Abstract
An insertion-deletion (indel) polymorphism within the 3′ untranslated region (UTR) of HLA-C has been shown to be involved in the regulation of HLA-C expression. Individuals who carry a deletion at this position exhibit increased HLA-C expression, which associates with lower viral set point in HIV-1 infected individuals. This 263 indel (rs67384697) is reported to be in strong linkage disequilibrium (LD) with a single nucleotide polymorphism (SNP) 35 kilobases upstream of HLA-C (-35T/C; rs9264942) in Caucasian individuals, making this SNP a potential marker for both HLA-C expression and HIV-1 disease progression. We therefore examined genetic variation within the HLA-C 3′ UTR of 265 Black and Caucasian South Africans by direct sequencing and identified haplotypes encompassing the 263 indel and another indel at position 230 in both populations. Concomitant evaluation of variability at the −35 SNP revealed this polymorphism to be an inappropriate marker for the 263 indel in these populations. These findings provide important insights into genetic variability within the regulatory regions of HLA-C that have potential implications for our understanding of the regulation of HLA-C expression and its impact on HIV-1 disease progression.
Introduction
Recently, there has been an increased interest in the role of HLA-C in HIV-1 infection. A single nucleotide polymorphism (SNP) 35 kb upstream of HLA-C has been shown to be associated with differences in HIV-1 viral set point [1] - a key early determinant of the rate of HIV-1 disease progression. This SNP (−35T/C; rs9264942) also strongly associates with differences in HLA-C mRNA [1], [2] and cell surface expression levels [3]. However, while these associations have consistently been shown in Caucasian cohorts [1], [3]–[7], the association between the -35 SNP (rs9264942) and HIV-1 viral set point has not been shown to be significant in African American cohorts - despite the presence of this SNP in this population [7]–[10]. This has lead to the suggestion that the -35 SNP is not the causative variant responsible for the alteration in HLA-C expression and viral set point, but rather acts as a marker in Caucasian populations for another polymorphism.
This view is supported by the findings of Kulkarni et al. [11], who identified a single base pair insertion-deletion (indel) polymorphism at position 263 of the HLA-C 3′ untranslated region (UTR) that has been shown to be in strong linkage disequilibrium (LD) with the -35 SNP in Caucasian individuals. They found that this variant (263 indel; rs67384697) potentially affects the binding of a regulatory microRNA (miRNA148a) to the 3′ UTR, with a deletion at this position (263del) abolishing miRNA148a binding and leading to increased HLA-C expression. They also found an overrepresentation of the 263del allele in HIV-1 controllers relative to non-controllers in a cohort of HIV-infected individuals of European ancestry, suggesting that the change in HLA-C expression associated with this allele could provide the basis for long-term protection against HIV-1 disease progression [11]. However, these data are again only representative of individuals of Caucasian ancestry and have yet to be confirmed in other populations.
That differences exist in patterns of genetic variation, and specifically in patterns of LD, between Black and Caucasian populations has become increasingly apparent. A recent study described HLA class I diversity in both Black and Caucasian South African populations [12], outlining the patterns of LD that characterise this region and highlighting the key differences in HLA-C allelic representation between these two population groups. However, no data are yet available regarding the -35 SNP or describing variation in the HLA-C 3′ UTR in these populations. Here we report the first description of these data in 265 unrelated South Africans from both the Black and Caucasian population groups.
Materials and Methods
Study Population
A total of 265 HIV-1 negative, unrelated South African individuals were used to describe genetic variation and patterns of LD within and between the coding and regulatory regions of the HLA-C locus. These 168 Black and 97 Caucasian South Africans were selected from a larger previously described cohort [12] on the basis of a non-reactive HIV enzyme-linked immunosorbent assay (ELISA) test (Genscreen HIV1/2 version 2; Bio-Rad, Marnes-La-Coquette, France). Informed consent was obtained from all study participants and the study was approved by the University of the Witwatersrand Committee for Research on Human Subjects.
The DNA used for genotyping was extracted from buffy coat samples using the PEL-FREEZ DNA Isolation Kit (DYNAL Invitrogen Corporation, Carlsbad, California, USA).
HLA-C Genotyping
HLA-C genotyping was performed at both low and high resolution as previously described [12]. However, because the genotyping was performed prior to 2005 using a single specific primer-polymerase chain reaction (SSP-PCR) genotyping method, it was possible that alleles that had not yet been identified at the time of genotyping may have been present within the sample population. Misclassification of these alleles could potentially confound LD analyses. HLA-C*02∶10 has previously been found at relatively high frequencies in other Black populations [13], however, it was not observed in the Black South African population during the prior genotyping [12]. HLA-C*02∶02 and HLA-C*02∶10 differ by only a single amino acid (T211C) in exon 4 [14], a difference that would not be detected by the SSP-PCR genotyping kit previously employed to genotype these samples. Therefore, all Black individuals who had originally been typed as having HLA-C*02∶02 alleles; as well as all individuals who were initially typed as homozygous at the HLA-C locus, were re-genotyped using the AlleleSEQR HLA-C PLUS Sequence-Based Typing (SBT) Kit (Abbot Molecular, Des Plaines, Illinois, USA) according to the manufacturer's instructions. Sequencing analysis and allele assignment were performed using Assign™ SBT v3.5.1 software (Conexio Genomics, Fremantle, Western Australia), with the IMGT/HLA July 2011 (v3.6.0) references. Because at least one of the alleles in any genotype combination was already known (based on the previously available genotyping data), none of the retyped samples were regarded as ambiguous.
HLA-C 3′ UTR DNA Sequencing
The HLA-C 3′ UTR was amplified from genomic DNA using the PCR primers described by Kulkarni et al. [11] and the following thermocycling conditions: 94°C for 2 minutes, followed by 30 cycles of 94°C for 15 seconds, 65°C for 30 seconds, 72°C for 90 seconds and 72°C for 7 minutes. The amplicons were sequenced in both directions by capillary electrophoresis using an ABI 3100 DNA analyzer (Applied Biosystems, Foster City, California, USA) and the sequencing primers: 5′-GTGAGATTCTGGGGAGCTGA-3′ and 5′-TCTGGAAGGTTCTCAGGTC-3′. The chromatograms obtained were analysed using Sequencher v4.2 (Genes Codes Corporation, Ann Arbor, Michigan, USA) and sequences were aligned with an available reference sequence (GenBank Accession Number NG_029422) to identify polymorphic positions.
-35 SNP Genotyping
A real-time PCR assay was designed to genotype the -35 SNP (rs9264942). PCR amplicons were amplified from genomic DNA using a common forward primer (5′-GCCCATACCTGTTTATACATCCA-3′) and allele-specific reverse primers (5′-CAGAAAGTCCCACAGTGCCTG-3′ and 5′-CAGAAAGTCCCACAGTGCCTA-3′). Both allele-specific primers were designed with a lock nucleic acid (LNA) modified 3′-end base. The assay was performed using the Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, Foster City, California, USA), under the following thermocycling conditions: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 15 seconds and 70°C for 1 minute. Reactions were performed in a 10 µl volume, containing 1x Maxima SYBR Green/ROX qPCR Master Mix (Fermentas, Burlington, Canada), 10 pmol of each primer and 20–60 ng of DNA. PCR analysis was based on calculation of ΔCT (difference in cycle threshold; the difference between the CT of the -35T and -35C reactions). Heterozygous individuals had ΔCT values of between 0 and 0.3, while homozygous TT and CC individuals had ΔCT values of ≥4 or ≤−4, respectively.
Computational and Statistical Analyses
Allele frequencies at all polymorphic positions were determined by direct counting [15] and comparisons of these frequencies between the Black and Caucasian population groups were performed using a two-sided Fisher exact test (SISA: Simple Interactive Statistical Analysis). Deviations from Hardy-Weinberg equilibrium (HWE) were assessed using the Markov chain exact test for HWE [16]. The Excoffier-Laval-Balding (ELB) algorithm [17] was used to estimate the gametic phase of the genotypic data generated at all polymorphic positions, allowing linkage disequilibrium (LD) to be quantified as the LD coefficients [18], [19] D' and r2. The significance of pairwise LD between all polymorphic positions was estimated using an exact test for LD [20]. All HWE, LD and haplotypic analyses were implemented through Arlequin v3.5.1.2 [21]. All statistical measures were considered significant at p<0.05.
Results
HLA-C Allele Representation
Although HLA-C genotyping data was already available for all 265 of the individuals included in this study [12]; the development of new genotyping methods, coupled with the identification new HLA-C alleles in the period since the initial genotyping had been performed, made it prudent to retype approximately 20% of the samples to avoid confounding subsequent LD analyses. While all the Caucasian individuals genotyped were confirmed to be in possession of the HLA-C*02∶02 allele, the majority of the Black individuals who were initially thought to be in possession of HLA-C*02∶02 were subsequently reclassified as having HLA-C*02∶10. Additionally, an individual from the Black population group was found to be homozygous for a putative new allele (herein referred to as C*15:xx), which had previously been classified as C*15∶05. This putative new allele was found to be identical to C*15∶05:01 across exons 2, 3 and 4, with the exception of a G>A transition at nucleotide position 728 in exon 4. The alleles and their frequencies in both populations are given in Table S1.
Genetic Variation within the HLA-C 3′ UTR
Direct sequencing of the 3′ UTRs of the HLA-C alleles present in the Black and Caucasian South African population revealed this region contained 33 polymorphisms within the approximately 400 bp of sequence analysed, including the indel at position 263 (Table S2). Thirty-one positions (including the 263 indel) were found to be polymorphic in both the Black and the Caucasian population groups (Table S2). Minor allele frequencies differed significantly between the two population groups at ten of these positions; however, these did not include the 263 indel. Positions 84 (84G/A; rs139211788) and 285 (285ACTT/-; rs60637457) were only polymorphic within the Black population. Seven other SNPs previously reported in this region were not observed in either population.
Several of the polymorphisms present in the 3′ UTR were found to only be associated with specific HLA-C alleles (Table 1 and 2). For the majority of alleles, these allele-specific polymorphisms were associated with the same HLA-C allele (or alleles) in both population groups (when the given HLA-C allele was present in both population groups). However, this was not always the case – as can be seen for C*02∶02 and C*02∶05. The Black population group deviated significantly from HWE at positions 46 (p = 0.026), 110 (p<0.001), 138 (p = 0.001), 267 (p = 0.023) and 303 (p<0.001), while positions 263 (p = 0.033), 266 (p = 0.032), 294 (p = 0.032), 299 (p = 0.031), 300 (p = 0.032), 307 (p = 0.024), 345 (p = 0.032) and 346 (p = 0.032) deviated significantly from HWE in the Caucasian population group.
Table 1. Genetic variation within HLA-C 3′ UTR sequences of the HLA-C alleles observed in the Black South African population group.
| Polymorphic Position1 | |||||||||||||||||||||||||||||||||
| 46 | 84 | 92 | 93 | 101 | 110 | 125 | 133 | 138 | 146 | 179 | 224 | 230 | 256 | 259 | 261 | 263 | 266 | 267 | 278 | 285 | 294 | 299 | 300 | 303 | 307 | 324 | 345 | 346 | 347 | 356 | 375 | 379 | |
| Allele | |||||||||||||||||||||||||||||||||
| 02∶02 2 | C | G | R | T | T | Y | R | K | S | Y | Y | R | I/D | M | C | T | I | C | R | R | I | A | G | T | R | C | K | A | G | S | R | T | R |
| 02∶05 | C | G | A | T | T | T | A | T | G | T | T | G | I 4 | C | C | T | I | C | A | G | I | A | G | T | A | C | G | A | G | C | G | T | A |
| 02∶10 2 | Y3 | R | A | T | T | Y | R | K | S | Y | Y | R | I/D | M | Y | Y | I/D | Y | R | R | I/D | M | R | W | R | Y | K | R | R | S | R | T | R |
| 03∶02 | C | G | A | T | T | T | G | G | C | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | G | C | T | A | G | G | A | T | G |
| 03∶03 | C | G | A | T | T | T | G | G | C | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | G | C | T | A | G | G | A | T | G |
| 03∶04 2 | C | G | A | T | T | T | G | G | S | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | R | C | T | A | G | G | A | T | G |
| 04∶01 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | C | T | A | G | G | A | T | G |
| 04∶04 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | C | T | A | G | G | A | T | G |
| 05∶01 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 06∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 06∶06 | C | G | A | T | T | C | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 06∶11 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 07∶01 2 | C | G | G | T | T | Y | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶02 | C | G | A | C | T | T | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶04 | C | G | A | T | T | T | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶06 | C | G | A | T | T | C | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶18 | C | G | A | T | T | C | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 08∶01 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 08∶02 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 08∶04 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 12∶03 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 14∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | G | T | A | G | G | A | T | G |
| 15∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 15∶05 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 15:xx | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 16∶01 2 | C | G | A | T | Y | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | C | G |
| 17∶01 | C | G | A | T | T | T | A | T | G | T | T | G | I | C | C | T | I | C | A | G | I | A | G | T | A | C | G | A | G | C | G | T | A |
| 18∶01 2 | C | G | A | T | T | T | G | G | S | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | C | T | A | G | G | A | T | G |
| 18∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | C | T | A | G | G | A | T | G |
positions are given relative to the start of the HLA-C 3′ UTR.
HLA-C alleles C*02∶02, C*02∶10, C*03∶04, C*07∶01, C*16∶01 and C*18∶01 were found to have more than one 3′ UTR sequence.
where more than one allele has been observed alleles are reported using standard IUB ambiguity codes.
I refers to an insertion and D to a deletion.
Table 2. Genetic variation within HLA-C 3′ UTR sequences of the HLA-C alleles observed in the Caucasian South African population group.
| Polymorphic Position1 | |||||||||||||||||||||||||||||||||
| 46 | 84 | 92 | 93 | 101 | 110 | 125 | 133 | 138 | 146 | 179 | 224 | 230 | 256 | 259 | 261 | 263 | 266 | 267 | 278 | 285 | 294 | 299 | 300 | 303 | 307 | 324 | 345 | 346 | 347 | 356 | 375 | 379 | |
| Allele | |||||||||||||||||||||||||||||||||
| 01∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D4 | C | C | T | I | C | G | A | I | A | G | T | A | C | T | A | G | G | A | T | G |
| 02∶02 2 | C | G | A | Y3 | T | Y | G | G | S | C | C | R | I/D | M | C | T | I | C | R | A | I | A | G | T | R | C | K | A | G | S | A | T | R |
| 02∶05 | C | G | A | T | C | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | C | G |
| 03∶03 | C | G | A | T | T | T | G | G | C | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | G | C | T | A | G | G | A | T | G |
| 03∶04 2 | C | G | A | T | T | T | G | G | S | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | G | C | T | A | G | G | A | T | G |
| 03∶16 | C | G | A | T | T | T | G | G | C | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | G | C | T | A | G | G | A | T | G |
| 04∶01 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | C | T | A | G | G | A | T | G |
| 04∶08 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | C | T | A | G | G | A | T | G |
| 05∶01 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 06∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 06∶11 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 07∶01 | C | G | G | T | T | T | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶02 | C | G | A | C | T | T | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶04 | C | G | A | T | T | T | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶06 | C | G | A | T | T | C | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 07∶18 | C | G | A | T | T | C | G | G | G | C | C | G | I | A | C | T | I | C | A | A | I | A | G | T | A | C | G | A | G | C | A | T | A |
| 08∶01 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 08∶02 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 08∶04 | T | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 12∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 12∶03 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 14∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | C | T | I | C | G | A | I | A | G | T | A | G | T | A | G | G | A | T | G |
| 15∶02 | C | G | A | T | T | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | T | G |
| 16∶01 | C | G | A | T | C | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | C | G |
| 16∶02 | C | G | A | T | C | T | G | G | G | C | C | A | D | C | T | C | D | T | A | A | I | C | A | A | A | T | T | G | A | G | A | C | G |
| 17∶01 | C | G | A | T | T | T | A | T | G | T | T | G | I | C | C | T | I | C | A | G | I | A | G | T | A | C | G | A | G | C | G | T | A |
positions are given relative to the start of the HLA-C 3′ UTR.
HLA-C alleles C*02∶02 and C*03∶04 were found to have more than one 3′ UTR sequence.
where more than one allele has been observed alleles are reported using standard IUB ambiguity codes.
I refers to an insertion and D to a deletion.
Estimation of LD Across the HLA-C 3′ UTR
In order to further investigate the relationships between the genetic variants identified within the HLA-C 3′ UTR, the ELB algorithm [17] was used to estimate the gametic phase of the genotyping data generated at all the polymorphic positions within this region. This allowed pairwise LD between each of these variants and the HLA-C alleles observed in each population group to be quantified, allowing the 3′ UTR sequences for each of the HLA-C alleles observed in both the Black and Caucasian population groups to be inferred. The HLA-C 3′ UTR sequences observed for each specific HLA-C allele are given in Table 1 (for the Black population group) and Table 2 (for the Caucasian population group). While the majority of alleles in both population groups were represented by a single 3′ UTR sequence, this was not always the case – with the Cw*02 alleles displaying multiple 3′ UTR sequences in both population groups. Similarly, the 3′ UTR sequence for a given allele was not necessarily the same in both population groups; with the Cw*02 alleles again providing the best example.
Given the observation that C*02∶10 could be associated with both an insertion and a deletion at position 263, we sought to establish the extent of LD between the 263 indel and the HLA-C alleles present in both population groups. These data are given in Table 3. Seventeen of the HLA-C alleles in the Black population group were in significant LD with the 263 indel. C*02∶02, C*03∶04, C*04∶01, C*07∶01, C*07∶02, C*07∶06, C*07∶18, C*17∶01 and C*18∶02 were in complete linkage with the insertion at position 263 (263ins); while C*05∶01, C*06∶02, C*08∶02, C*08∶04, C*12∶03, C*15∶05 and C*16∶01 were in complete linkage with the deletion at this position (263del). Although linkage was not complete, C*02∶10 was also found to be significantly associated with the 263del allele. Fifteen of the HLA-C alleles in the Caucasian population group were found to be in significant LD with the 263 indel. C*02∶02, C*03∶03, C*03∶04, C*04∶01, C*07∶01 and C*07∶02 were in complete linkage with 263ins, while C*05∶01, C*06∶02, C*08∶02, C*08∶04, C*12∶02, C*12∶03, C*15∶02, C*16∶01 and C*16∶02 were in complete linkage with 263del.
Table 3. Linkage disequilibrium between the 263 indel and the HLA-C alleles present in the Black and Caucasian South African population groups.
| Black Individuals (n1 = 168) | Caucasian Individuals (n1 = 96) | |||||||
| HLA-C Allele | 3’ UTR Allele | Frequency2 | D’3 | p-value4 | 3’ UTR Allele | Frequency2 | D’3 | p-value4 |
| 01∶02 | – | – | – | – | Ins | 0.0208 (45) | 1.00 | NS |
| 02∶02 | Ins | 0.0089 (5) | 1.00 | <0.05 | Ins | 0.0625 (12) | 1.00 | <0.05 |
| 02∶05 | Ins | 0.0030 (1) | 1.00 | NS | Del | 0.0052 (1) | 1.00 | NS |
| 02∶10 | Del | 0.0327 (11) | 0.33 | <0.05 | – | – | – | – |
| 03∶02 | Ins | 0.0149 (5) | 1.00 | NS | – | – | – | – |
| 03∶03 | Ins | 0.0030 (1) | 1.00 | NS | Ins | 0.0625 (12) | 1.00 | <0.05 |
| 03∶04 | Ins | 0.0446 (15) | 1.00 | <0.01 | Ins | 0.0573 (11) | 1.00 | <0.05 |
| 03∶16 | – | – | – | – | Ins | 0.0052 (1) | 1.00 | NS |
| 04∶01 | Ins | 0.1250 (42) | 1.00 | <0.01 | Ins | 0.0885 (17) | 1.00 | <0.01 |
| 04∶04 | Ins | 0.0030 (1) | 1.00 | NS | – | – | – | – |
| 04∶08 | – | – | – | – | Ins | 0.0052 (1) | 1.00 | NS |
| 05∶01 | Del | 0.0089 (3) | 1.00 | <0.05 | Del | 0.0521 (10) | 1.00 | <0.01 |
| 06∶02 | Del | 0.1488 (50) | 1.00 | <0.01 | Del | 0.0781 (15) | 1.00 | <0.01 |
| 06∶06 | Ins | 0.0030 (1) | 1.00 | NS | – | – | – | – |
| 06∶11 | Del | 0.0030 (1) | 1.00 | NS | Del | 0.0052 (1) | 1.00 | NS |
| 07∶01 | Ins | 0.0714 (24) | 1.00 | <0.01 | Ins | 0.1771 (34) | 1.00 | <0.01 |
| 07∶02 | Ins | 0.0625 (21) | 1.00 | <0.01 | Ins | 0.1458 (28) | 1.00 | <0.01 |
| 07∶04 | Ins | 0.0149 (5) | 1.00 | NS | Ins | 0.0104 (2) | 1.00 | NS |
| 07∶06 | Ins | 0.0387 (13) | 1.00 | <0.01 | Ins | 0.0208 (4) | 1.00 | NS |
| 07∶11 | Del | 0.0030 (1) | 1.00 | NS | – | – | – | – |
| 07∶18 | Ins | 0.0417 (14) | 1.00 | <0.01 | Ins | 0.0156 (3) | 1.00 | NS |
| 08∶01 | Del | 0.0030 (1) | 1.00 | NS | Del | 0.0052 (1) | 1.00 | NS |
| 08∶02 | Del | 0.0149 (5) | 1.00 | <0.01 | Del | 0.0208 (4) | 1.00 | <0.01 |
| 08∶04 | Del | 0.0298 (10) | 1.00 | <0.01 | Del | 0.0104 (2) | 1.00 | <0.05 |
| 12∶02 | – | – | – | – | Del | 0.0104 (2) | 1.00 | <0.05 |
| 12∶03 | Del | 0.0149 (5) | 1.00 | <0.01 | Del | 0.0208 (4) | 1.00 | <0.01 |
| 14∶02 | Ins | 0.0060 (2) | 1.00 | NS | Ins | 0.0104 (2) | 1.00 | NS |
| 15∶02 | Del | 0.0060 (2) | 1.00 | NS | Del | 0.0260 (5) | 1.00 | <0.01 |
| 15∶05 | Del | 0.0089 (3) | 1.00 | <0.05 | – | – | – | – |
| 15:xx | Del | 0.0060 (2) | 1.00 | NS | – | – | – | – |
| 16∶01 | Del | 0.0714 (24) | 1.00 | <0.01 | Del | 0.0677 (13) | 1.00 | <0.01 |
| 16∶02 | – | – | – | – | Del | 0.0104 (2) | 1.00 | <0.05 |
| 17∶01 | Ins | 0.1131 (38) | 1.00 | <0.01 | Ins | 0.0052 (1) | 1.00 | NS |
| 18∶01 | Ins | 0.0149 (5) | 1.00 | NS | – | – | – | – |
| 18∶02 | Ins | 0.0298 (10) | 1.00 | <0.01 | – | – | – | – |
the total number of individuals genotyped in each population group.
the observed frequency of each two-locus haplotype.
Lewontin's D' measure of linkage disequilibrium [19].
p-values are calculated using an exact test for linkage disequilibrium [20], and are significant at p<0.05.
the number of chromosomes on which the two-locus haplotype was found to occur.
Haplotype Structure within the HLA-C 3′ UTR
LD analysis also identified the presence of two highly conserved, overlapping haplotypes involving multiple positions across the HLA-C 3′ UTR. The larger of these haplotypes involved ten positions (Figure 1; indicated in pink), spanning approximately 90 bases, and encompassing the indel at position 263. Another involved five positions (Figure 1; indicated in blue), spanning 155 bases of the HLA-C 3′ UTR, and encompassed the indel at position 230. Only positions with both D’ and r2 measures of pairwise LD equal to 1 were included within these haplotypes.
Figure 1. The haplotypes identified within the HLA-C 3′ UTR.
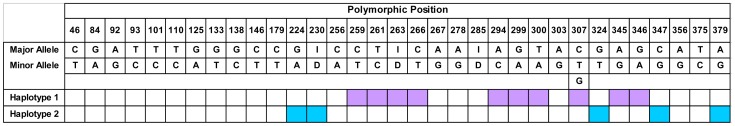
The positions involved in the two haplotypes identified are indicated in colour. The haplotype encompassing the 263 indel is shown in pink, while the haplotype encompassing the 230 indel is shown in blue. The major and minor alleles at each position are also indicated. Positions were only included in the haplotypes if both D' and r2 measures of pairwise LD were equal to 1. Polymorphic positions are indicated by their position relative to the start of the HLA-C 3′ UTR.
Because of complete linkage between the indels at positions 230 and 263, these haplotypes were found to be present in three possible combinations within all of the HLA-C alleles present in both population groups. These haplotypic combinations thus produced three “classes” of HLA-C 3′ UTR sequences, each defined by a specific combination of the two haplotypes observed. Class I corresponded to sequences with haplotypes defined by insertions at both 230 and 263, class II to sequences with a deletion at 230 and an insertion at 263 and class III to sequences with deletions at both 230 and 263.
Linkage Disequilibrium between Specific HLA-C Alleles and the -35 SNP
Genotyping of the -35 SNP (rs9264942) was also performed for 168 Black and 97 Caucasian South African individuals. The frequencies of the 35C allele in the Black (0.348) and Caucasian (0.284) population groups were not significantly different (p = 0.13), and neither population group was found to deviate significantly from HWE at this position. This SNP has previously been shown to be in strong LD with a number of HLA-C alleles in individuals of Caucasian descent [3], [11], therefore the extent of LD between the -35 SNP and the HLA-C alleles present in both population groups was also evaluated (Table 4).
Table 4. Linkage disequilibrium between the -35 SNP and the HLA-C alleles present in the Black and Caucasian South African population groups.
| Black Individuals (n1 = 168) | Caucasian Individuals (n1 = 97) | |||||||
| HLA-C Allele | -35 SNP Allele | Frequency2 | D’3 | p-value4 | -35 SNP Allele | Frequency2 | D’3 | p-value4 |
| 01∶02 | – | – | – | – | C | 0.0155 (35) | 0.65 | <0.05 |
| 02∶02 | C | 0.0149 (5) | 1.00 | <0.05 | C | 0.0723 (14) | 1.00 | <0.01 |
| 02∶05 | T | 0.0030 (1) | 1.00 | NS | C | 0.0052 (1) | 1.00 | NS |
| 02∶10 | T | 0.0655 (22) | 0.59 | <0.05 | – | – | – | – |
| 03∶02 | C | 0.0149 (5) | 1.00 | <0.01 | – | – | – | – |
| 03∶03 | C | 0.0030 (1) | 1.00 | NS | T | 0.0619 (12) | 1.00 | <0.05 |
| 03∶04 | T | 0.0446 (15) | 1.00 | <0.01 | T | 0.0567 (11) | 1.00 | <0.05 |
| 03∶16 | – | – | – | – | T | 0.0052 (1) | 1.00 | NS |
| 04∶01 | T | 0.1250 (42) | 1.00 | <0.01 | T | 0.0876 (17) | 1.00 | <0.01 |
| 04∶04 | T | 0.0030 (1) | 1.00 | NS | – | – | – | – |
| 04∶08 | – | – | – | – | T | 0.0052 (1) | 1.00 | NS |
| 05∶01 | C | 0.0060 (2) | 0.49 | NS | T | 0.0412 (8) | 0.28 | NS |
| 06∶02 | C | 0.1488 (50) | 1.00 | <0.01 | C | 0.0773 (15) | 1.00 | <0.01 |
| 06∶06 | C | 0.0030 (1) | 1.00 | NS | – | – | – | – |
| 06∶11 | C | 0.0030 (1) | 1.00 | NS | C | 0.0052 (1) | 1.00 | NS |
| 07∶01 | T | 0.0506 (17) | 0.16 | NS | T | 0.1753 (34) | 1.00 | <0.01 |
| 07∶02 | T | 0.0595 (20) | 0.86 | <0.01 | T | 0.1443 (28) | 1.00 | <0.01 |
| 07∶04 | T | 0.0149 (5) | 1.00 | NS | T | 0.0103 (2) | 1.00 | NS |
| 07∶06 | C | 0.0387 (13) | 1.00 | <0.01 | C | 0.0206 (4) | 1.00 | <0.01 |
| 07∶11 | T | 0.0030 (1) | 1.00 | NS | – | – | – | – |
| 07∶18 | C | 0.0417 (14) | 0.89 | <0.01 | C | 0.0155 (3) | 1.00 | <0.01 |
| 08∶01 | T | 0.0030 (1) | 1.00 | NS | T | 0.0052 (1) | 1.00 | NS |
| 08∶02 | T | 0.0119 (4) | 0.42 | NS | C | 0.0103 (2) | 0.31 | NS |
| 08∶04 | C | 0.0149 (5) | 0.24 | NS | C | 0.0103 (2) | 1.00 | <0.05 |
| 12∶02 | – | – | – | – | C | 0.0103 (2) | 1.00 | <0.05 |
| 12∶03 | C | 0.0149 (5) | 1.00 | <0.01 | C | 0.0206 (4) | 1.00 | <0.01 |
| 14∶02 | C | 0.0060 (2) | 1.00 | <0.05 | C | 0.0103 (2) | 1.00 | <0.05 |
| 15∶02 | T | 0.0060 (2) | 1.00 | NS | T | 0.0258 (5) | 1.00 | NS |
| 15∶05 | T | 0.0089 (3) | 1.00 | NS | – | – | – | – |
| 15:xx | T | 0.0060 (2) | 1.00 | NS | – | – | – | – |
| 16∶01 | T | 0.0655 (22) | 0.76 | <0.01 | T | 0.0670 (13) | 1.00 | <0.05 |
| 16∶02 | – | – | – | – | T | 0.0103 (2) | 1.00 | NS |
| 17∶01 | T | 0.1131 (38) | 1.00 | <0.01 | T | 0.0052 (1) | 1.00 | NS |
| 18∶01 | T | 0.0149 (5) | 1.00 | NS | – | – | – | – |
| 18∶02 | T | 0.0298 (10) | 1.00 | <0.05 | – | – | – | – |
the total number of individuals genotyped in each population group.
the observed frequency of each two-locus haplotype.
Lewontin's D' measure of linkage disequilibrium [19].
p-values are calculated using an exact test for linkage disequilibrium [20], and are significant at p<0.05.
the number of chromosomes on which the two-locus haplotype was found to occur.
Of the 30 HLA-C alleles present in the Black population, 14 were found to be in significant LD with the -35 SNP. C*02∶02, C*03∶02, C*06∶02, C*07∶06, C*07∶18, C*12∶03 and C*14∶02 were in complete linkage with the -35C allele, while C*03∶04, C*04∶01, C*17∶01 and C*18∶02 were in complete LD with the -35T allele (Table 4). While not complete, C*02∶10 (D’ = 0.59; p<0.05), C*07∶02 (D’ = 0.86; p<0.01) and C*16∶01 (D’ = 0.76; p<0.01) were also found to be in significant LD with the -35T allele. Of the 26 HLA-C alleles present in the Caucasian population, 15 were found to be in significant LD with the -35 SNP. C*02∶02, C*06∶02, C*07∶06, C*07∶18, C*08∶04, C*12∶02, C*12∶03 and C*14∶02 were in complete linkage with the -35C allele, while C*03∶03, C*03∶04, C*04∶01, C*07∶01, C*07∶02 and C*16∶01 were in complete LD with the -35T allele (Table 4). Although not complete, C*01∶02 was also found to be in significant LD with the -35C allele (D' = 0.65, p<0.05).
Linkage Disequilibrium between the -35 SNP and the 263 Indel
Finally, to further investigate the relationship between the -35 SNP and the 263 indel, LD between these polymorphisms was quantified in both the Black and Caucasian population groups. The -35 SNP was only weakly associated with the 263 indel in both the Black (D' = 0.46; p<0.001) and the Caucasian (D' = 0.34; p<0.001) population groups. These polymorphisms have previously been shown to be in relatively strong LD (D’ = 0.75) in other Caucasian population groups [11]. In both population groups, the -35C allele was associated with 263 del and the -35T allele with 263 ins.
Discussion
We examined genetic variation within the 3′ UTRs of the HLA-C alleles present in the Black and Caucasian South African population groups and investigated the relationship between these variants and a SNP 35 kb upstream of the HLA-C locus. The data confirmed the presence of the 263 indel (rs67384697) in both population groups, as well as that of other polymorphisms previously identified within this region in other populations [11], [22], [23]. The persistence of strong LD was observed between particular 3′ UTR polymorphisms in both population groups and the underlying haplotypic structure of the region was described for each HLA-C allele. Finally, these data demonstrated that the -35 SNP (rs9624942) is not in strong LD with the 263 indel in either the Black or the Caucasian South African population; and as such, is not an appropriate marker for this indel in either of these groups. These findings have important implications for our understanding of the regulation of HLA-C expression and the resulting impact on HIV-1 disease progression.
Previous investigations of genetic variability within the HLA-C 3′ UTR in other populations have identified numerous polymorphisms other than the 263 indel in this region [11], [22], [23]. Consistent with their findings, our analysis of the same region identified 33 of these polymorphisms in the Black South African population. The variation identified within the Caucasian South African population represented only a subset of that seen in the Black population group. However, the additional polymorphisms observed in the Black population group were not unique to this population, having been previously described in other studies [11], [22], [23]. A number of these polymorphisms were found to deviate from HWE in both population groups - although different polymorphisms were seen to deviate from HWE between the Black and Caucasian population groups. While deviation from HWE can often be attributed to genotyping errors, this is unlikely to be the case in this instance, as all genotyping was performed by direct sequencing. In light of the finding that a subset of these polymorphisms are unique to specific HLA-C alleles and given the differences observed in the HLA-C allele distributions between the two population groups, these deviations from HWE most likely reflect the influence of random genetic drift.
When we examined the variants present within the 3′ UTR sequences from Caucasian individuals in terms of their association with specific HLA-C alleles, we again observed a number of similarities with previous descriptions of variability within the region [11], [22], [23]. Each HLA-C allele was found to be in complete linkage with a specific 263 allele, although eleven of these associations did not reach statistical significance. Given the high number of HLA-C alleles observed in this population, this lack of statistical significance was most likely a consequence of the low frequencies of these alleles within the population. Similarly, all but one of the HLA-C alleles observed in the Black population were in complete LD with a specific 263 allele. Several of these associations also did not reach statistical significance, again most likely as a result of the low frequencies of these alleles within the population.
As previously observed [11], [22], [23], the majority HLA-C alleles observed within the Caucasian population group were represented by a single, distinctive 3′ UTR sequence. However, contrary to prior observations, several different sequences were observed for the C*02∶02 allele in this population group – none of which corresponded to sequences previously reported for this allele. Nonetheless, despite this variability, none of the 3′ UTR sequences observed for C*02∶02 differed with respect to their 263 indel allele. A similar pattern was observed for the 3′ UTR sequences of the HLA-C alleles present in the Black population group – with C*02∶02 again showing a high degree of variability within this population group, but without differing with respect to the 263 indel allele observed. However, one allele present only in the Black population group (HLA C*02∶10), was found to be associated with both the 263ins and 263del alleles. Coupled with the finding that the 3′ UTR sequence observed for C*02∶05 differed between the two population groups, these data may suggest that a diversity of genetic variability exists within the Cw 02 alleles that cannot be adequately described using the current commonly used HLA-C SBT genotyping methods.
LD analysis between all the polymorphisms present in the HLA-C 3′ UTR revealed the 263 indel to be at the centre of a large haplotype involving the same ten polymorphic positions in both the Black and Caucasian populations. This indel is thought to regulate differential HLA-C expression by disrupting a putative miRNA binding site [11]. It has previously been shown to be in complete linkage with the SNPs at positions 256, 261 and 266; and as none of the SNPs produced any significant change in luciferase activity when analysed independently, the indel is thought to be responsible for any differences seen in HLA-C expression [11]. However, all analyses of alterations in luciferase activity involving the indel included concomitant modifications at all four positions, and all four polymorphisms occur within the putative miR-148a/miR-148b binding site [11], allowing for the possibility that concomitant changes at these positions may act synergistically to disrupt miRNA binding.
Similarly, the SNP at position 307 was also previously shown to be potentially disruptive of a putative miRNA binding site, but when analysed independently did not produce any significant changes in luciferase activity [11]. However, if analysed in haplotypic combination with the SNPs at positions 299, 300 and 303, which occur within the same putative mi-657 binding site [11]; a significant difference in expression may potentially be observed, as the presence of multiple mismatched bases within the same binding site would most likely increase the probability of disruption of miRNA binding relative to modification at a single position. Additionally, the observation that these variants always occur in combination with each other, and could thus collectively potentially prevent binding of more than one miRNA, would further suggest that the presence of the haplotype (rather than any one single polymorphism) is responsible for the differential expression of HLA-C alleles observed. However, further functional studies would be required to test this hypothesis.
A second large haplotype encompassing the indel at position 230 and four other SNPs was also observed in both populations. The presence of an insertion at position 230, coupled with a guanine at 224, introduces at additional putative miR-181a binding site into the HLA-C 3′ UTR [11]. As a result, when both indels are present in a population, the two haplotypes overlap to produce three separate “classes” of HLA-C 3′ UTR sequences that are all potentially subject to varying degrees of miRNA-mediated gene regulation. HLA-C 3′ UTR sequences with deletions at both positions 230 and 263 would therefore potentially disrupt the binding of four miRNAs (miR-181a, miR-148a/miR-148b, miR-657 and miR-181a* [11]), while those sequences with insertions would be inhibited at the same positions. Sequences with a deletion at position 230 and insertion at 263 would disrupt two putative miRNA binding sites (miR-148a/miR-148b and miR-657 [11]), and thus potentially exhibit a phenotype of intermediate expression. These three separate lineages could thus account for the variances previously observed in HLA-C expression [3], [11], [24]. Again, however, further functional studies are necessary to test this hypothesis.
The suggestion that the overlap of the two haplotypes described could potentially account for observed variances in HLA-C expression is consistent with the previous findings of Corrah et al. [24], who prior to the identification of the 263 indel, used monoclonal antibodies to examine HLA-C expression in relation to -35 SNP genotype [24]. They attributed the association between the -35T allele and higher HIV-1 viral set point observed in Caucasian individuals[1], [3]–[7] to the especially low expression of HLA-Cw*07 alleles, which are particularly common in individuals of European descent [13]. This association is not seen in African-American populations[7]–[9], where HLA-Cw*07 is less prevalent. All the HLA-Cw*07 sequences analysed during the course of our investigation had insertions at both indel positions, and would thus (under the aforementioned model) be subject to especially strong miRNA-mediated inhibition. This could also partially account for variances in the distribution of HLA-C expression levels observed in other functional studies [10], [11].
This association between the -35 SNP and HLA-C expression [3], was previously thought to be responsible for the differences in HIV-1 viral set point observed in Caucasian individuals. However, the 263 indel is now considered to be the actual variant responsible for the differential regulation of HLA-C expression, which, in turn, is thought to be responsible for the variation seen in HIV-1 viral set point [11]. The association observed between the -35 SNP and in HIV-1 viral set point has since been attributed to strong LD between the SNP and indel [11].
Examination of the extent of LD between the -35 SNP and 263 indel in both the Black and Caucasian populations revealed LD between them (and all the other SNPs in the haplotype) to be weak in both groups. However when linkage between the -35 SNP and specific HLA-C alleles was examined, LD was found to be complete and significant for fourteen alleles in the Caucasian population. This was consistent with previous reports in other Caucasian populations [3], [11]. LD was not found to be significant for more than 50% of the HLA-C alleles in the Black population group. This is consistent with observations in African-American populations, where common HLA-C alleles show no significant LD with the -35 SNP; and for alleles that do show significant LD, linkage is not complete [10].
Thus while the -35 SNP may be indicative of specific HLA-C alleles in the Caucasian population, it cannot be regarded as an effective marker for the 263 indel in either the Black or Caucasian South African population groups. The -35 SNP was also not in strong LD with the 230 indel in either population group (data not shown). Thus, as has been shown in the African-American population [10], this SNP is unlikely to associate with either HLA-C expression or HIV-1 viral set point in these groups. However, this is yet to be confirmed. Whether the -35 SNP associates with any other as yet unidentified polymorphisms that influence HIV-1 disease progression by alternative mechanisms may warrant further investigation.
In conclusion, these data provide the first description of variation in the regulatory regions of HLA-C for the Black and Caucasian South African populations. Furthermore, the data from the Black population are the first for a sub-Saharan African population. These data allow for a description of the haplotypic patterns within the HLA-C 3′ UTR and identify two overlapping haplotypes within this region - which we hypothesize may act independently and synergistically to influence miRNA regulation of HLA-C expression. Concomitantly, we demonstrate that the -35 SNP is not in strong LD with either haplotype (in either population) and as such is unlikely to be an appropriate marker for HLA-C expression in either of these populations. Even in the absence of supporting expression data, these findings provide important insights into genetic variability within the regulatory regions of HLA-C, that have potential implications for our understanding of the regulation of HLA-C expression and its impact on HIV-1 disease progression in the populations occupying the region worst affected by this epidemic.
Supporting Information
The HLA-C alleles present in both the Black and Caucasian population groups and their frequencies. The observed frequency of each allele was calculated according to the formula n/2n, where n refers to the number of times each allele was observed within the given population group and 2n refers to the total number of chromosomes considered within each population group. The allelic frequencies are representative of 168 Black individuals and 97 Caucasian individuals.
(PDF)
The polymorphic positions within the HLA-C 3′ UTR and their minor allele frequencies in the Black and Caucasian population groups. All positions are given relative to the start of the HLA-C 3′ UTR. The allelic frequencies are representative of 168 Black individuals and 96 Caucasian individuals. The p-values given are for a two-sided Fisher’s exact test and only significant values (p<0.05) shown.
(PDF)
Funding Statement
This work was funded in part by the National Research Foundation and University of the Witwatersrand’s Medical Faculty Research Endowment Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, et al. (2007) A Whole-Genome Association Study of Major Determinants for Host Control of HIV-1. Science 317: 944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, et al. (2005) Genome-Wide Associations of Gene Expression Variation in Humans. PLoS Genet 1: e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomas R, Apps R, Qi Y, Gao X, Male V, et al. (2009) HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nature Genetics 41: 1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, et al. (2009) Common Genetic Variation and the Control of HIV-1 in Humans. PLoS Genet 5: e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dalmasso C, Carpentier W, Meyer L, Rouzioux C, Goujard C, et al. (2008) Distinct Genetic Loci Control Plasma HIV-RNA and Cellular HIV-DNA Levels in HIV-1 Infection: The ANRS Genome Wide Association 01 Study. PLoS ONE 3: e3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Manen D, Kootstra NA, Boeser-Nunnink B, Handulle MAM, van't Wout AB, et al. (2009) Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. AIDS 23: 19–28. [DOI] [PubMed] [Google Scholar]
- 7. The International HIVCS (2010) The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science 330: 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han Y, Lai J, Barditch-Crovo P, Gallant JE, Williams TM, et al. (2008) The role of protective HCP5 and HLA-C associated polymorphisms in the control of HIV-1 replication in a subset of elite suppressors. AIDS 22: 541–544. [DOI] [PubMed] [Google Scholar]
- 9. Shrestha S, Aissani B, Song W, Wilson CM, Kaslow RA, et al. (2009) Host genetics and HIV-1 viral load set-point in African-Americans. AIDS 23: 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Apps R, Qi Y, Carlson JM, Chen H, Gao X, et al. (2013) Influence of HLA-C Expression Level on HIV Control. Science 340: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, et al. (2011) Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472: 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paximadis M, Mathebula TY, Gentle NL, Vardas E, Colvin M, et al. (2012) Human leukocyte antigen class I (A, B, C) and II (DRB1) diversity in the black and Caucasian South African population. Human Immunology 73: 80–92. [DOI] [PubMed] [Google Scholar]
- 13. Middleton D, Menchaca L, Rood H, Komerofsky R (2003) New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens. 61: 403–407. [DOI] [PubMed] [Google Scholar]
- 14. Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, et al. (2011) The IMGT/HLA database. Nucleic Acids Research 39: D1171–D1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ceppellini R, Siniscalco M, Smith CAB (1955) The estimation of gene frequencies in a random-mating population. Annals of Human Genetics 20: 97–115. [DOI] [PubMed] [Google Scholar]
- 16. Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48: 361–372. [PubMed] [Google Scholar]
- 17. Excoffier L, Laval G, Balding D (2003) Gametic phase estimation over large genomic regions using an adaptive window approach. Human genomics 1: 7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. TAG Theoretical and Applied Genetics 38: 226–231. [DOI] [PubMed] [Google Scholar]
- 19. Lewontin RC (1964) The interaction of selection and linkage. I. general considerations; heterotic models. Genetics 49: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Slatkin M (1994) Linkage Disequilibrium in Growing and Stable Populations. Genetics 137: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 22. Xu Y, Deng Z, O'hUigin C, Wang D, Gao S, et al. (2011) Characterization and polymorphic analysis of 4.5 kb genomic full-length HLA-C in the Chinese Han population. Tissue Antigens 78: 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'hUigin C, Kulkarni S, Xu Y, Deng Z, Kidd J, et al. (2011) The molecular origin and consequences of escape from miRNA regulation by HLA-C alleles. The American Journal of Human Genetics 89: 424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corrah TW, Goonetilleke N, Kopycinski J, Deeks SG, Cohen MS, et al. (2011) Reappraisal of the relationship between the HIV-1-protective single-nucleotide polymorphism 35 kilobases upstream of the HLA-C gene and surface HLA-C expression. Journal of Virology 85: 3367–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The HLA-C alleles present in both the Black and Caucasian population groups and their frequencies. The observed frequency of each allele was calculated according to the formula n/2n, where n refers to the number of times each allele was observed within the given population group and 2n refers to the total number of chromosomes considered within each population group. The allelic frequencies are representative of 168 Black individuals and 97 Caucasian individuals.
(PDF)
The polymorphic positions within the HLA-C 3′ UTR and their minor allele frequencies in the Black and Caucasian population groups. All positions are given relative to the start of the HLA-C 3′ UTR. The allelic frequencies are representative of 168 Black individuals and 96 Caucasian individuals. The p-values given are for a two-sided Fisher’s exact test and only significant values (p<0.05) shown.
(PDF)


