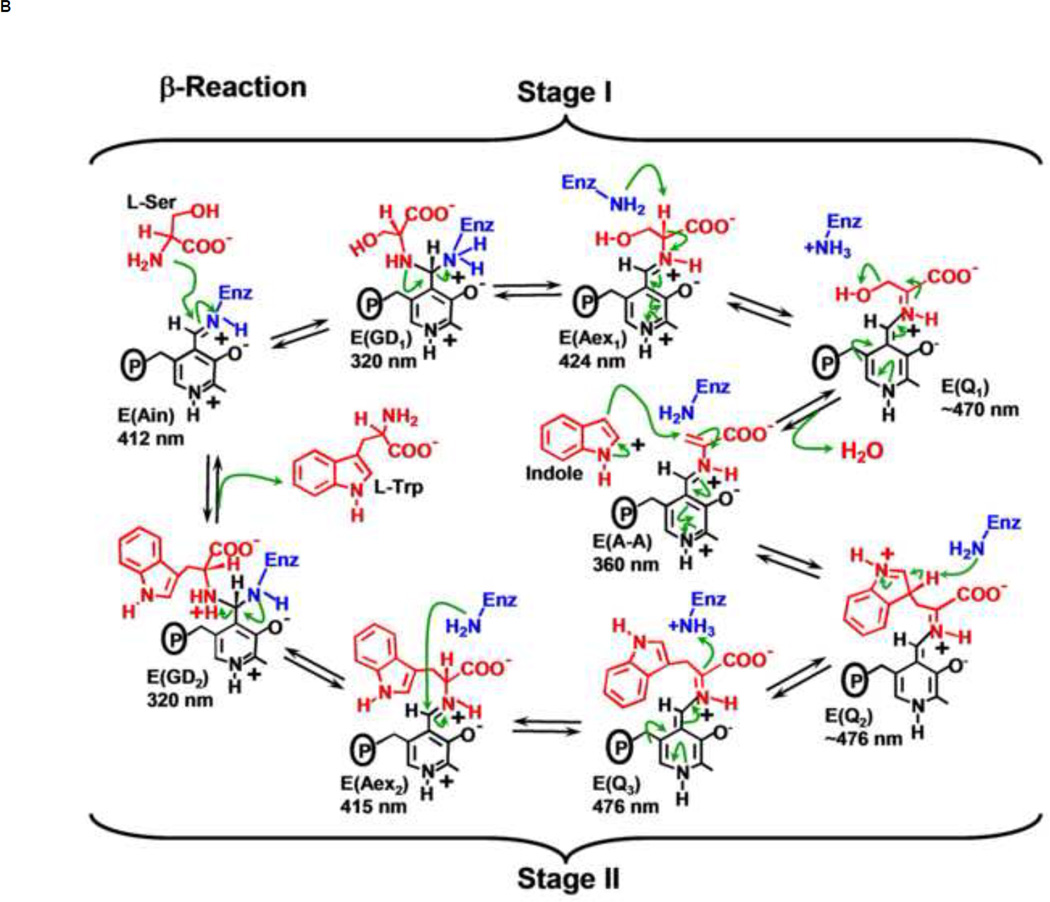
(B) Organic chemistry of Stages I and II of the β-reaction. The α-aminoacrylate intermediate, E(A–A), is formed in Stage I. Indole formed in the α-reaction reacts with E(A–A) in Stage II to give L-tryptophan.
The history of tryptophan synthase catalysis and regulation is very rich and it begins very early. In 1946, Gunsalus and his coworkers [4] discovered that pyridoxal 5’-phosphate (PLP) is essential for coupling indole and serine to give tryptophan. By 1955, Yanofsky [5] had begun to characterize the enzyme system responsible for this synthesis. In 1958 it already was recognized that 3-indole-D-glycerol 3’-phosphate (IGP) and L-serine (L-Ser) proceed to give L-Trp and G3P (Figure 1A, eqs. 1 and 2) without releasing indole into the medium [6, 7]. The work of Yanofsky and coworkers led to many early discoveries concerning the catalytic mechanism [2]. Michel Goldberg [8], Gordon Hammes [9], Sheldon York [10], and Kasper Kirschner [11, 12] also made important contributions regarding the catalytic mechanism of the β-subunits involving the detection of intermediates designated as “pale”, “aqua”, and “amber” during the reaction of L-Ser and indole with the enzyme. During the late 1970s and early 1980s, further mechanistic advances were achieved by Lane and Kirschner [13–15] and by my laboratory on the chemistry and mechanism of the β-reaction [16, 17], while Edith Miles’ laboratory was engaged in detailed protein chemical modification studies to identify amino acid residues important to the structure and mechanism of tryptophan synthase [3]. Miles et al. [18] also worked out the stereo chemistry of the proton transfers in the β-reaction. By 1986, essentially all of the chemical intermediates believed to be on the reaction path had been directly observed (principally by UV/Vis spectroscopy and rapid kinetics and by the use of substrate analogues) and were assigned chemical structures through comparison with the spectra of small molecule model compounds and deuterium kinetic isotope effects (Figure 1B, stages I and II) [13–17, 19, 20]. At this juncture, advances derived from x-ray crystallography [21, 22], rapid kinetic techniques [23], and site-directed mutagenesis converged [22, 24, 25], allowing tryptophan synthase structure-function studies to advance to the next level.
Since the 1980s, there has been a continuing refinement of our understanding of the relationship between protein structure, chemical transformations at the α- and β-sites, and the allosteric interactions that regulate substrate channeling in αβ-dimeric units of the α2β2 bienzyme complex [23, 26–30].
It is now clear that the chemical transformations at the β-site and ligand binding to the α-site are an integral part of the allosteric regulation of substrate channeling in tryptophan synthase [26–30]. This has been made apparent especially by the combination of solution kinetic studies with x-ray crystal structural results [26–28]. Consequently, a description of the α- and β-site catalytic mechanisms is essential for understanding the mechanism of allosteric regulation in tryptophan synthase. Therefore, a selective review of the catalytic mechanism literature is woven into the following allosteric regulation narrative.
Background: Discovery of the Bienzyme Complex, and Characterization of the Reactions Catalyzed by Tryptophan Synthase
In 1958, Crawford and Yanofsky demonstrated that the E coli tryptophan synthase consists of two protein components which they named A and B (later designated as α and β), that in vivo are responsible for catalyzing conversion of IGP and L-Ser to L-Trp, G3P and a water molecule (Figure 1A).
The α-site converts IGP to indole and G3P in a reverse aldolytic cleavage reaction (Figure 1A, eq. 1). The β-site carries out a substitution reaction wherein the Ser hydroxyl is replaced by Indole, giving L-Trp and a water molecule (Figure 1A, eq. 2). It now is well-established that the β-reaction takes place via the chemical intermediates shown in Stages I and II of Figure 1B.
This pathway is comprised of a series of at least 9 reactions involving the PLP C-4’, the Ser C-α and C-β carbons, and the indole ring C-3. Consequently, there are C-N, C-O, C-C, C-H, and O-H bond breaking and bond forming steps, all of which must be catalyzed by a single active site.
The PLP moiety of the internal aldimine, E(Ain), is covalently linked to the β-site via a Schiff Base linkage to the Nε of βLys87 (Figure 1B). Stage I of the β-reaction (Figure 1B) is initiated by nucleophilic attack of the L-Ser Nα on the E(Ain) C-4’ to give the gem diamine intermediate, E(GD1). There follows a transimination reaction at C-4’ to give the external aldimine, E(Aex1). This intermediate undergoes deprotonation at C-α to give the L-Ser quinonoid species E(Q1). Then elimination of the β-hydroxyl of the L-Ser quinonoid species gives the quasi-stable α-aminoacrylate intermediate, E(A–A).
Stage II (Figure 1B) is initiated by the transfer of indole from the α-site to the β-site via the interconnecting tunnel. The enamine functionality of indole makes possible C-C bond formation via the nucleophilic attack of the indole C-3 on the C-β of the E(A–A) to give the L-Trp quinonoid intermediate, E(Q2). E(Q2) is then converted to E(Q3) by proton extraction from the C-3 of the indole moiety. Protonation at C-α of this quinonoid intermediate leads to the L-Trp external aldimine species, E(Aex2), which undergoes a second transimination reaction involving the Nε of the active site βLys87 to give intermediate E(GD2). The reaction cycle is completed by release of L-Trp and regeneration of E(Ain).
Early Evidence of Substrate Channeling
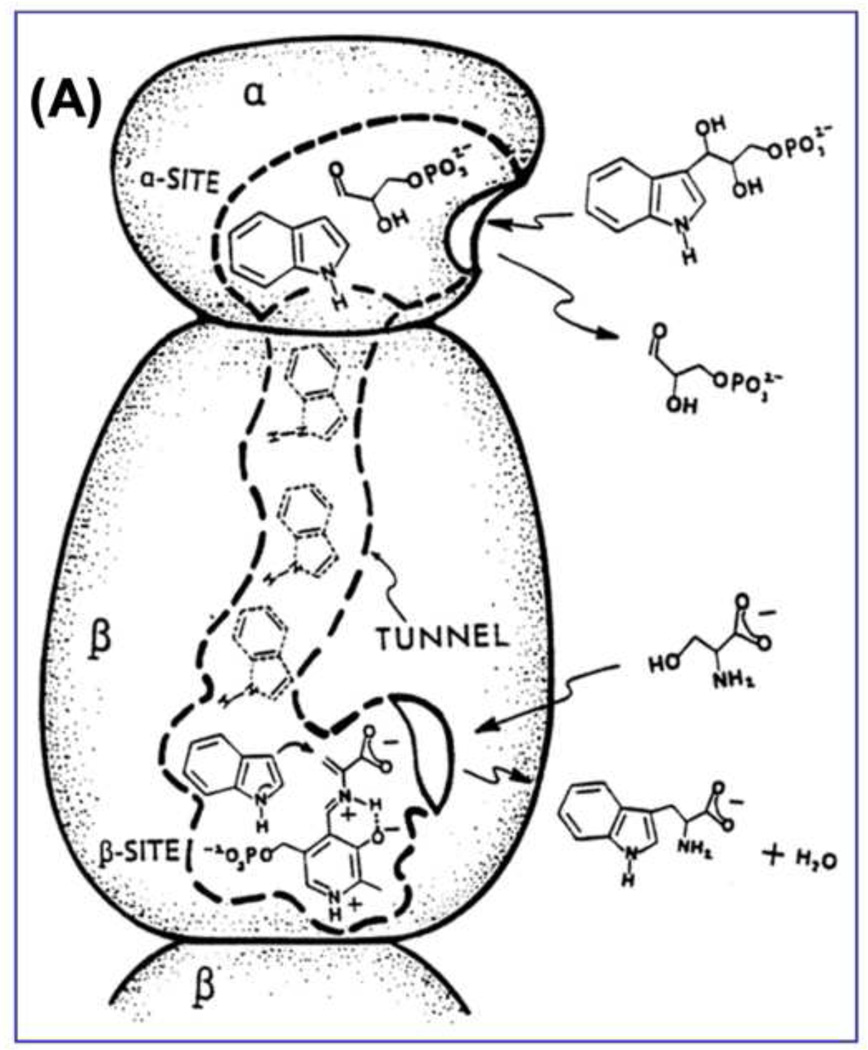
Because tryptophan synthase can synthesize L-Trp from L-Ser and indole, Yanofsky and Rachmeler [2] (1958) recognized that the synthesis of L-Trp from the physiologically important substrates IGP and L-Ser almost certainly involves indole as an intermediate on the chemical pathway. However, their experiments to detect free indole showed that indole is not released into solution during the synthesis of L-Trp from IGP and L-Ser, indicating that indole must be channeled between the α- and β-sites. Subsequent work in the ’60s and ‘70s by DeMoss [31], Creighton [32] and Matchett [33] further investigated the substrate channeling hypothesis for tryptophan synthase. All concluded that indole must be transferred via some (unknown) physical-chemical mechanism from the α-site to the β-site that does not allow release of indole into solution. Consequently, it was proposed that the transfer likely occurs either via a tunnel connecting the active sites, or by the close juxtaposition of the sites across the α-βsubunit interface [7, 31–33].
Evidence for a Protein Tunnel Connecting the α and β-Sites
In the ‘70s and early ‘80s, efforts by several groups to obtain diffraction quality crystals of the E. coli enzyme ended in failure. The discovery that the enzyme from Salmonella readily crystallized provided the breakthrough resulting in the determination of the first two tryptophan synthase structures [34] and most of the subsequent structures that have been published. Thus, in 1988, the first two structures of tryptophan synthase, E(Ain), and the complex of E(Ain) with the IGP analogue, indole 3-propanol phosphate (IPP), bound to the α-sites (Figure 2), were published by David Davies’ group at NIH in collaboration with Edith Miles’ group [21]. This achievement is recognized as a landmark for enzymology. Spectroscopic and kinetic tests showed that the E. coli and S. typhimurium enzymes exhibited very similar kinetic properties, consistent with the close similarity of the primary sequences [24]. Therefore, most subsequent structural and mechanistic studies have been performed with the S. typhimurium enzyme.
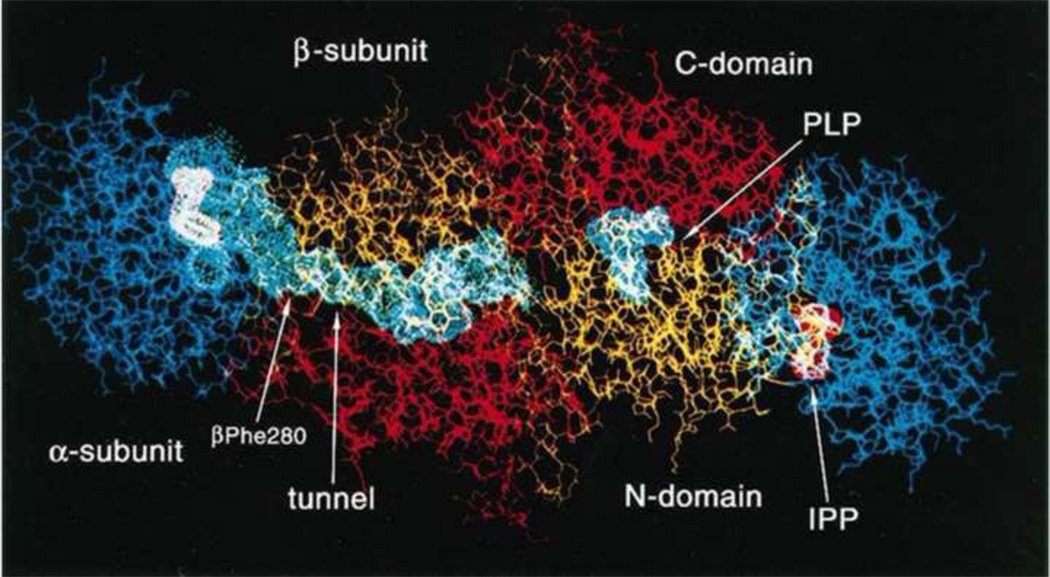
Figure 2.
Structural model for the tryptophan synthase α2β2 bienzyme complex showing important features of the subunits, sites, coenzyme PLP and the interconnecting tunnel. The IGP analogue, indole 3-propanol phosphate (IPP) is bound to the active sites of the α-subunits. Figure modified from Hyde et al. [21].
The Hyde et al. paper [21] presented the first structure of an enzyme complex involving substrate channeling where the channel was identified. This structure generated great interest in the nature of the interconnecting tunnel and the components responsible for the allosteric linkages between the α- and β-sites. The first two structures of the S. typhimurium enzyme [21] showed the presence of (i) the interconnecting channel; (ii) open conformations for both the α- and the β-subunits; (iii) details of the catalytic sites; (iv) the folding motifs of the subunits; and (v) the linear αββα arrangement of the subunits, which had been predicted by Wilhelm et al. [35] via small angle X-ray scattering in solution.
The α-Reaction Mechanism
In the ’80s and ’90s Edith Miles and her colleagues at NIH took the lead in applying site-directed mutagenesis to the determination of which amino acid residues play catalytic roles and which play allosteric roles in the catalysis and regulation of the α- and β-reactions [24, 25, 36, 37, 38, 39]. This effort was guided by the results of the x-ray crystal structure. For example, the site-directed mutagenesis work showed that αGlu49 and αAsp60 are essential for α-catalysis [25, 38], while βGlu109 [39] and βLys87 [40] are essential for the β-site. In collaboration with the Miles laboratory, we were able to demonstrate that the α-subunit switches between open and closed conformations, and that this switching was critically important both for catalysis at the β-site and for the allosteric regulation of channeling [41, 42]. This switching between open and closed conformations was inferred from rapid kinetic studies which showed that the reactions of indole and indole analogues with E(A–A) at the β-site are strongly inhibited by the binding of the α-site substrate analogue, D,L- α-glycerol 3-phosphate (GP) [41, 42].
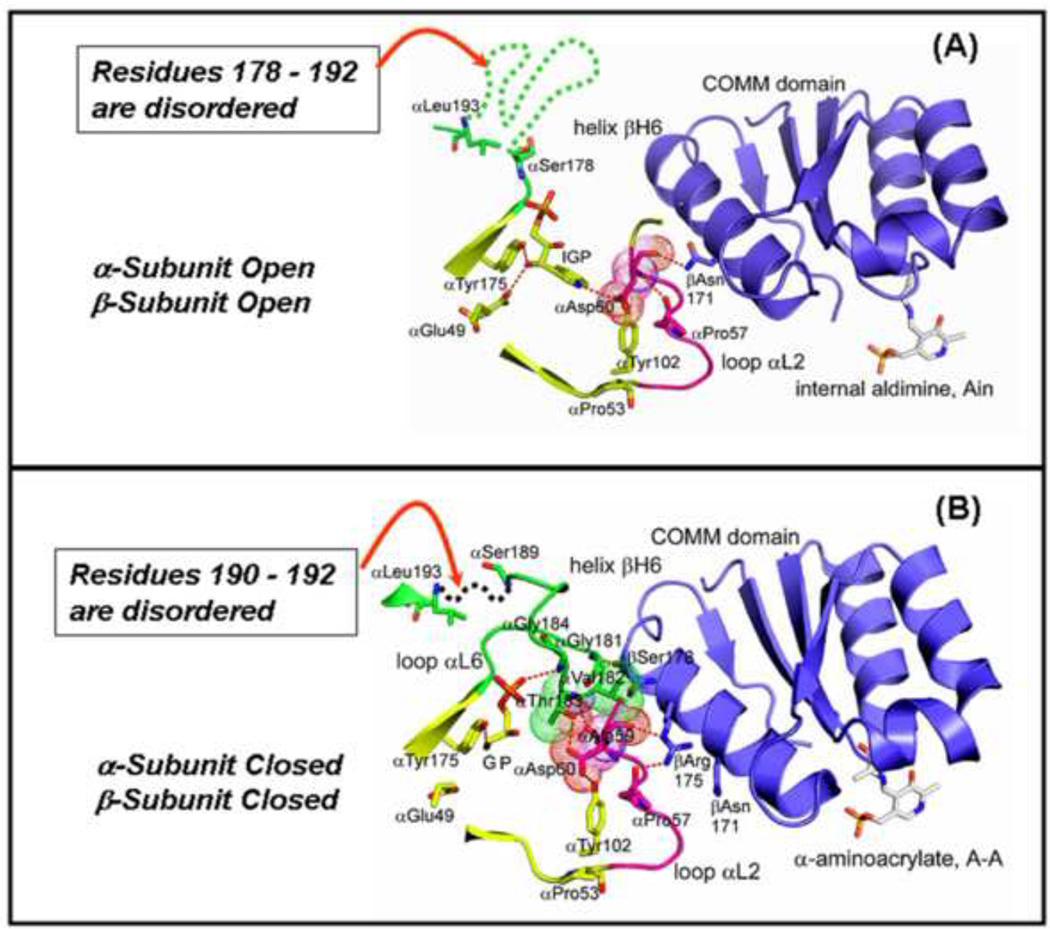
From x-ray crystallographic studies and experiments with α-site mutants, we now know that the α-subunit can switch from an open to a closed conformation upon binding substrate IGP or analogues of IGP [26, 43–49]. The open conformation is characterized by extensive disorder in 11 residues of Loop αL6, residues α179-α193 (Figure 3A) (here depicted by the dashed green line in the IGP complex). Upon formation of the closed conformation of the αβ-dimeric unit, all but two or three of these residues become well ordered [46, 47, 49]. This closed state of the α-subunit depends on the formation of a closed β-subunit conformation, here shown in the α-aminoacrylate structure with GP bound to the α-site (Figures 3B,C) [46].
Figure 3.
Structural models depicting components of the conformational transition from the open to the closed αβ-dimeric unit of tryptophan synthase. (A) The αβ-dimeric units of the E(Ain) complex (PDB ID: 1KFK) have the open conformation, whereas, the αβ-dimeric units of the E(A–A) complex have the closed conformation (PDB ID: 2J9X). In (A) and (B), loop αL6 (residues α-176 to α-193) is shown in green. Loop αL2 is shown in magenta. The α-site ligands and residues αE49, αP53, αY102, and αY175 are shown in yellow. van der Waals dot structures are drawn around αA59 and αD60 in (A) and around αA59, αD60, αV182 and αT 183 in (B). Comparison of the structural models in (A) and (B) show that residues α178–192 of loop αL2 are disordered (dashed green line) in the E(Ain) complex (A). In the E(A–A) complex (B), only residues α190–192 of αL2 (dashed green line) are disordered. (C) Cartoon depicting the ligand-mediated interconversion of the open and closed conformations of the αβ-dimeric unit. The combination of IGP binding to the α-site and L-Ser reaction to give E(A–A) at the β-site stabilize the closed conformation of the αβ-dimeric unit. Redrawn from Dunn et al. [27]
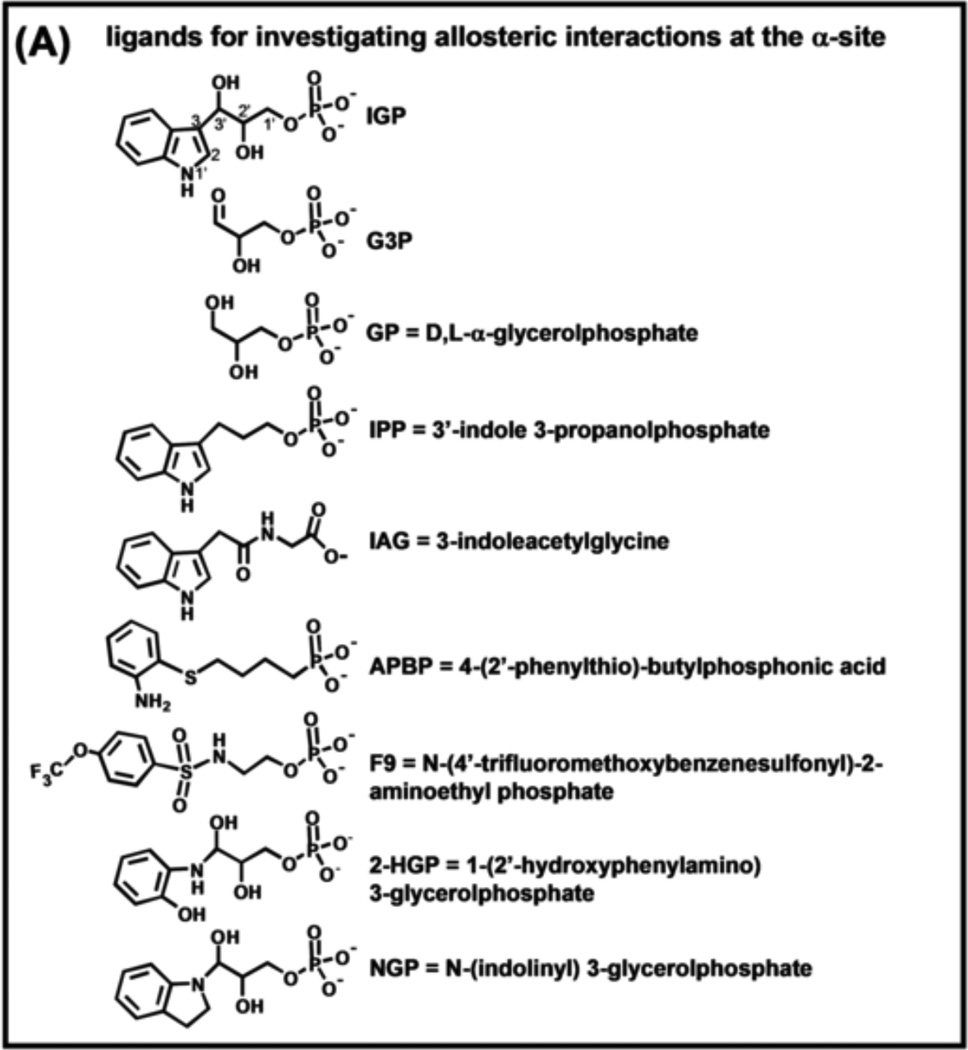
Advances in the understanding of the relationship between chemical events at the α- and β-sites and the allosteric regulation of substrate channeling have been greatly aided by the synthesis of substrate analogues and alternative substrates for tryptophan synthase and by the characterization of the interactions of these analogues with the α- and β-sites [11, 45, 47, 49–54]. The structures of some of the more well-studied analogues are shown in Figure 4A.
Figure 4.
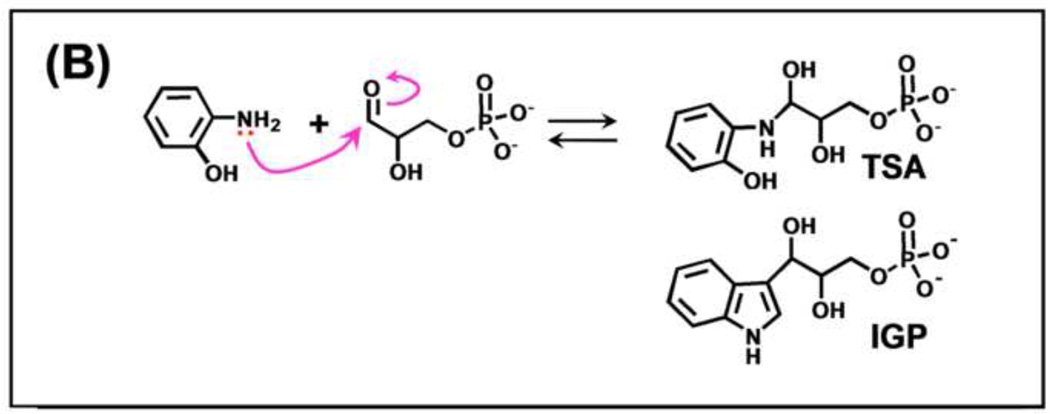
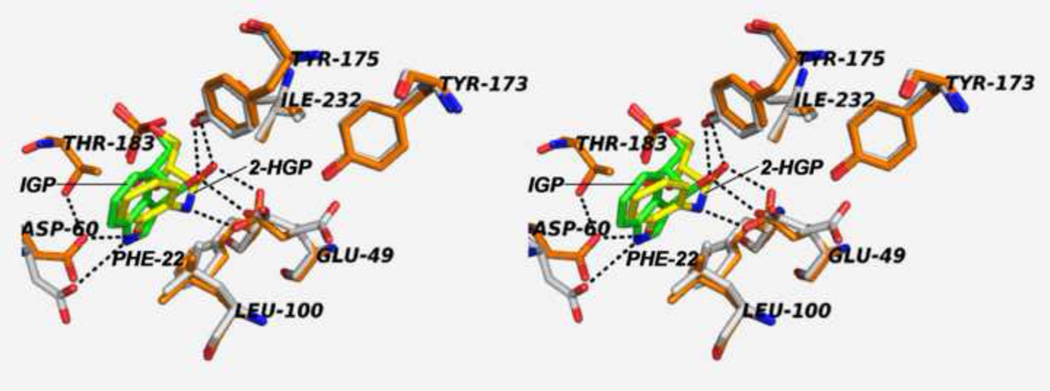
(A) Organic structures and names of some useful α-site ligands. APBP [52], 2-HGP [55] and NGP [27, 47] have been proposed to be transition state analogues for the α-reaction. (B) Reaction of 2-aminophenol with G3P to give the proposed transition state analogue, 2-HGP at the α-site [54]. (C) Stereo structural diagram comparing important features of the α-sites of the complex of 2-HGP (PDB ID: 1TJP) with the complex of IGP (PDB ID: 1QOQ). Color scheme: protein residues of the 2-HGP complex are shown with CPK coloring; 2-HGP, C yellow, N, O, and P in CPK colors. The protein residues of the IGP complex are shown with C orange, N, and O in CPK colors; IGP, C green, N, O, and P in CPK colors. Proposed hydrogen-bonding interactions are show as black dashed lines. Notice that the αD49 carboxylate takes up two orientations in the IGP complex, one with a hydrogen bond between the αD49 carboxylate and the 3-hydroxyl of IGP, the other with the carboxylate rotated by approximately 100° away from IGP.
Schlichting’s group was able to make two putative transition state analogues for the α-site, 2-HGP and NGP. The x-ray crystal structures of these analogues (Figures 4A–C [47, 54]), establish that these analogues are formed by the reaction of bound G3P at the α-site with either 2-hydroxyaniline (giving bound 2-HGP; see PDB ID 1TJP), or indoline (giving bound NGP, see PDB ID 3CEP). However, these adducts are unstable in aqueous solution and spontaneously revert to G3P and 2-aminophenol or indoline, respectively. Consequently, the affinities of these adducts for the α-site have not been determined. The x-ray structures of these complexes provide useful insights into how catalysis at the α-site takes place (Figure 4C, Figure 5) [27, 28, 47, 49].
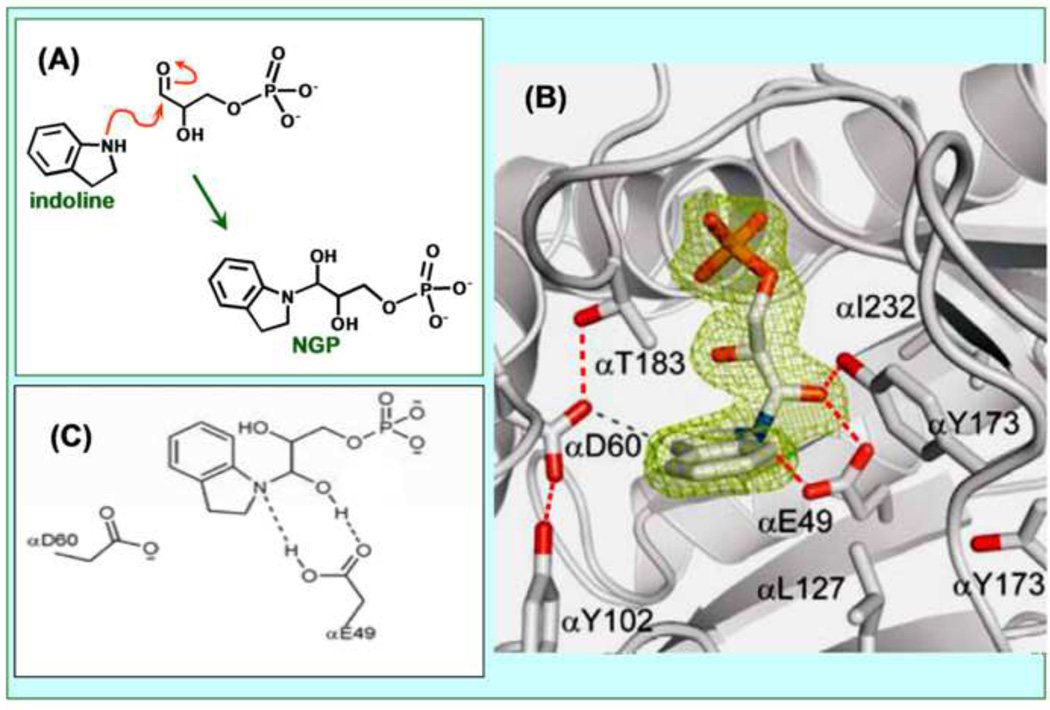
Figure 5.
(A) Reaction of indoline with G3P at the α-site to form the proposed transition state analogue NGP. (B) Structural model showing NGP fit to the electron density at the α-site (PDB ID: 3CEP). Hydrogen-bonding interactions between active site residues αE49, αD60, αY102 and αT183 and to the amino group of NGP are shown as dashed lines. The α-subunit is shown as a gray cartoon ribbon, active site resides and NGP are shown as sticks with CPK colors. (C) The organic structure of NGP and the hydrogen-bonding interactions to αE49. Redrawn from Dunn et al. [27]. (D) Stereo model depicting the microenvironment of the αE49 carboxyl group in the complex with NGP (PDB ID: 3CEP). Notice that a water molecule (wat291) forms a hydrogen-bonded bridge between the phenolic hydroxyl of αY173 and one oxygen of αE49.
The most interesting of these structures (Figures 5A,B) [47] involves the reaction of indoline at the α-site with bound G3P to give the species shown in Figure 5C as NGP (PDB ID: 3CEP). The X-ray structure (Figure 5B) shows that indoline forms a covalent adduct through the nitrogen of indoline and the carbonyl carbon of G3P. The active site acid-base catalyst, αGlu49, forms two hydrogen-bonds with this adduct, one to the C-3 hydroxyl, the other to the indoline ring nitrogen, as depicted in Figures 5B and 5C.
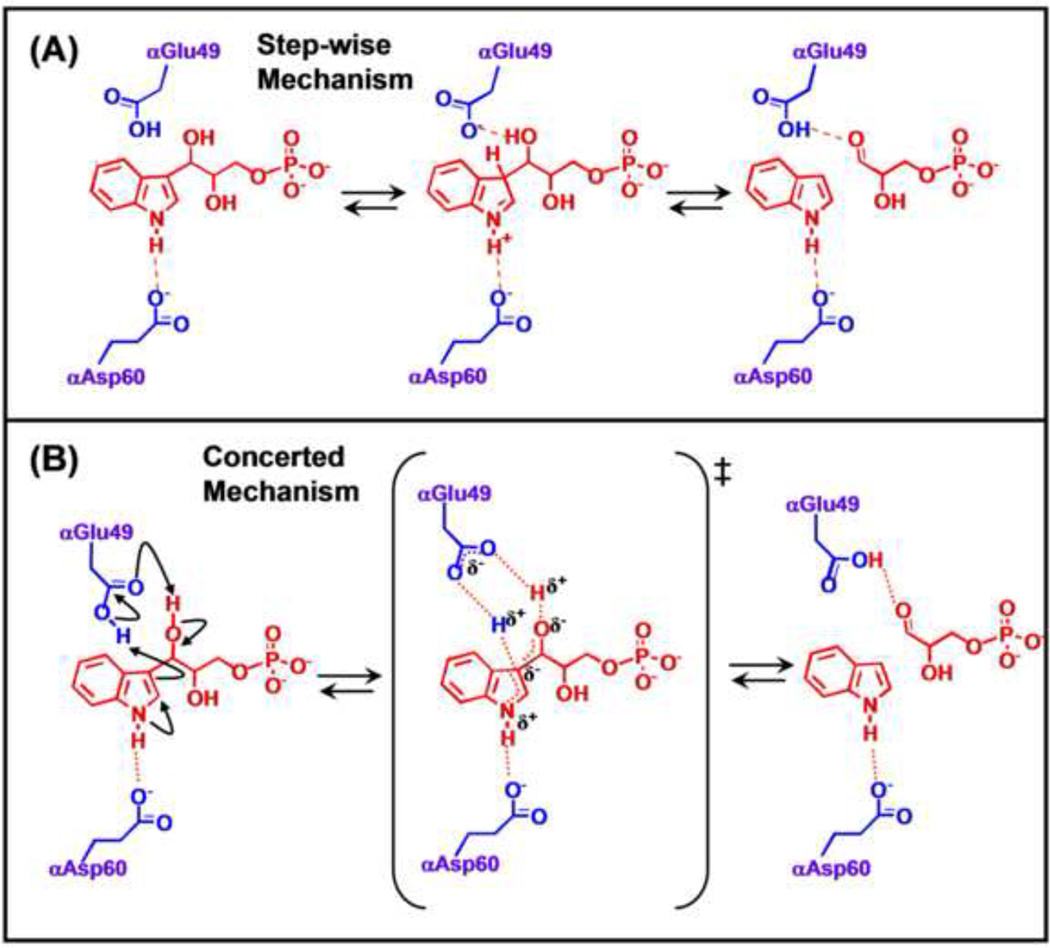
There are two mechanisms of interest for the aldolytic cleavage of IGP. One involves a step-wise series of proton transfers with discrete, charged intermediates (Figure 6A) wherein the first step involves protonation at C3 of the indole ring by αGlu49 as the proton donor. Following this protonation, the αGlu49 carboxylate deprotonates the glycerolyl hydroxyl of IGP, triggering C-C bond cleavage to give indole and G3P.
Figure 6.
The step-wise and concerted mechanisms proposed for the α-reaction are presented in (A) and (B) respectively. In both mechanisms, αE49 plays the role of acid-base catalyst for the proton transfers required for scission of the IGP C-C bond, while αD60 plays a charge-charge role in stabilizing charge development on the indole ring N.
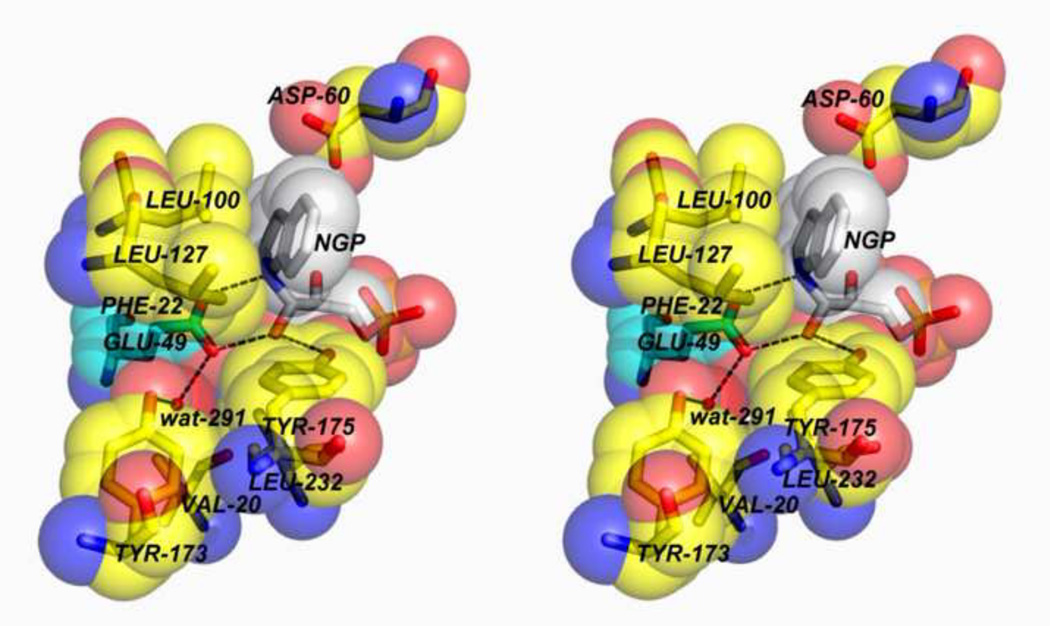
The alternative mechanism involves a concerted acid-base process (Figure 6B). Because the microenvironment surrounding the carboxylic acid group of αGlu49 is hydrophobic, a concerted acid-base mechanism has been proposed that avoids the buildup of charged species [27]. The αGlu49 carboxylic acid group is situated in a pocket defined by the side chains of αPhe22, αLeu100, αLeu1 27, αIle232, αTyr175 and a water molecule (Figure 5D). Interatomic distances indicate the water molecule is hydrogen-bonded to the phenolic hydroxyl of αTyr173 and one oxygen of αGlu49. At issue here is whether or not the microenvironment at the site is sufficiently polar to accommodate charged intermediates. The essentially hydrophobic microenvironment surrounding the cleavage site does not appear conducive to the formation of charged intermediates or to the ionization of the αGlu49 side chain to the carboxylate, therefore the concerted mechanism may be more likely [27].
The carboxylate of αAsp60 forms a hydrogen-bond to the indole ring N-H and therefore is positioned to provide a Coulombic charge-charge interaction that stabilizes the development of positive charge density on the indole ring during the cleavage of IGP, as required by either mechanism shown in Figure 6 [21, 27, 40, 55]. The β-hydroxyl of αThr183, a component of loop αL6, also forms a hydrogen bond to one carboxylate oxygen of αAsp60. Through site directed mutagenesis, x-ray structure determination and rapid kinetic measurements, Kulik et al. [55] were able to show that this hydrogen bonding interaction plays an important role in stabilizing the position of the αAsp60 carboxylate (Figure 4C). The αT183V variant exhibits a greatly impaired α-site activity, strongly impaired allosteric communication between the β- and α-sites, and the inability of loop αL6 to form a closed α-site structure [54, 56].
The β-Reaction Mechanism
The individual steps in the proposed pathway for the β-reaction are shown in Figure 1B. Notice there are 9 chemical steps with 9 chemical states and 9 transition-states in this cycle. The arrows reflect the a priori mechanistic chemist’s imagination for how these transformations occur. According to this scheme, βLys87 is involved in Schiff base formation, and in several proton transfers – clearly βLys87 is a key catalytic residue [40].
From 1988 to 2007, efforts to determine a detailed chemical mechanism for the β-site were frustrated because all the available x-ray structures published for the wild-type enzyme had the open/partially open conformation, i. e., the inactive (open) conformation, of the β-subunit. Consequently, these structures were of limited use in determining the roles of site residues.
In 2007, Ilme Schlichting’s group at the Max Planck Institute, in collaboration with my laboratory, published the first structure of the wild-type enzyme with the closed conformation of the β-subunit. This was the structure of the E(A–A) intermediate (PDB ID: 2J9X) [46]. Two closed structures for the quinonoid intermediate formed by reaction of the indole analogue, indoline, with E(A–A) soon followed (PDB IDs: 3CEP and 3PR2) [47, 49]. Both the E(A–A) structure and the E(Q)indoline structure provide critically important advances for the understanding of the β-subunit catalytic mechanism. Inspection of the closed conformation of the β-subunit made it obvious that βAsp305 can not be a catalytic group; rather βAsp305 plays an important structural role with βArg141 in forming the closed conformation. The conformational transition to the closed state positions the side chains of βGlu109 and βLys87 where proton transfers are needed for the chemical transformations and where charge-charge interactions are needed for catalysis. The key elements of the open and closed β-subunits are shown in Figures 7A–7D. The formation of the closed conformation of the β-subunit (Figures 7A–7C) is accompanied by the movement of the communication domain (COMM domain), and small changes in the αβ-subunit interface (not shown). The COMM domain moves relative to the α-subunit and rotates. This motion closes the opening from solution into the β-site and facilitates formation of the closed conformation of the α-subunit.
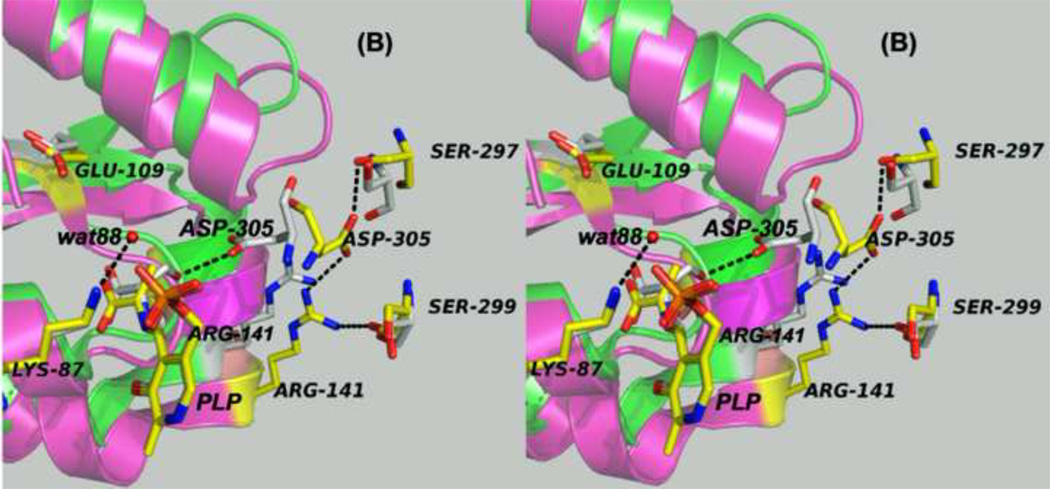
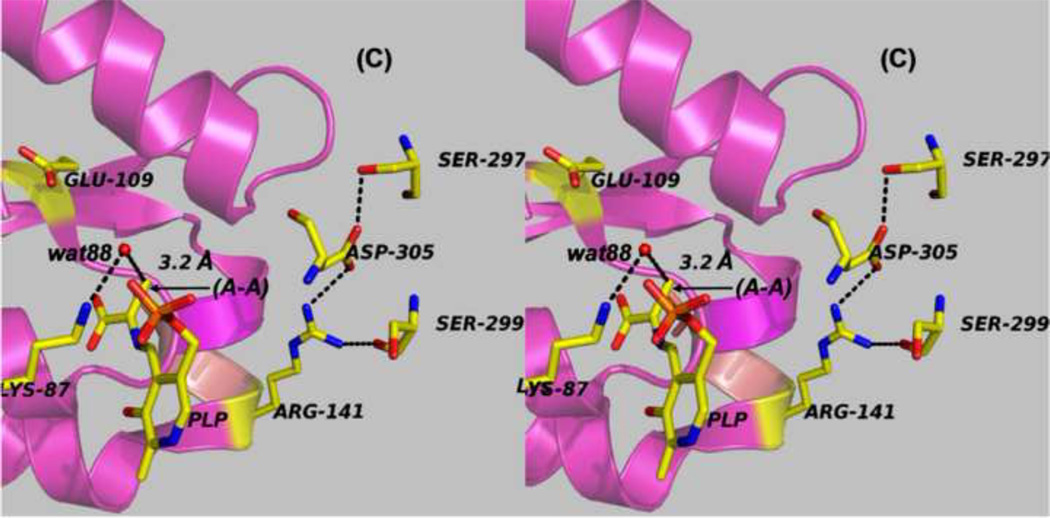
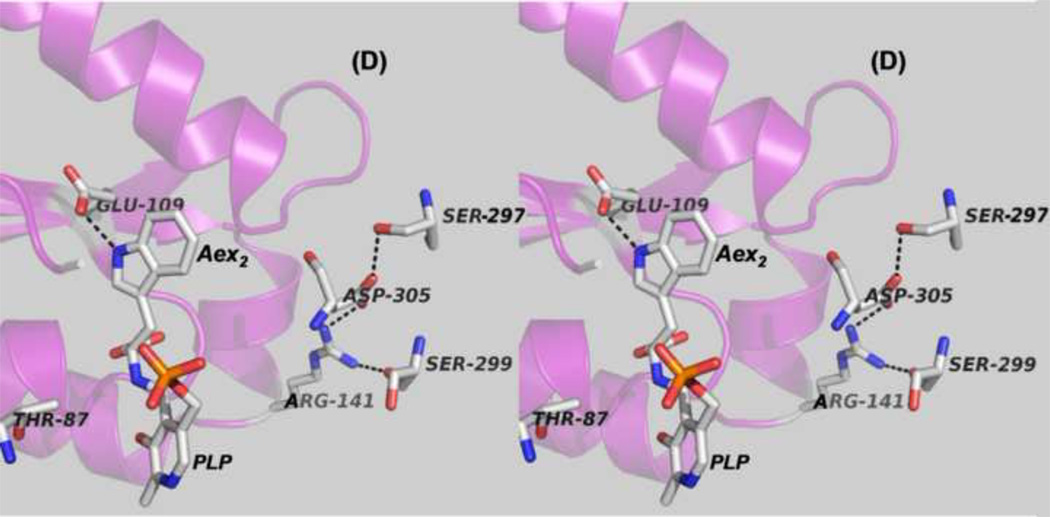
Figure 7.
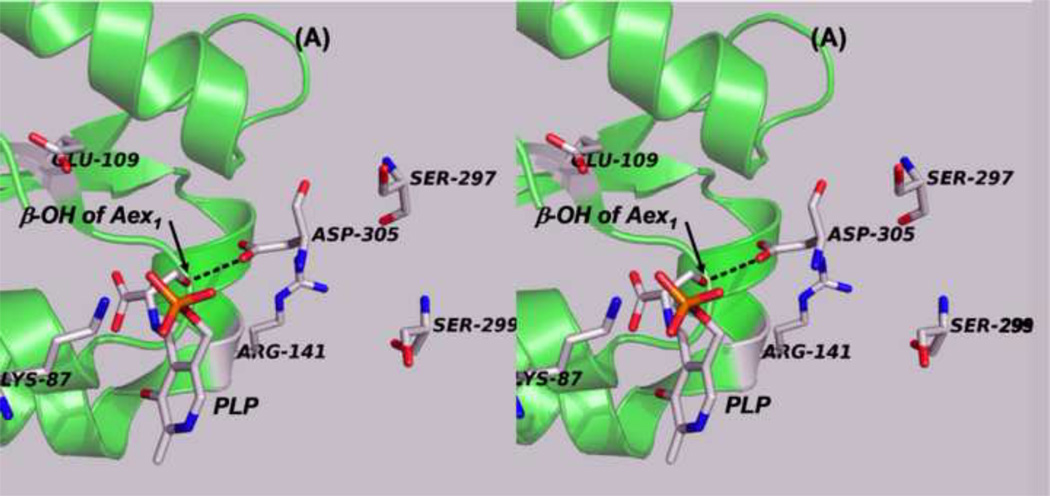
Panels (A) – (D) present stereo diagrams comparing structural models for the β-active site and the COMM domain (residues β102-β189) of the open and closed β- subunit conformations. (A) Open conformation of the E(Aex1) complex (PDB ID: 2CLL). The β-subunit COMM domain of E(Aex1) is shown as a cartoon ribbon (green), active site residues and the L-Ser moiety (Aex1) bound to PLP are shown as sticks with CPK coloring. In this structure [45] the β-hydroxyl of the Aex1 moiety (black arrow) is hydrogen-bonded (black dashed line) to the carboxylate of βD305. This interaction stabilizes the inactive (open) conformation of E(Aex1) by making removal of the proton chemically more difficult and by preventing bonding interaction(s) with the acid-base catalytic groups at the site, βK87 and βE109. The models in (B) compare the open structure of E(Aex1) from (A) with the closed structure of the α-aminoacrylate intermediate, E(A–A) (PDB ID: 2J9X). The closed structure of the E(A–A) complex is superimposed on the E(Aex1) structure. The COMM domain of E(A–A) is shown as a magenta cartoon ribbon with the active site residues and PLP-bound α-aminoacrylate depicted as stick structures with carbons yellow, and with O, N, and P in CPK colors. Water molecule, wat88, is shown as a red ball, located near the β-carbon of the α-aminoacrylate moiety. Dashed black lines indicate hydrogen bonds. For clarity, the structure of the closed conformation of the E(A–A) complex from (B) is shown in (C). The motion of the COMM domain in the transition from the open to the closed conformation brings βR141 and βD305 together to form a salt bridge along with new hydrogen bonding interactions to βS297 and βS299 [46]. Hydrogen bonds and the distance between the water molecule and the β-carbon of the α-aminoacrylate moiety, (A–A), are indicated by black dashed lines. The βR141-βD305 salt bridge provides a physical barrier which prevents the transfer of ligands between solution and the β- site. This closed conformation also positions βE109 and βK87 for the proton transfers required in the catalytic steps which take place in Stage II of the β- reaction [27, 46, 47, 49].
(D) Stereo structural model showing the closed β-site of the βK87T variant with the L-Trp external aldimine, E(Aex2), bound to the β-site. Notice that the carboxylate of βE109 is hydrogen-bonded to the indole ring N of Aex2 (black dashed line) (PDB ID: 2TYS).
Figure 7B compares the open structure of E(Aex1) with the closed structure of E(A–A). In the open E(Aex1) structure, the βAsp305 carboxylate hydrogen-bonds to the hydroxyl of the Ser moiety. This hydrogen-bond provides an electrostatic interaction that chemically stabilizes E(Aex1) and holds the hydroxyl away from the acid-base catalytic groups, βGlu109 and βLys87, giving an inactive complex. When the subunit switches to the closed conformation, the motion of the COMM domain causes βArg141 to move ~4 Å toward (βAsp305. The hydrogen-bond between the side chain of βAsp305 and the β-hydroxyl of the L-Ser moiety of E(Aex1) is broken, and the side chain of βAsp305 rotates by about 100°. This translation of βArg141 and rotation of the βAsp305 carboxylate allows formation of the extensively hydrogen-bonded network between βArg141 and βSer299 and between βAsp305 and βSer297 and formation of the salt bridge between βArg141 and βAsp305. Scission of the hydrogen-bond between the β-hydroxyl of Aex1 and the carboxylate of βAsp305 would allow the hydroxyl group to rotate around the Cα-Cβ single bond to a new orientation placing it in the proximity of the βLys87 Nε. The βArg141-βAsp305 salt bridge blocks the opening to the β-site and prevents the escape of indole and indole analogues from the β-site into solution. Via site-directed mutation and kinetic studies Ferrari et al. [57, 58] confirmed that βArg141 and βAsp305 play important structural roles, and that mutation of either of these residues to Ala alters the reaction specificity of the β-site.
The comparison of open and closed structures in Figure 7 shows that the Nε of βLys87 is located such that it could function as the base catalyst for conversion of the E(Aex1) intermediated to E(Q1) and then as the acid catalyst for conversion of E(Q1) to E(A–A). In the closed structures (viz., Figures 7C and 7D), the carboxyl group of βGlu109 is too far away to play either of these roles.
The x-ray structure of E(A–A) [46] (Figures 7B and 7C) appears to provide a snapshot of the product complex formed by elimination of the hydroxyl from the L-Ser quinonoid intermediate (which then appears in the structure as a water molecule, wat88). Notice that βLys87 is positioned to facilitate the nucleophilic attack of wat88 through base catalysis for conversion back to the L-Ser quinonoid intermediate (e.g., reaction in the reverse direction).
The x-ray structures of E(Q)indoline [47, 49] and the structure of the L-Trp external aldimine bound to the βK87T mutant [40] (Figure 7D) establish that the β-site has a well-defined sub-site for the indole moieties of the intermediates involved in Stage II of the β-reaction. This sub-site is defined by the side chains of several residues, including βPhe306, βHis115, βGly189, βThr190, and βGlu109. We postulated that when indole is transferred from the α-site to the β-site, the binding of indole to this sub-site displaces wat88 and positions the indole C-3 adjacent to the E(A–A) C-β so that nucleophilic attack to form the C-C bond can occur [46, 47, 49]. The closed β-subunit conformation was first seen in the x-ray structures of the βLys87Thr variant [40]. However, because this mutant is catalytically inactive, the mechanistic implications of these closed structures were not obvious. In retrospect, it is now clear that the structures of the complexes of the βK87T variant provide useful insights into the roles played by βGlu109, βArg141 and βAsp305. For example, all of the closed structures exhibit the salt bridge between βArg141 and βAsp305, the signature of the closed β-subunit conformation. The E(Aex2) complex shows the carboxylate of βGlu109 hydrogen-bonded to the indole ring NH (Figure 7D), the interaction expected if βGlu109 plays the charge-charge stabilization role in the attack of indole on the α-aminoacrylate [27, 39, 40, 47].
As anticipated from the detailed investigations of other PLP-requiring enzymes, all of the kinetic, structural, and mutagenic studies of the β-reaction implicate the Nε of βLys87 as the acid-base catalyst responsible for proton transfers at the C-α of the reacting amino acid (L-Ser in Stage I and L-Trp in Stage II of the β-reaction) as indicated in Figure 1B. The x-ray structures of the closed conformations of E(A–A) and E(Q)indoline indicate that the Nε of βLys87 also is the acid-base group involved in the proton transfers needed to catalyze elimination of the β-hydroxyl from E(Q1) and the proton transfer involved in the conversion of E(Q2) to E(Q3) (viz., Figure 1B).
Origins of Allosteric Interactions in Tryptophan Synthase α-β-Site-Site Interactions
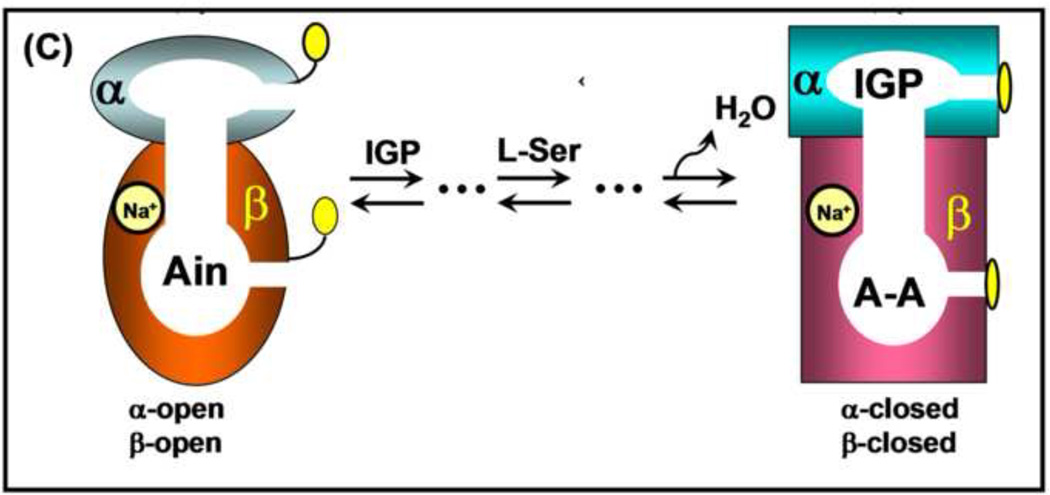
Early work from Kirschner’s laboratory [11–13] established that binding of ligands to the α-site altered the spectra of PLP intermediates bound to the β-site, suggesting site-site interactions. The x-ray structure results [21] showing the α- and β-sites are separated by ~ 25 Å (Figure 2) brought about renewed interest in the relationship of the allosteric properties of tryptophan synthase to the mechanism of channeling. We initiated a series of experiments to investigate the allosteric linkages between catalysis at the two active sites and the conformational switching between open and closed states [41, 42, 59, 60–64]. One effort involved a collaboration with Edith Miles to identify the protein scaffolding involved in the transmission of the allosteric interactions using site-directed mutagenesis and rapid kinetic measurements [41, 42]. These studies established that modification of residues in either loop αL6 or αL2 impaired closure of the α-subunit, and consequently interfered with activation of the α-site mediated by L-Ser reaction to form E(A–A) at the β-site. At about the same time, the laboratories of Kasper Kirschner and Karen Anderson also undertook efforts to understand substrate channeling in tryptophan synthase [65, 66]. Kirschner et al. [65] showed that the reaction of L-Ser with the β-site activates the α-site. The experiments of Anderson et al. [66] also provided further evidence showing a functional role for the tunnel in the channeling of indole and further confirming that reaction of L-Ser activates the α-site. We established that the tunnel indeed functions to transfer Indole between the active sites [41–44, 59, 60, 62, 63, 67, 68], and that it is the conversion of the L-Ser external aldimine to the α-aminoacrylate (via the Ser quinonoid intermediate) that activates the α-subunit [61], whereas conversion of the L-Trp quinonoid to the L-Trp external aldimine switches the α-subunit off [63]. These findings led to the proposal that the switching of subunits between open and closed conformations switches the activity of the α-site on and off. This conformational switch causes synchronization of the activities of the α- and β-sites, and prevents the escape of indole by forming αβ dimeric units with closed conformations that prevent the dissociation of indole [41–44, 61].
In 2007, Ngo et al. [45, 46] showed that the binding of α-site ligands activates the conversion of E(Aex1) to E(A–A) at the β-site by ~ 10-fold. Thus, chemical transformations in the β-site catalytic cycle trigger the switching of the α-site between active and inactive states, while IGP binding to the α-site almost certainly activates the rate determining step in the formation of E(A–A), the intermediate poised to react with indole generated by IGP cleavage at the α-site (Figure 1B).
Intermediate Trapping within the αβ-Dimeric Unit
The experiments of Dunn et al. [62] gave the first indication that ligand binding to the α-site and chemical reaction at the β-site could switch the αβ-dimeric unit to a closed structure that prevents exchange of indole or indole analogues between solution and the interior cavity defined by the α-site, the interconnecting tunnel and the β-site. The transfer of indole analogues to and from this interior space was further explored by using the intense absorption band of the indoline quinonoid intermediate (ε476 > 40000 M−1 cm−1) to examine the effects of α-site ligands on the escape of indoline from the E(Q)indoline complex [43, 44]. These experiments established that the escape of indoline, as measured by the disappearance of the E(Q)indoline absorption band, is strongly retarded by the binding of ligands to the α-site, and that the rate of escape was inversely related to the affinity of the α-site ligand. These results are consistent with a mechanism where the escape of indoline is limited by the rate of dissociation of the α-site ligand.
Discovery of Allosteric Interactions arising from Monovalent Cation interactions at the-β-Site
In 1995, Andera Mozzarelli’s group in Parma, Italy, discovered that the bienzyme complex has an essential requirement for the binding of a monovalent cation (MVC) cofactor [69]. We independently came to the same discovery [70–73]. The Miles laboratory also made important contributions in this area [74–76]. Rhee et al. [74] showed that the MVC site is buried in the β-subunit about 8 Å from the catalytic site and that, depending on ionic radius, the coordination of monovalent cations involves up to six peptide carbonyl oxygens βVal231, βGly232, βGly268, βLeu304, βPhe306, and βSer308) in a loop structure [30]. Together, these efforts showed that the binding of an MVC to the β-subunit is essential both for β-site catalysis and for allosteric communication between the α- and β-sites. A variety of MVCs will satisfy this requirement, including Na+, K+, NH4+, Cs+, and Rb+ [48, 69, 70–73] and, surprisingly, the guanidinium cation [30, 75, 76]. The catalytic activity of the β-site is strongly impaired by removal of all MVCs from the enzyme; the α-aminoacrylate intermediate is rendered essentially unreactive toward indole and indole analogues [48]. α-Site ligands and MVCs act synergistically to modulate the conformational switching between open states of low activity and closed states of high activity [41, 42, 26, 27, 77]. Comparison of the structures of Na+ and Cs+ forms of the enzyme show that the Cs+-bound enzyme, (Cs+)E, exhibits a preformed sub-site for indole, whereas in the Na+ form of the enzyme, (Na+)E, the indole sub-site is collapsed, and too small to bind indole [48].
Consequently, the (Cs+)E favors intermediates with the closed β-subunit conformation when the indole sub-site is occupied, while the (Na+)E stabilizes the open β-subunit conformation. These effects appear to arise from the differences in coordination at the MVC site due to differences in MVC ionic radius [30]. For example, Na+ is coordinated to βGly232, βPhe306, and βSer308 and two waters in the (Na+)E complexes, while Cs+ is coordinated to all six carbonyls and no waters in the (Cs+)E complexes [74].
Equilibrium binding and kinetic analyses of the MVC cofactor effect [48, 69–74, 77], have shown that under physiological conditions the bound cation is a structural element which functions to selectively stabilize active conformations of certain enzyme intermediates. Recently, Phillips et al. [77] have extended investigation of the MVC cofactor effect by employing pressure jump perturbation of equilibrium studies to quantify the effects of both α-site ligands and MVCs on the energetics of the conformational switch.
With reference to the metal-ion-free state, cation binding drives the protein from the MVC-free inactive (or less active) conformation to a conformation with high activity, this switching almost certainly involves interconversion of the open (low activity) and closed (high activity) states [77]. It has been shown that the bound metal ion interacts more strongly with the active conformation, biasing the distribution of states in favor of the active forms [70–72, 78]. Because catalysis at the β-site involves the interconversion of at least nine covalent intermediates (viz. Figure 1B), the β-site must adapt to the structure of each activated complex along the β-reaction pathway (Figure 1B) [16, 17, 48, 70–73, 77, 78]. That is, according to transition state kinetic theory, the site must undergo structural/electronic changes so that the site stabilizes each activated complex. Therefore, we hypothesize that the monovalent cation interacts specifically to lower the activation energies of select steps by interacting more strongly (i.e., binding with higher affinity) to the conformation of the protein that is complementary to the structure of the transition state for that step [71, 72, 78]. That this selectivity occurs is born out by the observation that different distributions of enzyme-bound intermediates are observed both with different MVC forms of the enzyme and with the cation-free enzyme [70, 71–73, 77, 78].
The Function of Allosteric Interactions in Tryptophan Synthase: The Mechanism of Substrate Channeling
There is a consensus among investigators that substrate channeling in the tryptophan synthase bienzyme complex evolved to prevent the loss of indole (via hydrophobic interactions) to other components of the cell. The preservation of this channeling function in bacteria, yeasts, molds and plants underscores the importance of this phenomenon for the synthesis of L-Trp by these organisms. There are at least two functional roles played by allosteric interactions in tryptophan synthase: (a) The synchronization of the α- and β-reactions to achieve the efficient utilization of the indole moiety of IGP for the biosynthesis of L-Trp via substrate channeling, and (b) the facilitation of chemical steps along the catalytic path [78]. In vivo, an MVC cofactor (likely K+) plays an allosteric role in the lowering of the activation energies of selected steps in the catalytic cycle of the β-site, as described above, and alters the energetics of the conformational switch between open and closed states [23, 27, 30, 79].
Substrate binding to the α-site modulates reaction at the β-site, and substrate reaction at the β-site activates and deactivates reaction at the α-site. Therefore, the binding of the monovalent cation to the β-subunit also is essential both for the activity of the β-site and for the allosteric communication between the α- and β-sites. In the MVC-activated enzyme, chemical transformations specific to the β-catalytic cycle trigger the interconversion between inactive and active states of the α-site, and the switching between open and closed conformation states. Finally, the binding of IGP activates the rate determining step in stage I of the β-reaction [46].
Considerable effort has gone in to the determination of the structural scaffolding that transmits the allosteric signals [23, 26, 29, 36, 38, 41, 42, 54–58, 73]. However, it is not yet clear exactly how the allosteric signaling between sites is transmitted. What appears essential for transmitting the allosteric signal which activates the α-site is the formation of a well-ordered, closed structure of loop αL6, including especially the formation of the hydrogen-bonding interaction between the carboxylate of αAsp60 and the hydroxyl of αThr183 [23, 26, 29, 36, 38, 41, 42, 54–58, 73]. When this interaction is compromised, the activity of the α-site is strongly impaired, as is the allosteric signaling between the subunits [55].
Another aspect of the scaffolding which is not well understood is the network of salt bridges within the β-subunit and across the subunit interface. No clear pattern has emerged from the x-ray structure data base. However, site directed mutations of these residues indicates that the salt bridging structural elements are important for maintaining the balance between open and closed structures [23, 38,.41, 42, 57,58, 73, 74].
The motion of the COMM domain (Figure 7B) is an integral part of the allosteric transition. The translation of the COMM domain relative to other portions of the structure (the α-subunit and other structural components of the β-subunit) severs the hydrogen bond between the β-hydroxyl of Aex1 and the carboxylate of βAsp305, brings βArg141 and βAsp305 together in a salt bridge, and closes the portal for entry into the β-site from solution. During the β-site catalytic cycle, this conformational change traps indole inside, preventing its escape. Alteration of the α-β-subunit interface by the motion of the COMM domain stabilizes the closure of loop αL6.
Recent work shows that molecular dynamics simulations of the conformational transitions can provide critically important information regarding the linkages between conformational transitions in the protein and the allosteric regulation of function for tryptophan synthase. Fatmi et al. [79] have investigated the synergistic regulation of the ligand-mediated conformational transitions in tryptophan synthase via detailed dynamics calculations using molecular dynamics simulations. These simulations indicate that loop αL6 samples both open and closed conformations in the ligand-free state but shifts to the fully closed state when substrates are bound. The motion of the COMM domain is coupled to the motion of the α-subunit, and fluctuations in loop αL6 have a preferential directionality which likely facilitates formation of the fully closed α-β dimeric unit.
Conclusions: Allosteric regulation of channeling in tryptophan synthase
The preceding sections of this review reveal that the allosteric regulation of substrate channeling in tryptophan synthase is a sophisticated, highly nuanced process. The salient features of this process include: ligand binding at the α-site, the action of a monovalent cation cofactor in the β-subunit, and covalently triggered allosteric interactions acting over a distance of 20 to 25 Å. These interactions choreograph the switching of the α- and β-subunits between open conformations of low activity and closed conformations of high activity that also regulate access of ligands and intermediates to the surrounding milieu. The allosteric effects of these interactions are part of a delicate balancing of the thermodynamics of the interconversion between open and closed allosteric states. The interactions arising from MVC binding to the MVC site play roles both in mediating the transmission of allosteric signals between the sites and the modulation of activation energies for select steps along the β-site catalytic path. The accumulated information on the relationship between the physical and chemical properties and the three dimensional structure of tryptophan synthase are combined into a structure-function hypothesis for the regulation of substrate channeling that is discussed in the following paragraphs.
For efficient channeling in a bienzyme complex, the following constraints must be met [23]:
“The architecture of the multienzyme complex must provide a physical structure with dynamic properties that constrain the degrees of freedom of the common metabolite [i.e., indole] so that transfer from one site to next is assured.
Catalysis at the two active sites must be coupled such that turnover at each site occurs in phase.”
The switching to a closed αβ-dimeric unit during the catalytic cycle and the presence of an interconnecting tunnel fulfills the first constraint (Figures 2 and 8A). The activation and deactivation of the α-site by chemical events at the β-site allows synchronization of the α- and β-catalytic cycles and therefore satisfies the second constraint.
Figure 8.
(A) Cartoon for the αβ-dimeric unit of tryptophan synthase depicting the binding and dissociation of ligands to the α- and β-sites and the transfer of intermediate indole from the α-site to the β-site (redrawn from [62]).
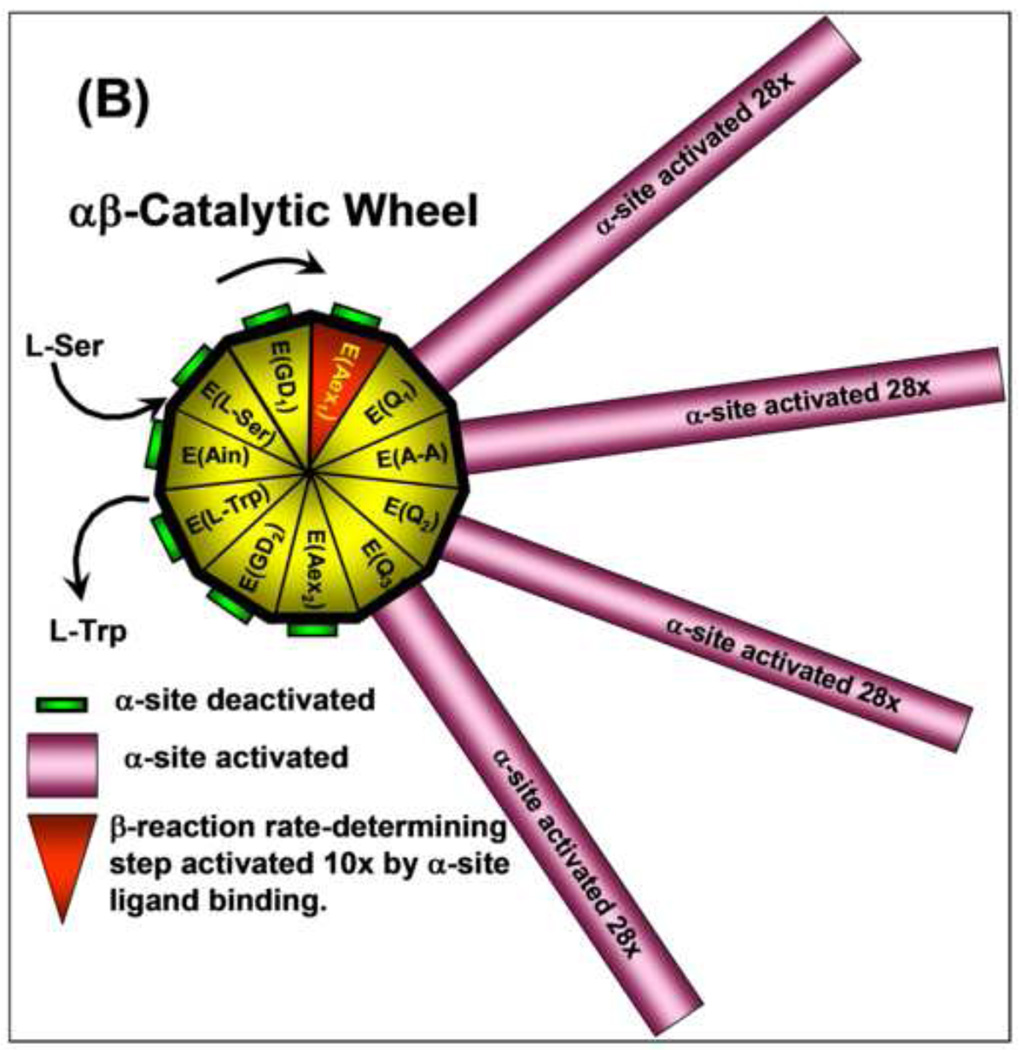
(B) Catalytic wheel summarizing the interplay of ligand binding and reaction with the allosteric signaling between the α- and β-sites during the catalytic cycles of the α- and β-reactions. The gold and red triangular segments of the wheel hub represent the different known chemical states of the β-catalytic cycle (see Figure 1). The green or magenta bars extending out from each triangular segment have lengths proportional to the relative activity of the α-site for each chemical state of the β-site. As the E(Ain) species is converted by reaction with L-Ser to E(GD1) and E(Aex1) the α-site remains in a catalytically inactive state. When E(Aex1) is converted to E(A–A), via E(Q1), the α-site is activated at least 28-fold [61], and remains activated until E(Q3) is converted to E(Aex2) when the α-site is switched off again [63]. As the β-reaction continues from E(Aex2) to the complex of E(Ain) with L-Trp, the α-site remains deactivated. The triangular segment designated as E(Aex1) is colored red to signify activation of the conversion of this intermediate to E(A–A) by IGP binding to the α-site [27]. Modulation of the switching of the α- and β-sites between open states of low activity and closed states of high activity by ligand binding and reaction provides a mechanism for both synchronizing the α- and β-reactions and preventing the escape of indole from the confines of the α- and β-sites and the interconnecting tunnel. This conformational switching makes possible the efficient channeling of indole during the αβ-catalytic cycle of tryptophan synthase. Figure redrawn from [27].
The catalytic wheel cartoon shown in Figure 8B combines into a single diagram the essential components of the channeling mechanism thus far revealed by the experimental observations discussed above in this review.
This catalytic wheel shows how the activities of the β- and α-sites are synchronized by the switching of the subunits between open conformations of low activity and closed conformations of high activity. The inner wheel depicts the chemical states of the β-site. The lengths of the spokes show how the activity of the α-site responds to each chemical state of the β-site. The catalytic cycle begins with the binding and reaction of L-Ser with E(Ain) at the β-site. The α-site of E(Ain) is inactivated and remains in this inactive state until E(Aex1) is converted to E(A–A) at the β-site. This transformation activates the α-site at least 28-fold by switching the αβ-dimeric unit to the closed conformation [41, 42, 46, 47, 49, 61]. Leja et al. [19] showed that the α-site is turned off when the E(Q3) species is converted to the E(Aex2) species. Pan and Dunn [64] showed that E(Aex2) is switched to the open conformation when the α-site is unoccupied. The α-site then remains deactivated as the β-reaction chemical cycle is completed. Ngo et al. [46] established that the binding of IGP activates the conversion of E(Aex1) to E(A–A) by about 10-fold; this is the slow step in Stage I of the β-catalytic cycle (Figure 1B). Thus the activation of the sites is a reciprocal process. This allosteric control acts to prevent the escape of indole from the bienzyme complex, ensures synchronization of the activities of the two active sites so that catalysis occurs in phase, and modulates activation energies for select steps in the β-site catalytic cycle. The interplay between ligand binding and covalent reaction in tryptophan synthase results in an ensemble of allosteric effects that make this system an efficient nanomachine for the synthesis of L-Trp.
Highlights.
Allosteric interactions control substrate channeling in tryptophan synthase.
Allosteric interactions are mediated by substrate binding and covalent reaction.
Binding of a monovalent cation cofactor also exerts allosteric interactions.
The allosteric transition switches the enzyme between open and closed states.
Efficiency is achieved by the synchronization of the α- and β-catalytic cycles.
Acknowledgments
Support of this work by NIH grant R01GM097569 is gratefully acknowledged. I thank the many undergraduate, graduate and postdoctoral scholars that contributed to the tryptophan synthase story in my laboratory over the past 31 years. It has been a delight to work with these individuals during a major portion of my career. I thank my many collaborators, especially Ilme Schlichting, Edith Miles, Gian Luigi Rossi, Andrea Mozzarelli, and Len Mueller for their many experimental and intellectual contributions. I also thank my scientific mentors, especially James Cox, Jr., Sidney A. Bernhard, and Poul Schack for their encouragement, patience and contributions as role models. Finally, I thank Charles Yanofsky for his early and continuing encouragement of our work on tryptophan synthase.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bertrand K, Korn L, Lee F, Platt T, Squires CL, Squires C, Yanofsky C. New features of the regulation of the tryptophan operon. Science. 1975;189:22–26. doi: 10.1126/science.1094538. [DOI] [PubMed] [Google Scholar]
- 2.Yanofsky C, Crawford IP. In: Tryptophan synthase. 3rd ed. Boyer PD, editor. New York: The Enzymes, Academic Press; 1972. pp. 1–31. [Google Scholar]
- 3.Miles EW. Tryptophan synthase: structure, function, and subunit interaction. Adv. Enzymol. Relat Areas Mol. Biol. 1979;49:127–186. doi: 10.1002/9780470122945.ch4. [DOI] [PubMed] [Google Scholar]
- 4.Umbreit W, Wood W, Gunsalus I. The activity of pyridoxal phosphate in tryptophan formation by cell-free enzyme preparations. J. Biol. Chem. 1946;165:731–732. [PubMed] [Google Scholar]
- 5.Yanofsky C. Tryptophan synthetase from Neutrospora. In: Colowick SP, Kaplan NO, editors. Methods of Enzymol. vol. II. N. Y.: Academic Press, Inc.; 1995. pp. 233–238. [Google Scholar]
- 6.Crawford I, Yanofsky C. On the separation of the tryptophan synthetase of E. coli into two protein components. Proc. Natl. Acad. Sci. U. S. 1958;44:1161–1170. doi: 10.1073/pnas.44.12.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanofsky C, Rachmeler AM. The exclusion of free indole as an intermediate in the biosynthesis of tryptophan in .Neorospora crassa. Biochem. et Biophys. Acta. 1958;28:640–641. doi: 10.1016/0006-3002(58)90533-x. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg ME, York S, Stryer L. Fluorescence studies of substrate and subunit interactions of the β2 protein of Escherichia coli tryptophan synthase. Biochemistry. 1968;7:3662–3667. doi: 10.1021/bi00850a045. [DOI] [PubMed] [Google Scholar]
- 9.Faeder EJ, Hammes GG. Kinetic studies of tryptophan synthase. Interaction of L-serine, indole, and tryptophan with the native enzyme. Biochemistry. 1971;10:1041–1045. doi: 10.1021/bi00782a016. [DOI] [PubMed] [Google Scholar]
- 10.York S. Kinetic spectroscopic studies of substrate and subunit interactions of tryptophan synthase. Biochemistry. 1972;11:2733–2740. doi: 10.1021/bi00764a029. [DOI] [PubMed] [Google Scholar]
- 11.Kirschner K, Wiskocil R, Foehn M, Rezeau L. The tryptophan synthase from Escherichia coli. An improved purification procedure for the α-subunit and binding studies with substrate analogues. Eur. J. Biochem. 1975;60:513–523. doi: 10.1111/j.1432-1033.1975.tb21030.x. [DOI] [PubMed] [Google Scholar]
- 12.Kirschner K, Weischet WO, Wiskocil RL. Ligand binding to enzyme complexes. In: Sund H, Blaver HG, editors. Protein-Ligand Interactions. Berlin: Walter de Gruyter; 1975. pp. 27–44. [Google Scholar]
- 13.Lane AN, Kirschner K. The mechanism of tryptophan binding to tryptophan synthase from Escherichia coli. Eur. J. Biochem. 1981;120:379–387. doi: 10.1111/j.1432-1033.1981.tb05715.x. [DOI] [PubMed] [Google Scholar]
- 14.Lane AN, Kirschner K. The Mechanism of binding of L-serine to tryptophan synthase from Escherichia coli. Euro. J. of Biochem. 1983;129:561–570. doi: 10.1111/j.1432-1033.1983.tb07086.x. [DOI] [PubMed] [Google Scholar]
- 15.Lane AN, Kirschner K. The catalytic mechanism of tryptophan synthase from E. coli. Eur. J. Biochem. 1983;129:571–582. doi: 10.1111/j.1432-1033.1983.tb07087.x. [DOI] [PubMed] [Google Scholar]
- 16.Drewe WF, Jr, Dunn MF. Detection and identification of intermediates in the reaction of L-serine with Escherichia coli tryptophan synthase via rapid-scanning ultraviolet-visible spectroscopy. Biochemistry. 1985;24:3977–3987. doi: 10.1021/bi00336a027. [DOI] [PubMed] [Google Scholar]
- 17.Drewe WF, Jr, Dunn MF. Characterization of the reaction of L-serine and indole with Escherichia coli tryptophan synthase via rapid-scanning ultraviolet-visible spectroscopy. Biochemistry. 1986;25:2494–2501. doi: 10.1021/bi00357a032. [DOI] [PubMed] [Google Scholar]
- 18.Miles EW, Houck DR, Floss HG. Stereochemistry of sodium borohydride reduction of tryptophan synthase of Escherichia coli and its amino acid Schiff bases. J. Biol. Chem. 1982;257:14203–14210. [PubMed] [Google Scholar]
- 19.Metzler CM, Cahill A, Metzler ADE. Equilibria and absorption spectra of schiff bases. J. Am. Chem. Soc. 1980;102:6075–6082. [Google Scholar]
- 20.Metzler CM, Metzler DE. Quantitative description of absorption spectra of a pyridoxal phosphate dependent enzyme using lognormal distribution curves. Anal. Biochem. 1987;166:313–327. doi: 10.1016/0003-2697(87)90580-x. [DOI] [PubMed] [Google Scholar]
- 21.Hyde CD, Padlan EA, Ahmed SA, Miles EW, Davies DR. Three dimensional structure of the tryptophan synthase multienzyme complex from Salmonella typhimurium. J. Biol. Chem. 1988;263:17857–71. [PubMed] [Google Scholar]
- 22.Miles EW. Structural Basis for Catalysis by Tryptophan Synthase. Adv. Enzymol. and Related Areas of Mol. Biol. 1991;64:93–172. doi: 10.1002/9780470123102.ch3. [DOI] [PubMed] [Google Scholar]
- 23.Pan P, Woehl E, Dunn MF. Protein architecture, dynamics and allostery in tryptophan synthase channeling. Trends Biochem Sci. 1997;22:22–27. doi: 10.1016/s0968-0004(96)10066-9. [DOI] [PubMed] [Google Scholar]
- 24.Miles EW, Bauerle R, Ahmed SA. Tryptophan synthase from Escherichia coli and Salmonella typhimurium. Methods In Enzymol. 1987;142:398–414. doi: 10.1016/s0076-6879(87)42051-x. [DOI] [PubMed] [Google Scholar]
- 25.Yutani K, Ogasahara K, Tsujita T, Kanemoto K, Matsumoto M, Tanaka S, Miyashita T, Matsushiro A, Sugino Y, Miles EW. Tryptophan synthase alpha subunit glutamic acid 49 is essential for activity. Studies with 19 mutants at position 49. J. of Biol. Chem. 1987;262:13429–13433. [PubMed] [Google Scholar]
- 26.Miles EW, Rhee S, Davies DR. . The molecular basis of substrate channeling, Journal of Biological Chemistry. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 27.Dunn MF, Niks D, Ngo H, Barends TRM, Schlichting I. Tryptophan synthase: the workings of a channeling nanomachine. Trends in Biochemical Sciences. 2008;33:254–264. doi: 10.1016/j.tibs.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Barends TRM, Dunn MF, Schlichting I. Tryptophan synthase, an allosteric molecular factory. Curr. Opin. in Chem. Biol. 2008;12:619–625. doi: 10.1016/j.cbpa.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Raboni S, Bettati S, Mozzarelli A. Tryptophan synthase: a mine for enzymologists. Cellular and Mol. Life Sci. 2009;66:2391–2403. doi: 10.1007/s00018-009-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn MF. The role of monovalent cations in tryptophan synthase catalysis and regulation of substrate channeling. Encyclopedia of Metalloproteins. 2011 in press. [Google Scholar]
- 31.DeMoss JA. Studies on the mechanism of the tryptophan synthase reaction. Biochem. Biophys. Acta. 1962;62:279–293. doi: 10.1016/0006-3002(62)90041-0. [DOI] [PubMed] [Google Scholar]
- 32.Creighton TE. A steady-state kinetic investigation of the reaction mechanism of the tryptophan synthase of Escherichia coli. Eur. J. Biochem. 1970;13:1–10. doi: 10.1111/j.1432-1033.1970.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 33.Matchett WH. Indole channeling by tryptophan synthase of Neurospora. J. Biol. Chem. 1974;249:4041–4049. [PubMed] [Google Scholar]
- 34.Ahmed SA, Miles EW, Davies DR. Crystallization and preliminary x-ray crystallographic data of the tryptophan synthase α2 β2 complex from Salmonella typhimurium. J. Biol. Chem. 1985;260:3716–3718. [PubMed] [Google Scholar]
- 35.Wilhelm P, Pilz I, Lane AN, Kirschner K. Small-angle x-ray scattering studies of tryptophan synthase and its α and β2 subunits. Eur. J. Biochem. 1982;129:51–56. doi: 10.1111/j.1432-1033.1982.tb07019.x. [DOI] [PubMed] [Google Scholar]
- 36.Miles EW, Kawasaki H, Ahmed SA, Morita H, Morita H, Nagata S. The β-subunit of tryptophan synthase -clarification of the roles of histidine-86 lysine-87, arginine-148, cysteine-170, cysteine-230. J. of Biol. Chem. 1989;264:6280–6287. [PubMed] [Google Scholar]
- 37.Yang L, Ahmed SA, Miles EW. PCR mutagenesis and overexpression of tryptophan synthase from Salmonella typhimurium: on the roles of β2 subunit Lys-382. Protein Expr. Purif. 1996;8:126–136. doi: 10.1006/prep.1996.0082. [DOI] [PubMed] [Google Scholar]
- 38.Nagata S, Hyde CC, Miles EW. The α subunit of tryptophan synthase. Evidence that aspartic acid 60 is a catalytic residue and that the double alteration of residues 175 and 211 in a second-site revertant restores the proper geometry of the substrate binding site. J. Biol. Chem. 1989;264:6288–6296. [PubMed] [Google Scholar]
- 39.Brzovic PS, Kayastha AM, Miles EW, Dunn MF. Substitution of glutamic acid 109 by aspartic acid alters the substrate specificity and catalytic activity of the β-subunit in the tryptophan synthase bienzyme complex from S. typhimurium. Biochemistry. 1992;31:1180–1190. doi: 10.1021/bi00119a030. [DOI] [PubMed] [Google Scholar]
- 40.Rhee S, Parris KD, Hyde CC, Ahmed SA, Miles EW, Davies ER. Crystal structures of a mutant (βK87T) tryptophan synthase α2 β2 complex with ligands bound to the active sites of the α- and β-subunits reveal ligand-induced conformational changes. Biochemistry. 1997;36:7664–7680. doi: 10.1021/bi9700429. [DOI] [PubMed] [Google Scholar]
- 41.Brzovic PS, Sawa Y, Miles EW, Dunn MF. Evidence that mutations in a loop region of the α-subunit inhibit the transition from an open to a closed conformation in the tryptophan synthase bienzyme complex. J. Biol. Chem. 1992;267:13028–13038. [PubMed] [Google Scholar]
- 42.Brzovic PS, Hyde CC, Miles EW, Dunn MF. Characterization of the functional role of a flexible loop in the α-subunit of tryptophan synthase from S. Typhimurium by rapid-scanning stopped-flow spectroscopy and site-directed mutagenesis. Biochemistry. 1993;32:10404–10413. doi: 10.1021/bi00090a016. [DOI] [PubMed] [Google Scholar]
- 43.Harris RM, Dunn MF. Intermediate trapping via a conformational switch in the Na+ -activated tryptophan synthase bienzyme complex. Biochemistry. 2002;41:9982–9990. doi: 10.1021/bi0255672. [DOI] [PubMed] [Google Scholar]
- 44.Harris RM, Ngo H, Dunn MF. Synergistic effects on escape of a ligand from the closed tryptophan synthase bienzyme complex. Biochemistry. 2005;44:16886–16895. doi: 10.1021/bi0516881. [DOI] [PubMed] [Google Scholar]
- 45.Ngo H, Harris R, Kimmich N, Casino P, Niks D, Blumenstein L, Barends TR, Kulik V, Weyand M, Schlichting I, Dunn MF. M. Synthesis and characterization of allosteric probes of substrate channeling in the tryptophan synthase bienzyme complex. Biochemistry. 2007;46:7713–7727. doi: 10.1021/bi700385f. [DOI] [PubMed] [Google Scholar]
- 46.Ngo H, Kimmich N, Harris R, Niks D, Blumenstein L, Kulik V, Barends TR, Schlichting I, Dunn MF. Allosteric regulation of substrate channeling in tryptophan synthase: Modulation of the L-serine reaction in stage I of the β-reaction by α-site ligands. Biochemistry. 2007;46:7740–7753. doi: 10.1021/bi7003872. [DOI] [PubMed] [Google Scholar]
- 47.Barends TRM, Domratcheva T, Kulik V, Blumenstein L, Niks D, Dunn MF, Schlichting I. Structure and mechanistic implications of a tryptophan synthase quinonoid intermediate. ChemBiochem. 2008;9:1024–1028. doi: 10.1002/cbic.200700703. [DOI] [PubMed] [Google Scholar]
- 48.Dierkers AT, Niks D, Schlichting I, Dunn MF. Tryptophan synthase: Structure and function of the monovalent cation site. Biochemistry. 2009;48:10997–11010. doi: 10.1021/bi9008374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai J, Niks D, Wang Y, Domratcheva T, Barends TRM, Schwarz F, Olsen RA, Elliott DW, Fatmi MQ, Chang CA, Schlichting I, Dunn MF, Mueller LJ. X-ray and NMR crystallography in an enzyme active site: The indoline quinonoid intermediate in tryptophan synthase. J. Am. Chem. Soc. 2011;133:4–7. doi: 10.1021/ja106555c. [DOI] [PubMed] [Google Scholar]
- 50.Roy M, Miles EW, Phillips RS, Dunn MF. Detection and identification of transient intermediates in the reactions of tryptophan synthase with oxindolyl-L-alanine and 2,3-dihydro-L-tryptophan. Evidence for a tetrahedral (gem-diamine) intermediate. Biochemistry. 1988;27:8661–8669. doi: 10.1021/bi00423a023. [DOI] [PubMed] [Google Scholar]
- 51.Roy M, Keblawi S, Dunn MF. Stereoelectronic control of bond formation in E. coli tryptophan synthase: Substrate specificity and the enzymatic synthesis of the novel amino acid, dihydroisotryptophan. Biochemistry. 1988;27:6698–6704. doi: 10.1021/bi00418a009. [DOI] [PubMed] [Google Scholar]
- 52.Sachpatzidis A, Dealwis C, Lubetsky JB, Liang PH, Anderson KS, Lois E. Crystallographic studies of phosphonate-based α-reaction transition-state analogues complexed to tryptophan synthase. Biochemistry. 1999;38:12665–12674. doi: 10.1021/bi9907734. [DOI] [PubMed] [Google Scholar]
- 53.Marabotti A, De Biase D, Tramonti A, Bettati S, Mozzarelli A. Novel allosteric effectors of the tryptophan synthase α2 β2 complex identified by computer-assisted molecular modeling. Biochim. Biophys. Acta. 2000;1476:287–299. doi: 10.1016/s0167-4838(99)00242-3. [DOI] [PubMed] [Google Scholar]
- 54.Kulik V, Hartmann E, Weyand M, Frey M, Gierl A, Niks D, Dunn MF, Schlichting I. On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes. J. Mol. Biol. 2005;352:608–620. doi: 10.1016/j.jmb.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Kulik V, Weyand M, Seidel R, Niks D, Arac D, Dunn MF, Schlichting I. On the role of αThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase. J. Mol. Biol. 2002;324:677–690. doi: 10.1016/s0022-2836(02)01109-9. [DOI] [PubMed] [Google Scholar]
- 56.Yang XJ, Miles EW. Threonine 183 and adjacent flexible loop residues in the tryptophan synthase α subunit have critical roles in modulating the enzymatic activities of the β subunit and the α2 β2 complex. J. Biol. Chem. 1992;267:1–9. [PubMed] [Google Scholar]
- 57.Ferrari D, Yang L-H, Miles EW, Dunn MF. M. F. The βD305A mutant of tryptophan synthase shows strongly perturbed allosteric regulation and substrate specificity. Biochemistry. 2001;40:7421–7432. doi: 10.1021/bi002892l. [DOI] [PubMed] [Google Scholar]
- 58.Ferrari D, Niks D, Yang L-H, Miles EW, Dunn MF. Allosteric communication in the tryptophan synthase bienzyme complex: Roles of the β-subunit aspartate 305-arginine 141 salt bridge. Biochemistry. 2003;42:7807–7818. doi: 10.1021/bi034291a. [DOI] [PubMed] [Google Scholar]
- 59.Houben KF, Kadima W, Roy M, Dunn MF. L-serine analogues form Schiff base and quinonoidal intermediates with Escherichia coli tryptophan synthase. Biochemistry. 1989;28:4140–4147. doi: 10.1021/bi00436a003. [DOI] [PubMed] [Google Scholar]
- 60.Houben KF, Dunn MF. Allosteric effects acting over a distance of 20–25 Å in the Escherichia coli tryptophan synthase bienzyme complex increase ligand affinity and cause redistribution of covalent intermediates. Biochemistry. 1990;29:2421–2429. doi: 10.1021/bi00461a028. [DOI] [PubMed] [Google Scholar]
- 61.Brzovic PS, Ngo K, Dunn MF. Allosteric interactions coordinate catalytic activity between successive metabolic enzymes in the tryptophan synthase bienzyme complex. Biochemistry. 1992;31:3831–3839. doi: 10.1021/bi00130a014. [DOI] [PubMed] [Google Scholar]
- 62.Dunn MF, Aguilar V, Brzovic P, Drewe WF, Jr, Houben KF, Leja CA, Roy M. The tryptophan synthase bienzyme complex transfers indole between the α- and β-sites via a 25–30 Å long tunnel. Biochemistry. 1990;29:8598–8607. doi: 10.1021/bi00489a015. [DOI] [PubMed] [Google Scholar]
- 63.Leja CA, Woehl EU, Dunn MF. Allosteric linkages between β-site covalent transformations and α-site activation and deactivation in the tryptophan synthase bienzyme complex. Biochemistry. 1995;34:6552–6561. doi: 10.1021/bi00019a037. [DOI] [PubMed] [Google Scholar]
- 64.Pan P, Dunn MF. β-site covalent reactions trigger transitions between open and closed conformations of the tryptophan synthase bienzyme complex. Biochemistry. 1996;35:5002–5013. doi: 10.1021/bi960033k. [DOI] [PubMed] [Google Scholar]
- 65.Kirschner K, Lane AN, Strasser AWM. Reciprocal communication between the lyase and synthase active-sites of the tryptophan synthase bienzyme complex. Biochemistry. 1991;30:472–478. doi: 10.1021/bi00216a024. [DOI] [PubMed] [Google Scholar]
- 66.Anderson KS, Miles EW, Johnson KA. Serine modulates substrate channeling in tryptophan synthase -A novel intersubunit triggering mechanism. J. Biol. Chem. 1991;266:8020–8033. [PubMed] [Google Scholar]
- 67.Dunn MF, Aguilar V, Drewe WR, Jr, Houben K, Robustell B, Roy M. The interconversion of E. coli tryptophan synthase intermediates is modulated by allosteric interactions. Indian J. of Biochem. and Biophys. 1987;24:44–51. [PubMed] [Google Scholar]
- 68.Dunn MF, Roy M, Robustell B, Aguilar V. Chemical probes of substrate channeling in the E. coli tryptophan synthase α2 β2 bienzyme complex. In: Korpela T, Christen P, editors. Biochemistry of Vitamin B6. Proceedings of the 7th International Congress on Chemical and Biological Aspects of Vitamin B6 Catalysis Birkauser; Basel. 1987. pp. 171–181. [Google Scholar]
- 69.Peracchi A, Mozzarelli A, Rossi GL. Monovalent cations affect dynamic and functional properties of the tryptophan synthase α2 β2 complex. Biochemistry. 1995;34:9459–9465. doi: 10.1021/bi00029a022. [DOI] [PubMed] [Google Scholar]
- 70.Woehl EU, Dunn MF. Monovalent metal ions play an essential role in catalysis and intersubunit communication in the tryptophan synthase bienzyme complex. Biochemistry. 1995;34:9466–9476. doi: 10.1021/bi00029a023. [DOI] [PubMed] [Google Scholar]
- 71.Woehl EU, Dunn MF. Mechanisms of monovalent cation action in enzyme catalysis: the first stage of the tryptophan synthase β-reaction. Biochemistry. 1999;38:7118–7130. doi: 10.1021/bi982918x. [DOI] [PubMed] [Google Scholar]
- 72.Woehl EU, Dunn MF. Mechanisms of monovalent cation action in enzyme catalysis: the tryptophan synthase α-, β-, and αβ-reactions. Biochemistry. 1999;38:7131–7141. doi: 10.1021/bi982919p. [DOI] [PubMed] [Google Scholar]
- 73.Weber-Ban E, Hur O, Bagwell C, Banik U, Yang L-H, Miles EW, Dunn MF. Investigation of allosteric linkages in the regulation of tryptophan synthase: The roles of salt bridges and monovalent cations probed by site-directed mutation, optical spectroscopy, and kinetics. Biochemistry. 2001;40:3497–3511. doi: 10.1021/bi002690p. [DOI] [PubMed] [Google Scholar]
- 74.Rhee S, Parris KD, Ahmed SA, Miles EW, Davies DR. Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase α2 β2 complex. Biochemistry. 1996;35:4211–4221. doi: 10.1021/bi952506d. [DOI] [PubMed] [Google Scholar]
- 75.Fan YX, McPhie P, Miles EW. Guanidine hydrochloride exerts dual effects on the tryptophan synthase α2 β2 complex as a cation activator and as a modulator of the active site conformation. Biochemistry. 1999;38:7881–7890. doi: 10.1021/bi990307e. [DOI] [PubMed] [Google Scholar]
- 76.Fan YX, McPhie P, Miles EW. Regulation of tryptophan synthase by temperature, monovalent cations, and an allosteric ligand. Evidence from Arrhenius plots, absorption spectra, and primary kinetic isotope effects. Biochemistry. 2000;39:4692–4703. doi: 10.1021/bi9921586. [DOI] [PubMed] [Google Scholar]
- 77.Phillips RS, McPhie P, Miles EW, Marchal S, Lang R. Quantitative effects of allosteric ligands and mutations on conformational equilibria in Salmonella typhimurium tryptophan synthase. Arch. Biochem. Biophys. 2008;470:8–19. doi: 10.1016/j.abb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 78.Woehl EU, Dunn MF. The roles of Na+ and K+ in pyridoxal phosphate enzyme catalysis Coord. Chem. Rev. 1995;144:147–197. [Google Scholar]
- 79.Qaiser Fatmi M, Ai R, Chang CA. Synergistic Regulation and ligand-induced conformational changes of tryptophan synthase. Biochemistry. 2009;48:9921–9931. doi: 10.1021/bi901358j. [DOI] [PubMed] [Google Scholar]