SUMMARY
Tank-binding kinase 1 (TBK1) is a serine/threonine protein kinase mediating innate antimicrobial immunity. TBK1 is involved in the signaling of TLRs, RLRs, and STING-mediated sensing of cytosolic DNA. Stimulation of these receptors results in the activation of TBK1, which phosphorylates interferon regulatory factor IRF-3. Phosphorylated IRF-3 translocates into the nucleus to initiate the transcription of the IFN-β gene. Here we show that TBK1 is activated by autophosphorylation at residue Ser172. Structures of TBK1 bound to two inhibitors showed that TBK1 has the IκB kinase fold with three distinct domains: the kinase domain, the ubiquitin like domain, and the scaffold and dimerization domain. However, the overall structures of TBK1 monomer and its dimer are different from IKKβ in the arrangements of the three domains and in dimer formation. Phosphorylation of IRF-3 by TBK1 in vitro results in its oligomerization, and phosphorylation of residue Ser386 plays a key role in IRF-3 activation.
INTRODUCTION
Viral or bacterial infection stimulates host innate immune responses through a wide spectrum of pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs) such as lipids, glycans, and nucleic acids (Barber, 2011; Keating et al., 2011; Takeuchi and Akira, 2010). For example, Toll-like receptor 3 (TLR3) recognizes viral dsRNA at the cell surface or in the endosome and stimulates the expression of interferons via the adaptor protein TRIF (Oshiumi et al., 2003; Yamamoto et al., 2003). The RIG-I like receptors recognize viral dsRNA in the cytosol and signal through the adaptor protein MAVS/IPS-1 (Kato et al., 2011; Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005). Microbial DNA in the cytosol activates the enzyme cGAS, which catalyzes the synthesis of cyclic-GAMP, a secondary messenger that regulates innate immune responses via the adaptor protein STING (Burdette and Vance, 2013; Sun et al., 2013; Wu et al., 2013). Stimulation of these receptors leads to the expression of several kinds of cytokines, including type I interferons, such as IFN-α and β (Takeuchi and Akira, 2009). Two protein kinases, TBK1 and IKKε, play critical roles in the signaling pathway of these immunoreceptors (Fitzgerald et al., 2003; Sharma et al., 2003). Both TBK1 and IKKε belong to the inhibitor κB (IκB) kinase (IKK) family and share about 30% sequence identity to the canonical IκB kinases IKKα and IKKβ (Hacker and Karin, 2006). Engagement of PAMPs by the PRRs leads to the recruitment and activation of TBK1 or IKKε in a variety of signaling complexes (Hacker and Karin, 2006). Activated TBK1 and IKKε in turn phosphorylate transcription factors IRF-3 and IRF-7 in the C-terminal regulatory domain (RD), resulting in their oligomerization (Fitzgerald et al., 2003; Hemmi et al., 2004; Sharma et al., 2003). Phosphorylated IRF-3 or IRF-7 then translocate to the nucleus, bind to CBP/P300, and initiate the transcription of type I interferon genes (Lin et al., 1998; Yoneyama et al., 1998). The adaptor protein TANK, which associated with TBK1, is needed for the activation of IRF-3 and NF-κB (Cheng and Baltimore, 1996; Goncalves et al., 2011).
The structural studies of the IKK family protein kinases have provided important insights into the structure and functions of these kinases. The 3.6 Å resolution structure of the first IKK family kinase Xenopus IKKβ was determined recently, revealing a novel tripartite structure of the IKK kinase (Xu et al., 2011). IKKβ contains an N-terminal kinase domain (KD), followed by an ubiquitin-like domain (ULD) and a scaffold and dimerization domain (SDD) (Xu et al., 2011). IKKβ forms an intimate dimer through interactions between the SDDs. The dimerization of IKKβ is not necessary for its kinase activity but is required for IKKβ activation (Xu et al., 2011). Crystal structures of the human TBK1 KD plus ULD in an inhibited conformation and the phosphorylated human TBK1 KD bound to the inhibitor BX795 were determined recently, providing insight into the mechanism of TBK1 activation and regulation (Helgason et al., 2013; Ma et al., 2012). While we were preparing this manuscript for submission, two reports describing the structures of human TBK1 including the SDD were published online (Larabi et al., 2013; Tu et al., 2013).
To elucidate the structure and function of TBK1 in innate immunity, we have expressed both full-length and truncated forms of mouse TBK1 (mTBK1) in insect cells and determined the structures of mTBK1 bound to two inhibitors, BX795 and SU6668 (Clark et al., 2009; Godl et al., 2005). TBK1 kinase assays showed that it is activated by autophosporylation at residue Ser172. In vitro phosphorylation assays of IRF-3 revealed that TBK1 phosphorylates a distinct set of residues in IRF-3 and results in the formation of various kinds of IRF-3 oligomers in solution. These results provided insight into in the mechanism of TBK1 activation and its roles in IRF-3 activation in innate immune responses.
RESULTS
TBK1 is activated by autophosphorylation
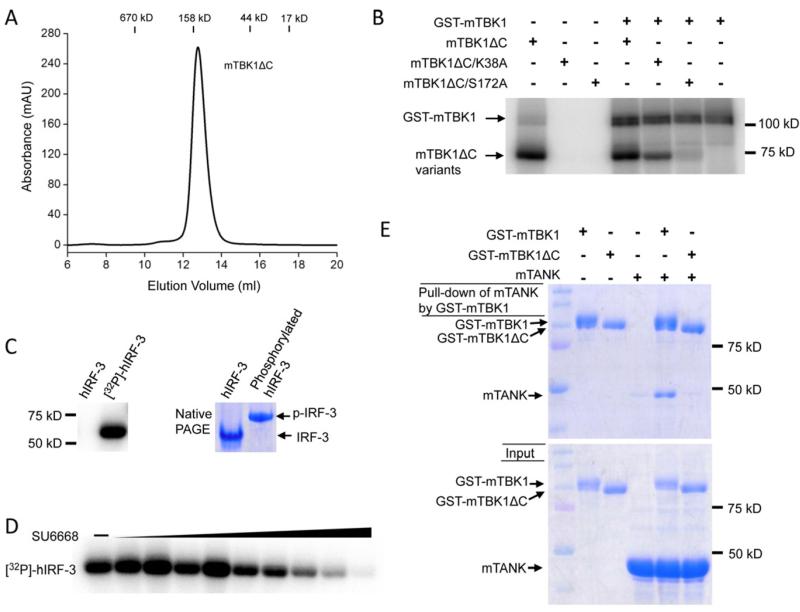
We have expressed GST-fusions of full-length mouse mTBK1, a C-terminal truncation form of mTBK1 (residues 1-657, mTBK1ΔC), and two kinase inactive mutants of mTBK1 (K38A and S172A in mTBK1ΔC constructs) in insect cells. Gel filtration chromatography showed that all the TBK1 proteins we purified formed dimers in solution after the cleavage of the GST tag (Figure 1A). To confirm that Ser172 is the sole phosphorylation site in autophosphorylated TBK1, we conducted in vitro phosphorylation assays of the kinase-inactive mutants S172A and K38A (without the GST tag) using GST-mTBK1. We found that the S172A mutant cannot autophosphorylate or be phosphorylated by GST-mTBK1 (Figure 1B). The mutation K38A abolished autophosphorylation of TBK1, but this mutant can still be phosphorylated by the wild type GST-mTBK1 (Figure 1B). These results demonstrated that Ser172 is the sole autophosphorylation site of mTBK1. Next we analyzed the kinase activity of TBK1 using full-length human IRF-3 as a substrate. We found that TBK1 phosphorylates IRF-3 and induces the oligomerization of IRF-3 (Figure 1C). In addition, we found that mTBK1 can no longer phosphorylated IRF-3 in the presence of 1-2μM of the inhibitor SU6668 (also known as TSU-68), an anticancer drug under phase III clinical trials for hepatocellular carcinoma (Kanai et al., 2011) (Figure 1D). Furthermore, GST pull-down assays of full-length mTBK1 with TANK showed that the two proteins interact with each other directly. Truncation of the TANK-binding domain (TBD, residues 658~729) abolished the interaction between TBK1 and TANK (Figure 1E). Consistent with this result, truncation of human TBK1 in the TBD abolished the binding of another TBK1 binding protein NAP1 (Larabi et al., 2013). Interestingly, this truncation does not affect TBK1 kinase activity or the phosphorylation of IRF-3 by TBK1.
Figure 1.
TBK1 is activated by autophosphorylation. (A). Gel filtration chromatography analysis of purified mTBK1. (B). Phosphorylation assays of wild type mTBK1 and its mutants. (C). TBK1 phosphorylates human IRF-3. The assay on the left used γ-32P-ATP in the reaction. The band shows phosphorylated hIRF-3. The assay on the right is a native PAGE analysis of unphosphoryalted and phosphorylated hIRF-3 (D). The inhibition of mTBK1 kinase activity by SU6668. The concentration of SU6668 used in the assay is from 8 nM to 2 μM. The bands show phosphorylated hIRF-3. (E). GST pull-down assays of full-length and C-terminal deletion mutant of mTBK1 with TANK.
Overall structure of mouse TBK1
To understand the structure and function of TBK1, we have determined the crystal structures of mTBK1ΔC (referred to as mTBK1 here after) bound to two inhibitors, SU6668 and BX795, at resolutions of 3.0 and 3.1 Å, respectively (Table 1, see also Figure S1). The refined models contain 644 residues of mTBK1. Residues 168 to 174 and 490 to 493 are not well defined in the electron density maps. Both crystals belong to the space group P3221 with similar unit cells. The crystallographic asymmetric unit contains one mTBK1 molecule. The two mTBK1 molecules in the two complexes are similar, with an r.m.s.d of 0.47 Å for all the Cα atoms. Two mTBK1 molecules in the crystals form an intimate dimer related by a crystallographic two-fold axis. The mTBK1 complex with SU6668 will be used to describe structures (see below), due to its slightly higher resolution and better-defined electron density map (see also Figure S1).
Table 1.
Data collection and refinement statistics for TBK1
| mTBK1/BX795 | mTBK1/SU6668 | |
|---|---|---|
| Data collection | ||
| Space group | P3221 | P3221 |
| Cell dimensions | ||
| a, b, c (Å) | 140.78, 140.78, 86.57 | 140.48, 140.48, 86.56 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 3.10 (3.15 to 3.10)* | 3.0 (3.18 to 3.0) |
| Rsym or Rmerge | 6.4% (>1.0) | 10.6% (>1.0) |
| I/σI | 40.0 (1.8) | 12.1 (1.7) |
| Completeness (%) | 99.9 (100.0) | 100.0 (100.0) |
| Redundancy | 7.2 (7.5) | 10.4 (10.3) |
| Refinement | ||
| Resolution (Å) | 3.1 | 3.0 |
| No. reflections | 18240 | 20015 |
| Rwork / Rfree | 22.7% (26.9%) | 23.4% (26.7%) |
| No. atoms | ||
| Protein | 5230 | 5230 |
| Ligand/ion | 34 | 23 |
| Water | 0 | 0 |
| B-factors | ||
| Protein | 108.2 | 110.0 |
| Ligand/ion | 80.3 | 72.1 |
| Water | ||
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.004 |
| Bond angles (°) | 0.683 | 0.826 |
One crystal was used to collect each of the dataset. Values in parentheses are for highest-resolution shell.
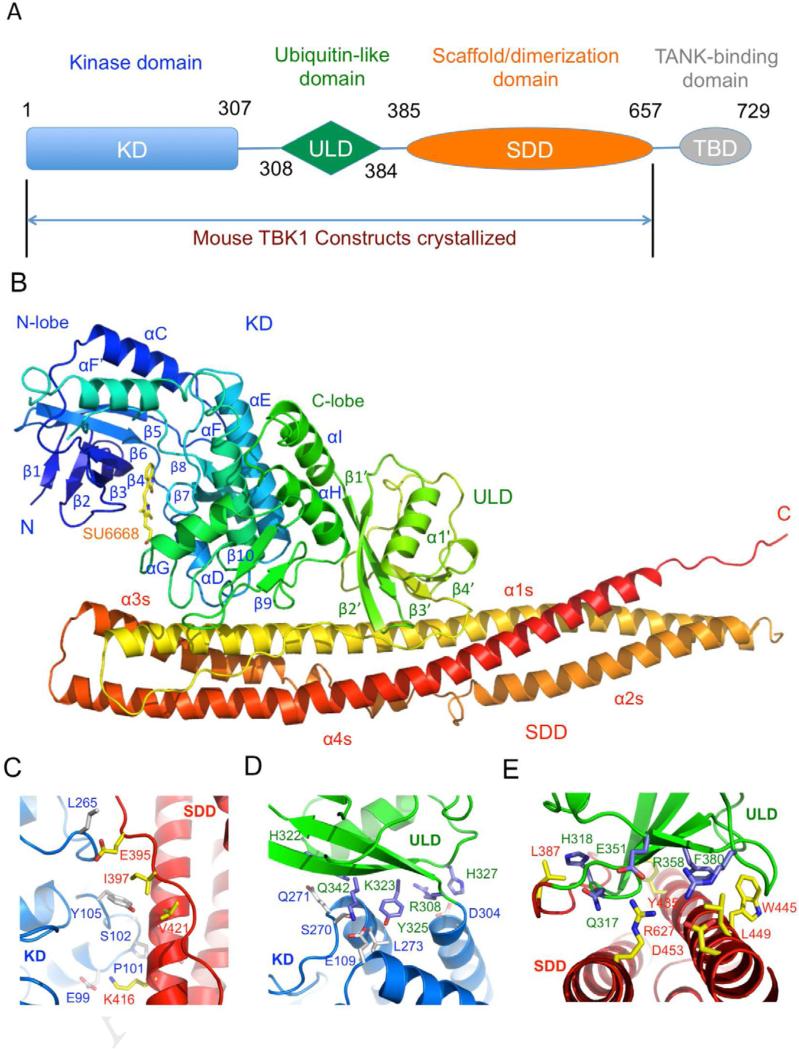
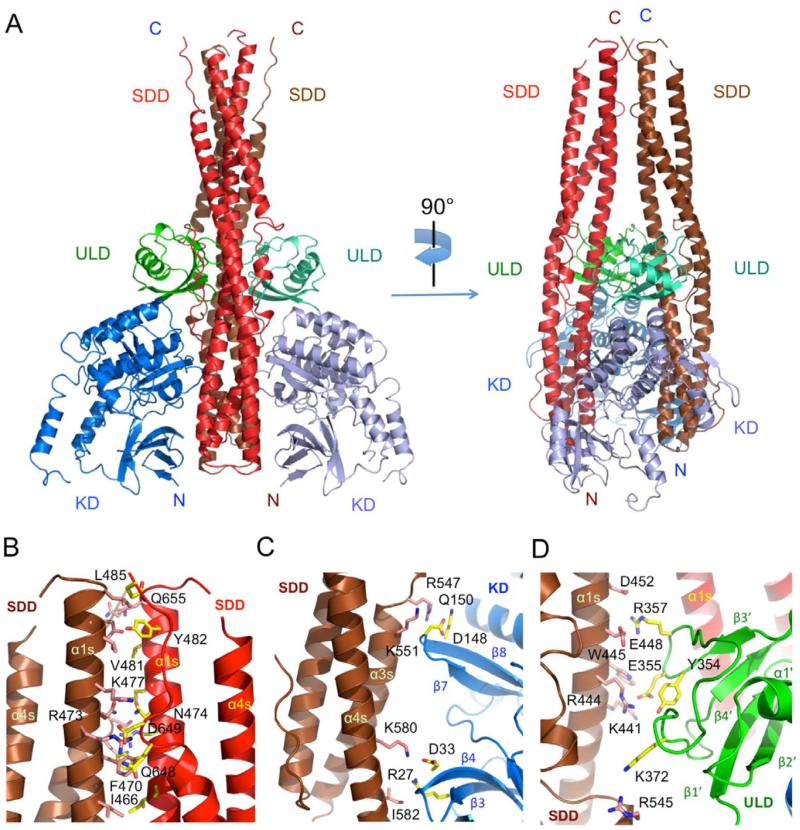
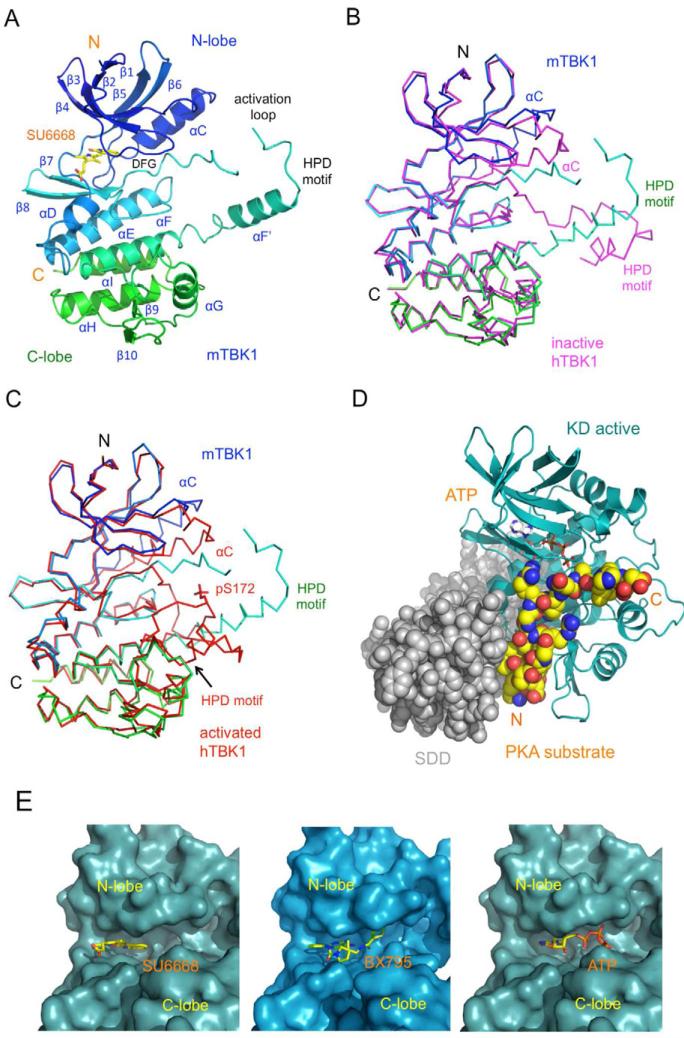
The overall structure of mTBK1 (Figure 2) is similar to the recently described inactive and phosphorylated human TBK1 structures (r.m.s.d of 1.0 Å compared to 4IM0 and 1.3 Å compared to 4IW0) (Larabi et al., 2013; Tu et al., 2013), with some differences in the activation loop and the C-terminus tail (see also Figure S2). Mouse TBK1 has three structurally distinct domains, the kinase domain (KD), the ubiquitin-like domain (ULD), and the scaffold and dimerization domain (SDD, Figure 2A and B). The KD of mTBK1 contains residues 1 to 307 and exhibits a typical bi-lobal protein kinase fold (Figure 2B). The KD is followed by the ULD (residues 308 to 384). The structure of the ULD in our TBK1 construct is similar to the isolated human TBK1 ULD structure (r.m.s.d 0.6 Å) determined previously (Li et al., 2012). The overall fold of the ULD is similar to human ubiquitin with slightly different conformations in the exposed loop connecting strands β3’ and β4’. The SDD of mTBK1 contains residues 385 through 656 (Figure 2A, B). The ULD is connected to the SDD via a 22-residue linker. Both of the KD and the ULD have extensive contacts with the SDD within the mTBK1 monomer (Figure 2B). The mTBK1 dimer resembles a pair of scissors with the KDs and the ULDs forming the handles, and the two SDDs forming the blades (Figure 3A). The overall structure of the mTBK1 dimer is triangular, with dimensions of approximately 100 Å × 120 Å × 55 Å. Unlike IKKβ, however, the KD and ULD of mTBK1 interact with the SDD of the other mTBK1 molecule in the dimer (Figure 3). The SDDs interact with each other through the C-terminal tip of the helical bundle (Figure 3B), promoting the formation of the TBK1 dimer.
Figure 2.
Crystal structure of mouse TBK1. (A). Domain organization of mTBK1, showing the boundaries of the kinase domain (KD), ubiquitin-like domain (ULD), the scaffold and dimerization domain (SDD), and the TANK-binding domain (TBD). The construct used for crystallization is truncated at the C-terminus of the SDD at residue Thr657. (B). Ribbon representation of the structure of the mTBK1 monomer colored rainbow from blue at the N-terminus to red at the C-terminus. The inhibitor SU6668 is shown by the yellow stick model. (C). Close up of the interface between the KD (blue) and the SDD (red). Residues mediating the interactions between the two domains are shown by the stick models. (D). Interactions between the KD (blue) and the ULD (green). (E). Close up of the interface between the ULD (green) and the SDD (red).
Figure 3.
Structure of the mTBK1 dimer. (A). Ribbon representations of two views of the mTBK1 dimer. The KDs are colored blue and light blue, the ULDs green and cyan, and the SDDs red and brown. (B). Interactions between the SDDs in the mTBK1 dimer. Residues at the SDD interface are shown by the pink and the yellow stick models. For clarity, only one set of residues are labeled in the symmetrical dimer. (C). Interactions between the KD (blue) and the SDD (brown) in the mTBK1 dimer. (D). Interactions between the ULD (green) and SDD (brown) in the mTBK1 dimer.
Interactions between the three domains of mTBK1
There are extensive interactions between the three domains within an mTBK1 molecule (Figure 2C to E). The KD interacts with the C-terminal region of the SDD through its C-lobe, burying a total surface area of ~830 Å2. The two loops connecting the helices αD and αE, helix αH and strand β10 interact with the helix α1s of the SDD and the linker between the ULD and the SDD (Figure 2C). Hydrophobic interactions between residues Ala104, Tyr105, and Pro101 in the KD, and residues Ile397, Ala 417, Val421 and Tyr424 in the SDD, make major contributions to these interactions (Figure 2C). Electrostatic interaction between residues Glu99 and Lys416 further contributes to the interactions between the KD and the SDD (Figure 2C). The ULD contacts both the KD and SDD, burying ~810 Å2 and ~2200 Å2 surface areas at the two interfaces, respectively. The interaction between the KD and ULD involves two pairs of charged residues, His327 and Asp304, Glu109 and Lys323 (Figure 2D). In addition, hydrophobic interaction between residues Tyr325, Leu113, and Leu273, also contributes to these interactions (Figure 2D). Moreover, the sidechain of Tyr325 forms a hydrogen bond with the sidechain of Glu109. The KD/ULD interface is conserved in both mouse and human TBK1 structures with or without the SDD, indicating the rigid structure of the linker between these two domains. The ULD binds at the middle of the SDD helical bundle (Figure 2B). The interface between the ULD and the SDD is extensive and hydrophobic. A large number of hydrophobic residues, such as Phe314, Leu316, Met319, Ala321, Ile353, and Phe380 from the ULD, residues Leu438, Met439, Leu387, Met439, Tyr435, Trp445, Leu449, Phe612, and Met620 from SDD, are involved in these interactions (Figure 2E). Electrostatic interactions between Glu351 and Arg627, and Arg358 and Asp453, make additional contributions to the ULD/SDD interaction (Figure 2E).
Structure of the mTBK1 scaffold and dimerization domain
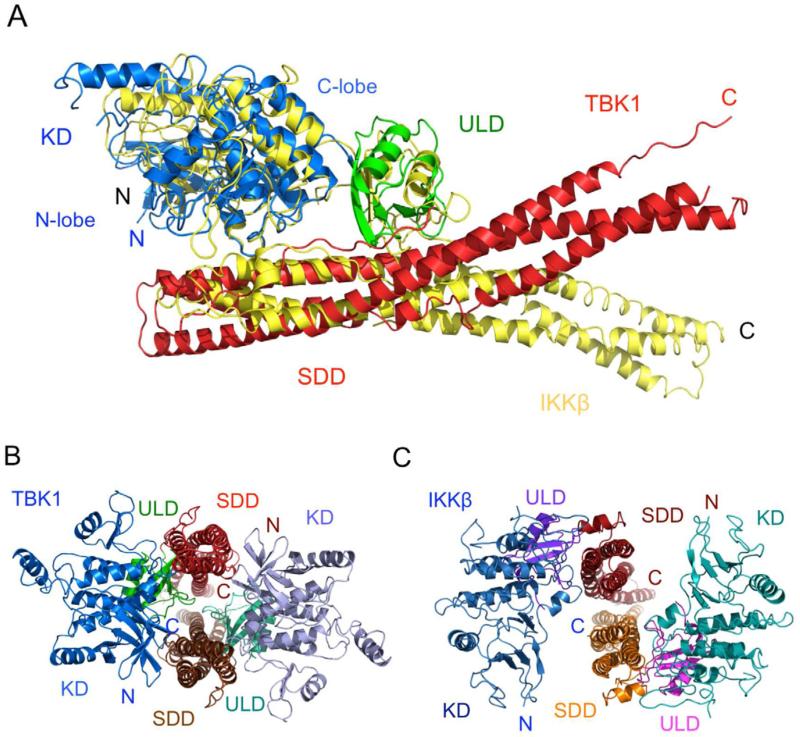
The SDD of mTBK1 forms a long twisted helical bundle of ~110 Å in length (Figure 2B). The two long continuous helices α1s and α4s contain 77 and 70 residues, respectively. The C-terminal 30 residues of helix α1s make major contributions to the TBK1 dimer formation (Figure 3A, B). The third helix is broken into two segments of 24 (α2s) and 31 (α3s) residues connected by a long loop of 29 residues (Figure 2B). Although the overall structure of mTBK1 is similar to that of IKKβ, the organization of the three domains in mTBK1 is dramatically different from that of IKKβ (Figure 4). When the structurally conserved kinase domains of mTBK1 and IKKβ are superimposed, the positioning of the ULDs and SDDs are obviously different in the two structures (Figure 4A). As reflected by the low sequence identity between the SDDs, the structures of the helical bundle in mTBK1 and IKKβ are quite different (r.m.s.d of 4.5 Å for 151 Cα atoms). Superposition of the two SDDs showed that only the third helices of the two structures match each other (see also Figure S3). The other helices, especially the first helices in the SDDs, are positioned differently in the two structures. These structural differences may contribute to the different functions of the two kinases in innate immunity, such as substrate specificity.
Figure 4.
Mouse TBK1 exhibits distinct domain organization compared to the canonical IκB kinase. (A). Superposition of mTBK1 (blue, green, and red) and Xenopus IKKβ (yellow) structures via the kinase domains. (B). Structure of the mTBK1 dimer viewed along the SDD helical bundles (red and brown). The C-termini of the SDDs point away from the reader. (C). Structure of Xenopus IKKβ in the same orientation as mTBK1, showing the different arrangement of its KD and ULD relative to the SDD.
Molecular interactions in the TBK1 dimer
Both human TBK1 KD and KD-ULD are monomers in solution (Ma et al., 2012). In contrast, mTBK1 with the SDD forms an intimate dimer in solution and in the crystal, suggesting the SDD-mediated dimer is likely functionally relevant. Small angle X-ray scattering (SAXS) and electron microscopy (EM) analysis of similar human TBK1 constructs confirmed this hypothesis (Larabi et al., 2013; Tu et al., 2013). All the three domains of mTBK1 are involved in the dimer formation (Figure 3). The total buried surface area at the mTBK1 dimer interface is ~4300 Å2, with the SDDs contributing ~1800 Å2, the ULDs contributing ~1200 Å2, and the KDs contributing ~1500 Å2. Electrostatic and hydrophobic interactions between the SDDs make major contributions to the dimer interface (Figure 3B). Four pairs of charged residues between Glu456 and His459, Asp649 and Lys479, are observed at the interface between the SDDs (Figure 3B). In addition, five pairs of hydrophobic residues, including Leu458, Val481, Phe470, Ile466 and Tyr482 from both of the SDDs, are buried at the SDD interface (Figure 3B). The kinase domain interacts with the neighboring SDD through two loops connecting strands β3/β4 and strands β7/β8 (Figure 3C). Six pairs of charged residues, including Asp148 and Arg547, Glu147 and Lys596, Asp33 and Lys589, are observed at the KD/SDD interface across the dimer (Figure 3C). In addition, three hydrophobic residues, Val554, Phe585, and Ile582, mediate van der Waals contacts between the KD and the SDD (Figure 3C). The extensive interactions between the KD and the SDD, both within the mTBK1 monomer and across the dimer interface, likely contribute to the overall structural rigidity of mTBK1. In addition, four pairs of charged residues, Glu355 and Arg444, Arg357 and Asp452, mediate the interactions between the ULDs and the SDDs in the mTBK1 dimer (Figure 3D). The bulky sidechains of Tyr354 and Trp455 buried at the dimer interface also contribute to the ULD/SDD interactions (Figure 3D). Mutations of residues at the dimer interface that disrupted the dimerization of TBK1 reduce the kinase activity and the autophosphorylation of human TBK1 (Larabi et al., 2013; Tu et al., 2013). In contrast to mTBK1, the dimer interaction between IKKβ is mediated exclusively by the SDDs (buried surface of ~3200 Å2), with extensive contacts at both ends of the SDDs (Figure 4C). Hydrophobic interactions near the C-terminal of the SDDs make major contributions to the IKKβ dimer formation. In addition, electrostatic interactions near the N-terminal of the SDDs make minor contributions to the dimer interface.
Structure of the mTBK1 kinase domain
The structure of the mTBK1 KD (Figure 5A) is similar to that of the previously determined human TBK1 KD in the inactive conformation (r.m.s.d 1.46 Å for 257 Cα atoms, Figure 5B). The overall structure of mTBK1 KD is also very similar to the phosphorylated human TBK1 KD (r.m.s.d 1.28 Å for 259 Cα atoms, Figure 5C) (Ma et al., 2012). The major structural differences between mTBK1 KD and the inactive or the active human TBK1 KD are obvious only at the activation loops and the αC helices (Figure 5B, C). It seems that the kinase domains of TBK1 excluding the activation loop and the αC helix are quite rigid even in the absence of the SDD. The backbone structures of the kinase domain are not affected by interactions with the ULD or the SDD as well as phosphorylation at Ser172 (Figure 5B, C). Although the activation loop structure of mTBK1 is quite different from that in the human TBK1 KD-ULD structure (Figure 5B), both of them form a domain swapped dimer in the crystals, interacting with a neighboring kinase domain using the same set of residues (residues 178 to 184 in mTBK1) containing the HPD motif (see also Figure S4). These residues are well defined in the electron density map, but the flanking residues are poorly defined in the mTBK1 structure, reflecting the structural flexibility of the activation loop in the inactive kinases. In contrast, the HPD motif was modeled as part of the KD within a TBK1 monomer in the inactive human TBK1 structures (see also Figure S2) (Larabi et al., 2013; Tu et al., 2013). Phosphorylation of human TBK1 at residue Ser172 results in the refolding the activation loop (Figure 5C). Interactions between p-Ser172 and three positively charged residues, Arg54, Arg134, and Arg163, stabilize the active conformation of the activation loop. Strikingly, residues 178 to 184 around the HPD motif within the active kinase displaced these residues from a neighboring kinase to form the active conformation of TBK1 (Figure 5C), suggesting that the interactions between residues 178 to 184 and KD in the inactive kinase may play an inhibitory role in the domain swapped dimer. The same short polypeptide also plays a key role in maintaining the active conformation of TBK1.
Figure 5.
Structure of the mTBK1 kinase domain. (A). Ribbon representation of the structure of mTBK1 KD bound to the inhibitor SU6668. The KD is colored rainbow from blue at the N-terminus to green at the C-terminus. The inhibitor SU6668 is shown by the yellow stick model. (B). Comparison of the mouse (blue to green) and human (magenta) TBK1 KD structures in the inactive conformations. The human TBK1 KD structure was determined without the SDD. (C). Comparison of the mouse TBK1 KD structure with the phosphorylated human TBK1 KD structure. The human TBK1 KD structure was determined without the ULD and the SDD. Phosphorylated Ser172 in the human TBK1 KD is shown by the red stick model. (D). Molecular model of phosphorylated mTBK1. The model was generated by superimposing of the phosphorylated human TBK1 KD (cyan ribbons) onto the KD of the mTBK1 structure. ATP (gray stick model) and the substrate of PKA (yellow space-filling model) were introduced into the model by superimposing of the PKA structure onto the TBK1 KD structure. The mTBK1 SDD were shown by the gray space-filling model, with the C-terminus of the SDD points away from the reader. (E). The SU6668 and BX795 binding sites of mTBK1. TBK1 is shown by the cyan and light blue surface representations. The inhibitors are shown by the yellow stick models. ATP was introduced to the mTBK1 structure by superimposing of the PKA/ATP complex structure onto the mTBK1 KD.
To understand roles of the SDD in TBK1, we generated a molecular model of active mTBK1 by superpositioning the crystal structure of phosphorylated human TBK1 KD (PDB, 4EUU) onto the structure of mTBK1. The backbone structure of the model is very close (r.m.s.d of 1.0 Å) to that of the 4.0 Å resolution structure of phosphorylated human TBK1, confirming that this model is valid for structural analysis (Larabi et al., 2013). To probe the substrate binding site and the active site of TBK1 in this model, we superimposed the protein kinase A (PKA) substrate complex structure (PDB, 1ATP) onto this model (Zheng et al., 1993). The model indicates that mTBK1 likely bind the substrate at a deep groove between the KD and the SDD (Figure 5D, see also Figure S5). It is likely the N-terminal section of helix α1 and the linker between the ULD and the SDD are involved in substrate recognition by TBK1 (Figure 5D, see also Figure S5). It has been suggested that the corresponding regions of IKKβ are likely involved in substrate recognition as well (Xu et al., 2011).
Furthermore, we observed that the two inhibitors of TBK1, BX795 and SU6668, bind to the ATP binding pocket between the N-lobe and the C-lobe of the kinase domains (Figure 5E). The aromatic rings of the two inhibitors occupy the binding pocket for the adenine base of ATP (Figure 5E). The inhibitors interact with TBK1 mainly through hydrophobic interactions and hydrogen bonds. Based on the structure of SU6668 bound to TBK1, it is possible to improve the specificity and affinity of this family of compounds to TBK1. The methyl group attached to C5 could be replaced by a hydrophilic hydroxyl or amine group that may form additional hydrogen bonds with the nearby carbonyl groups of residues Cys89 and Pro90. A hydroxyl group or amine group could be attached to C15 to form a hydrogen bond with the carbonyl of Glu87. It will also be favorable to replace the methyl group connected to C7 with a hydroxyl group that may form a hydrogen bond with the amine group of Gly16.
The mechanism of IRF-3 activation by TBK1
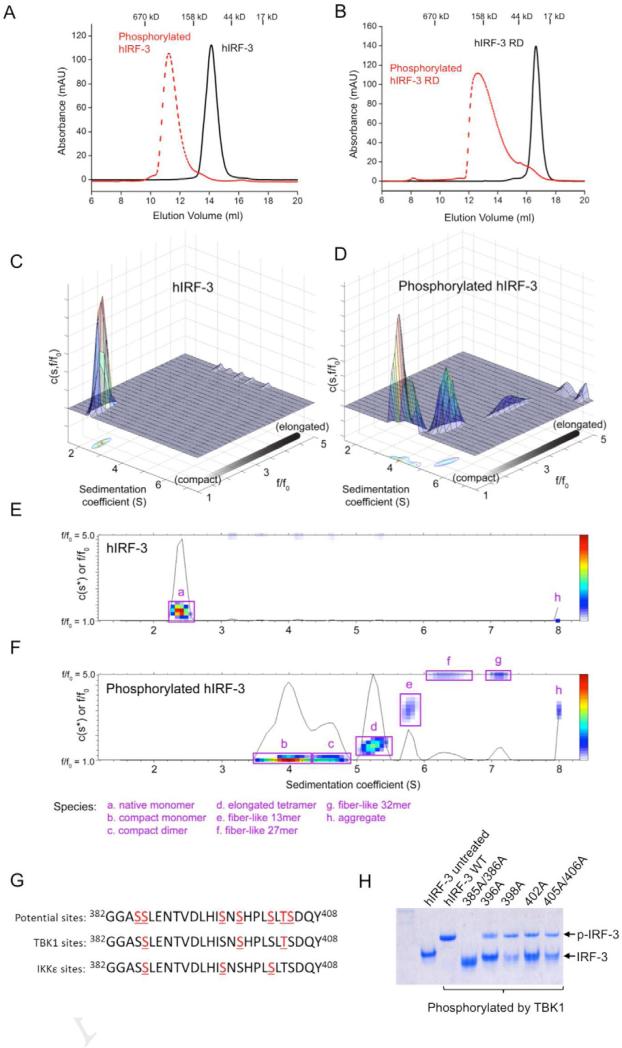
To elucidate the mechanism of IRF-3 activation by TBK1, we conducted in vitro phosphorylation assays of human IRF-3 (hIRF-3) using purified mTBK1. We found that hIRF-3 is potently phosphorylated by mTBK1 without the need of any additional proteins. Gel filtration chromatography analysis showed phosphorylation of IRF-3 induced the oligomerization of the protein (Figure 6A). The estimated mass of the hIRF-3 oligomer is about 300 kD (Figure 6A), which is much larger than an IRF-3 dimer (theoretical mass 95 kD). The phosphorylated hIRF-3 peak is broad and unsymmetrical (Figure 6A), indicating the existence of multiple species in the samples. In addition, phosphorylation of hIRF-3 regulatory domain (RD) by mTBK1 also induced the oligomerization of the protein (Figure 6B), demonstrating that the phosphorylated IRF-3 RD mediates the oligomerization of IRF-3. The estimated molecular mass of the phosphorylated hIRF-3 RD is about 130 kD, which is much larger than a dimer of the RD (theoretical mass 65 kD). This further supports that phosphorylation of IRF-3 by TBK1 induces the oligomerization of the protein.
Figure 6.
Phosphorylation of IRF-3 by TBK1 induces the oligomerization IRF-3. (A). Gel filtration chromatography analysis of phosphorylated full-length hIRF-3. (B). Gel filtration chromatography analysis of phosphorylated hIRF-3 regulatory domain (RD). (C). Analytical ultracentrifugation analysis of unphosphorylated IRF-3, shown as a 3-D shape and size distribution plot. Higher values of f/f0 correspond to elongated species; a f/f0 value of 1.0 corresponds to an ideal sphere. (D). 3-D shape and size distribution plot for phosphorylated IRF-3, revealing a combination of nearly spherical species (f/f0 near 1.0) and a series of elongated oligomeric forms. (E, F). Two-dimensional representation of panels (C) and (D), with the major peaks identified based on frictional ratio and predicted mass. (G). Phosphorylation sites in the RD of hIRF-3 identified by LC-MS/MS. (H). Mutations of hIRF-3 at the TBK1 phosphorylation sites affect phosphorylation-induced oligomerization of IRF-3. Wild type and mutants of hIRF-3 were phosphorylated with mTBK1. The phosphorylated samples were analyzed by native PAGE.
To determine the oligomerization state of the phosphorylated IRF-3, we analyzed the phosphorylated hIRF-3 by analytical ultracentrifugation (AUC, Figure 6C to F). Sedimentation velocity experiments revealed that native hIRF-3 sedimented predominantly as a single monomeric species with a frictional ratio of 1.5, consistent with a mildly elongated protein (Figure 6C and E). Upon phosphorylation, the hIRF-3 monomeric peak shifted to a combination of compact, faster-sedimenting species and a series of higher-order oligomers (Figure 6D and F). The compact species show very low frictional coefficients near 1.0 (which corresponds to an ideal sphere), with estimated masses consistent with monomer and dimer species that are in exchange over the time course of the sedimentation experiment. Thus, phosphorylation induces a conformational collapse of the hIRF-3 monomer into nearly-spherical monomeric and dimeric species. Furthermore, the phosphorylated hIRF-3 formed several higher-order oligomers, including a highly elongated putative tetramer with a frictional ratio above 3, and a series of fiber-like oligomers with estimated masses ranging from a 13-mer up to a 32-mer (Figure 6D and F). To our knowledge, these data are the first reported evidence of significant conformational change in full-length hIRF-3 and the formation of highly elongated oligomers and fiber-like species upon phosphorylation by TBK1. It is obvious from this study that phosphorylation of IRF-3 induced the oligomerization instead of merely dimerization of IRF-3. To investigate which oligomer of IRF-3 might be functional, we expressed the IRF-3 binding domain of CBP, purified the phosphorylated IRF-3 RD bound to CBP, and analyzed the complex by gel filtration chromatography. The IRF-3 RD and CBP complex is homogenous and shows a symmetrical peak with an estimated mass of ~98 kD (see also Figure S6). Based on this result, it is likely the IRF-3 RD dimer or trimer associate with CBP. As no higher order complex between CBP and IRF-3 were observed, it is likely that phosphorylated IRF-3 is in a equilibrium of various oligomers, but only one kind of these oligomers binds CBP.
To identify the TBK1 phosphorylation sites of IRF-3, we separated the phosphorylated hIRF-3 from mTBK1 by SDS-PAGE, and then analyzed the phosphorylated hIRF-3 by mass spectrometry (LC-MS/MS) after trypsin digestion. Over 80% of the IRF-3 sequence was covered in this analysis, which included the complete RD (see also Figure S7). Phosphorylation at residues Thr3, Ser14, Thr75 and Ser97 were observed in the N-terminal DNA binding domain of hIRF-3 (see also Figure S7). Phosphorylation of Thr180, Ser188, Thr237, Thr244, Thr253, Ser386, Ser398, Thr404, and Ser427 were observed in the hIRF-3 RD (see also Figure S7). The MS analysis clearly identified Ser386, Ser398, and Thr404 as the primary TBK1 phosphorylation sites in the RD of hIRF-3 (Figure 6G). To test the roles these phosphorylated residues in IRF-3 activation, we generated five mutants of hIRF-3 based on the phosphorylation sites identified in our MS analysis and previous reports (Suhara et al., 2000; Takahasi et al., 2010; Yoneyama et al., 2002). We have purified all the hIRF-3 mutants to homogeneity and conducted in vitro phosphorylation assays using purified mTBK1. Consistent with results from previous studies, we found that mutations of Ser386 to alanine abolished hIRF-3 dimerization after phosphorylation (Figure 6H). Mutations of other residues, such as Ser396, Ser398, Ser402 individually, or Thr404 together with Ser405, reduced the rate of IRF-3 oligomer formation (Figure 6H). These results showed that the phosphorylation of Ser386 in IRF-3 is necessary for the activation of IRF-3 by TBK1.
DISCUSSION
We have conducted comprehensive biochemical and structural characterizations of mTBK1. Our results suggest that TBK1 is activated by autophosphorylation in trans at residue Ser172. Although the truncation of TBD does not affect TBK1 kinase activity or the phosphorylation of IRF-3, the truncated TBK1 failed to associate with the adaptor protein TANK. In addition, the structures of TBK1 bound to the inhibitors revealed novel structure features of TBK1. Although TBK1 contains three similar domains similar to IKKβ (Xu et al., 2011), the relative orientation of the three domains and the way they interacts with each other are dramatically different in TBK1 and IKKβ. The structure of the SDDs of TBK1 and IKKβ are rather different. Furthermore, the in vitro phosphorylation experiments showed that TBK1 phosphorylates IRF-3 directly and induces the oligomerization of IRF-3. Mass spectrometry analysis of IRF-3 phosphorylated by TBK1 identified a distinct set of residues in both the DNA binding domain and C-terminal RD of TBK1. Interestingly, our mutagenesis analyses show that phosphorylation of residue Ser386 is critical for the activation of IRF-3 by TBK1. These results provided important structural insights into the biological functions of TBK1 in mediating the signaling of innate immune receptors.
The structures of human TBK1 KD+ULD in the inactive conformation and the phosphorylated KD provided insight into the mechanism of TBK1 activation and inhibition (Ma et al., 2012). Our structural study of nearly full-length TBK1 provided additional insight into structure and function of TBK1 (Figure 2B). The structure showed that mTBK1 forms an intimate dimer through extensive interactions between the SDDs, the KDs and ULDs with the SDDs (Figure 3). The extensive interactions between the three domains within the TBK1 monomer and dimer indicate that the TBK1 kinase domain has a relatively rigid structure, thus it is unlikely that two kinase domains within the TBK1 dimer would interact with each other or phosphorylate each other. Consistent with this observation, only a small fraction of TBK1 is phosphorylated when purified from the insect cells. Perhaps, this is a mechanism to prevent the premature activation of the enzyme without the stimulation of innate immune receptors. In contrast, TBK1 constructs without the SDD are extensively phosphorylated even when they were coexpressed with phosphatase (Ma et al., 2012), indicating the SDD is important in regulating the activation of TBK1. As shown by the domain-swapped dimer structure of TBK1 observed in the crystals (see also Figure S4), it is most likely that TBK1 autophosphorylates in trans, which is consistent with the model proposed previously based on the structural studies of human TBK1 without the SDD (Ma et al., 2012). Since TBK1 with truncations in the SDD is not functional in cells (Goncalves et al., 2011), it is obvious that the SDD plays a critical role in the function of TBK1. The C-terminal region of TBK1 is needed to interact with various kinds of adaptor proteins such as TANK and NAP1 (Goncalves et al., 2011). These adaptor proteins are likely mediating the recruitment of TBK1 to distinct signaling complexes (Goncalves et al., 2011). Consistent with these implications, we found that truncation of the TBD of TBK1 disrupted its interaction with TANK. Our structural analysis indicated that the SDD likely has additional functions. For example, TBK1 may bind its substrate at a deep groove between the SDD and KD, so the SDD may also contribute to substrate recognition by TBK1.
Another important question about the function of TBK1 is how TBK1 activates IRF-3 in response to microbial infection. An induced proximity model for TBK1 activation has been proposed previously (Ma et al., 2012). This model suggests that stimulation of immunoreceptors such as TLRs, RLRs, and STING results in the recruitment of TBK1 and IRF-3 to the signaling complex and the induced proximity of TBK1 leads to the activation of the kinase and subsequent phosphorylation of IRF-3 (Ma et al., 2012). Our in vitro IRF-3 phosphorylation experiments support this model. We found that at sufficiently high concentration, TBK1 autophosphorylates and activates IRF-3 without the need of any additional factors (Figure 1B and C). Consistent with these results, overexpression of TBK1 in cells induces the expression of IFN-β (Fitzgerald et al., 2003). The conformation change of STING upon ligand binding likely promotes the recruitment of TBK1 to the signaling complex (Huang et al., 2012). In addition, K63-linked ubiquitination of TBK1 at residues Lys30 and Lys401 seems also required for TBK1 activation and signaling (Tu et al., 2013). The polyubiquitin chains on these residues likely play a role in bringing the TBK1 dimers close to each other for autophosphoryltion (Larabi et al., 2013; Tu et al., 2013).
Our mass spectrometric analysis of in vitro phosphorylated IRF-3 by TBK1 showed that Ser386, Ser398, and Thr404 in the RD of IRF-3 are key phosphorylation sites by TBK1 (Figure 6G, see also Figure S6) that are critical for the phosphorylation-induced oligomerization of IRF-3. A comparison of phosphorylation sites identified previously with our data indicates that TBK1 and IKKε phosphorylate a distinct but overlapping set of residues (Fujii et al., 2010; Takahasi et al., 2010). Both TBK1 and IKKε specifically phosphorylate Ser386, but selectively phosphorylate other residues in the RD of IRF-3. Previous studies showed both sets of residues were observed in phosphorylated IRF-3 (Lin et al., 1998; Yoneyama et al., 1998). It is likely both TBK1 and IKKε are involved in the activation of IRF-3 and these two kinases may not be functionally redundant.
Analytical ultracentrifugation analysis of in vitro phosphorylated IRF-3 showed that phosphorylation of IRF-3 by TBK1 induces a conformational change and results in the formation of dimers, tetramers and oligomers of IRF-3 (Figure 6C to F). Consistent with the AUC results, gel filtration chromatography of phosphorylated IRF-3 shows a broad peak with estimated size much larger than an IRF-3 dimer (Figure 6A and B). These different forms of oligomers are likely resulted from different levels of phosphorylation. Consistent with this, ion exchange chromatography showed that IRF-3 RD coexpressed with IKKε also forms various kinds of monomers, dimers, and oligomers at different levels of phosphorylation (Takahasi et al., 2010). It is also possible that fully phosphorylated IRF-3 exists in a equilibrium of these oligomers. It is likely that the oligomers of IRF-3 are required for transcription initiation due to their increased avidity for DNA. Since TBK1 and IKKε may play complementary roles in IRF-3 activation, it will be interesting to test what happens when IRF-3 is phosphorylated by both TBK1 and IKKε.
There are still a number of unresolved questions concerning the structure and function of TBK1. For example, it is not clear how TBK1 mediates distinct signaling events from different receptors. Interactions of TBK1 with various kinds of adaptor proteins may contribute to its roles in different signaling events. TBK1 phosphorylates a wide range of substrates involved in immunity and other cellular functions (Barbie et al., 2009; Ou et al., 2011; Pilli et al., 2012; Wild et al., 2011; Xie et al., 2011), but it is not clear how TBK1 achieves such a wide range of substrate specificity. Structures of phosphorylated TBK1 in complex with various substrates or peptides from these substrates will provide insight into these important questions.
TBK1 is an important therapeutic target for antimicrobial and anticancer agents. The crystal structures of mTBK1 bound to the two inhibitors provided a structural basis for designing novel inhibitors of TBK1. Although both of BX795 and SU6668 bind at the ATP binding site, they interact with TBK1 in different ways. SU6668 has additional contacts with TBK1 on the opposite side of the triphosphate group, while BX795 has additional contacts near the triphosphate binding sites (Figure 5E). New inhibitors can be designed based on these two structures to improve their specificity for TBK1. Moreover, the HPD motif binding site may serve as another target for drug design. This motif interacts specifically with inactive TBK1 in the domain-swapped dimer and likely plays a role in suppressing the autophosphorylation of TBK1. Inhibitors based on this structure are likely highly specific for TBK1. The potential substrate-binding site between the KD and the SDD can also be a target for drug design. A short helical peptide is likely to bind to this site and block the access of the substrate. Therefore, our work may have important implications in designing novel antimicrobial or anticancer agents targeting TBK1.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Full-length and C-terminal truncation (residues1-657) of mouse TBK1 were cloned into the pAcGHLTc baculovirus transfer vector. The vector contains a glutathione S-transferase (GST) tag and a 6xHis tag upstream of the multiple cloning sites. The plasmids were transfected with Baculo-Gold bright linearized baculovirus DNA (BD Biosciences) into sf9 insect cells to generate recombinant baculovirus. The recombinant viruses were amplified for at least two rounds before being used for large-scale protein expression. To express the proteins, the insect cells were infected with the recombinant baculovirus in serum-free media (Thermo Scientific). The cells were cultured at 27°C and harvested 60 hrs after infection. The selenomethionine (Se-Met)-substituted mTBK1ΔC S172A was expressed in sf9 cells. These cells were initially grown in serum-free insect cell medium (Thermo Scientific). The viral stock was added to the cells at a density of 2.5 × 106 cells/ml, the infected cells were harvested by centrifugation and resuspended in the ESF921 methionine-free medium (Expression Systems) 24 hrs after infection. Se-Met (Arcos Organics) was added at 100 mg per liter of culture at 12 hrs and again at 36 hrs after medium change. The cells were harvested 72 hrs after infection. The cells were lysed in the lysis buffer (0.2M Tris-HCl, 150mM NaCl, 1mM PMSF, 1% NP-40, pH 8.0). The recombinant proteins in the lysate supernatant were purified by nickel affinity chromatography (batch method) followed by gel filtration chromatography. For crystallization or other assays, the GST-His tag was cleaved using thrombin before gel filtration chromatography.
Human IRF-3 (hIRF-3), human IRF-3 RD (residues 189-427, hIRF-3C), mouse TANK (mTANK), and human CBP (residues 2066 to 2111) were cloned into vector pET28-SUMO and expressed in Escherichia coli BL21(DE3) cells at 15 °C. The recombinant proteins were purified by nickel affinity chromatography followed by gel filtration chromatography. The 6xHis-SUMO tag was cleaved using SUMO protease before gel filtration chromatography. All these proteins were eluted from the gel filtration column using a buffer containing 20 mM Tris-HCl, 150 mM NaCl, at pH7.5 (buffer A), The samples were concentrated and snap frozen in liquid nitrogen and stored at -80 °C. All the mutants of TBK1 and IRF-3 were generated using the QuickChange™ site-directed mutagenesis Kit (Stratagene) according to the manufacturer's protocol. The mutant proteins were purified the same way was the wild type proteins.
Protein kinase assays
The in vitro kinase assay was performed as described previously with some modifications (Shu et al., 2012). For TBK1 autophosphorylation assays, in each 30 μl kinase assay reactions, 3 μg mTBK1 or mutants were incubated with or without 1 μg GST-mTBK1 in a buffer consisting of 20 mM HEPES (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 2 mM DTT, 100 μM ATP, and 0.5 μl [γ-32P] ATP for 2 hrs at room temperature. To analyze TBK1 inhibition by SU6668, in each 30 μl kinase assay reactions, 100 ng prephosphorylated mTBK1ΔC was incubated with a serial dilution (2x) of SU6668 in a buffer consisting of 10 μg hIRF-3, 20 mM HEPES (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 2mM DTT, 100 μM ATP, and 0.5 μl [γ-32P] ATP for 2 hrs at room temperature. The phosphorylated samples were resolved by SDS-PAGE using 10% gel and imaged using a storage phosphor imaging system. For IRF-3 gel shift assays, each 50 μl kinase assay reaction contains 36 μg hIRF-3 or its mutants and incubated with 6 μg prephosphorylated mTBK1 in a buffer containing 20 mM HEPES (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 2 mM DTT, 5 mM ATP for 40 min at room temperature. Phosphorylation induced oligomerization of WT hIRF-3 or its mutants was examined by native PAGE using 10% gels.
GST pull-down assays
In the GST pull-down assays, 100 μl recombinant GST-mTBK1 or GST-mTBK1ΔC with or without purified TANK were incubated with 40 μl glutathione–Sepharose beads (GE Healthcare) and incubated at 4°C for 1 hr. The beads were washed for four times with 200 μl of PBS buffer, boiled in SDS–PAGE loading buffer, and analyzed by SDS–PAGE using 10% gels.
Crystallization, data collection, and structure determination
Mouse TBK1 is partially phosphorylated when purified from the insect cells and does not crystallize. To generate homogeneous sample for crystallization, we treated mTBK1 with λ phosphatase and crystallized it in complex with BX795. The kinase inactive S172A mutant of mTBK1 was generated later on and crystallized under similar conditions as the dephosphorylated protein. To crystalize of mTBK1, native or Se-Met labeled mTBK1ΔC S172A was concentrated to ~10 mg/ml, the inhibitors were added to the samples at a concentration of 0.15 mM. The proteins were crystallized by hanging drop vapor-diffusion method at 4°C over a reservoir containing 0.1 M sodium citrate, 10%~14% isopropanol and 0.1 M HEPES buffer at pH 8.0. Large single crystals grew in about two weeks. The crystals were frozen in liquid nitrogen in a cryo-buffer containing 30% ethylene glycol and 10% PEG400 in the mother liquor. The diffraction data were collected at the Advance Light Source (ALS) beamline 5.1 and 5.2. The BX795 complex data were processed with the HKL2000 package (Otwinowski and Minor, 1997). The SU6668 complex data were processed with Mosflm and scaled with Scala in the CCP4 package (Winn et al., 2011). Statistics of data collection are shown in Table 1.
The structure of the mTBK1/BX795 complex was determined by molecular replacement using the kinase and ubiquitin like domain of human TBK1 (PDB, 1EUT) and the C-terminal domain of Xenopus IKKβ (PDB, 3QA8) as search model using Phaser in the Phenix package (Adams et al., 2010). The C-terminal domain of mTBK1 was completely remodeled due to low sequence homology and structural similarity with IKKβ. The amino acid sequence of mTBK1 was confirmed by an anomalous difference map (Dano-Φc map) from a Se-Met derivative crystal. Thirteen peaks (>4.8σ) were observed in the map of which 12 correspond to the Se sites and one corresponds to the iodine atom in BX795. The structures were refined using the Phenix package (Adams et al., 2010). The mTBK1/SU6668 complex structure was solved by molecular replacement using the refined mTBK1/BX795 complex structure as search model. After remodeling in O (Jones and Kjeldgaard, 1997), the structure was refined with the Phenix package. Statistics of structural refinement are shown in Table 1. Buried surface area and molecular contacts were evaluated with the CNS 1.3 package (Brunger et al., 1998). Comparison of the structures was carried out with the program Superpose in the CCP4 package. The graphics software Coot (Emsley et al., 2010) was used extensively during remodeling and refinement. All structural figures were generated with PyMol.
Gel filtration chromatography of phosphorylated IRF-3
Human IRF-3 at 1 mg/ml concentration was incubated for 16 hrs at 27 °C with 0.05 mg/ml GST-mTBK1 in a buffer containing 20 mM HEPES at pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.1 mM sodium vanadate, 5mM sodium fluoride, 5 mM DTT, and 5 mM ATP. The phosphorylated IRF-3 was purified on a Hiload 16/60 superdex200 column (GE Healthcare) eluted with buffer A. The purified sample was analyzed on a Superdex200 (10/300 GL) column (GE Healthcare) eluted with buffer A. The column was calibrated with a set of protein standards for gel filtration chromatography (Bio-Rad) to ensure accurate estimation of the molecular weight of the proteins.
Analytical ultracentrifugation analysis of IRF-3
Sedimentation velocity AUC analyses were conducted using a Beckman ProteomeLab XL-I with samples of full-length hIRF-3 in both native and phosphorylated states, each in a buffer of 20 mM pH 7.5 Tris-HCl, 150 mM NaCl, and 5 mM β-mercaptoethanol. Samples were spun overnight at 44,000 rpm in an An-60Ti rotor maintained at 10 °C and data were collected using absorbance optics. Raw sedimentation data were deconvoluted into sedimentation coefficient distributions using both c(s) and c(s,f/f0) approaches in the program Sedfit version 12.0c (Brown and Schuck, 2006). Data were examined at both 95% and 67% confidence intervals in the c(s) and c(s,f/f0) analysis. Both analyses revealed similar predicted molecular masses for the various species observed. The 67% confidence interval data are shown to better illustrate the component species in solution after hIRF3 phosphorylation. Values for buffer density and viscosity and partial specific volume of hIRF3 were calculated using Sednterp version 1.09.
Mass spectrometry of phosphorylated IRF3 and TBK1
The proteins in the SDS-PAGE gel bands were reduced with DTT to break disulfide bond and cysteine residues were alkylated by iodoacetamide to allow the recovery of Cys-containing peptides. The gel bands were washed, dried down in a speed vacuum, and rehydrated with a buffer containing trypsin to allow trypsin to enter the gel. The peptide samples were loaded on a nanoscale capillary reverse phase C18 column by a HPLC system (Waters ACQUITY UPLC), and eluted by a gradient (~30 min). The eluted peptides were ionized, and detected by an inline mass spectrometer (Thermo Q Exactive). The spectra were collected first (in ~0.5 sec) and the top 20 abundant ions or ions within pre-defined mass ranges (for targeted MS/MS) were sequentially isolated for MS/MS analysis (each in ~0.1 sec, totaling ~2 sec). This process (~2.5 sec) was cycled over the entire elution chromatography gradient, acquiring more than 14,000 MS/MS spectra during a 30-min elution. Database searches were performed using Sequest search engine in the SPIDERS in-house software package. All matched MS/MS spectra were filtered by mass accuracy and matching scores to reduce protein false discovery rate to less than 1%. The modification sites were determined by dynamically assigning related mass addition to all possible amino acid residues during database search. Modified residues were further validated by manual examination of raw spectra and confirmed on the basis of the unambiguous assignment of characteristic site-specific fragment ions.
Supplementary Material
Highlights.
TBK1 is activated by autophosphorylation at residue Ser172
TBK1 exhibits three structural domains similar to IκB kinase
The kinase domain of TBK1 exhibits rigid structure
Phosphorylation of IRF-3 by TBK1 results in the oligomerization of IRF-3
Acknowledgement
The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This work was partially supported by ALSAC (American Lebanese Syrian Associated Charities to A. M.). We are grateful to Dr. Xiuren Zhang and Dr. Ping He for their help with the protein kinase assays and Dr. Jun-yuan Ji for critical reading of the manuscript. Research in the Li lab is supported by the NIH grant 1R01AI087741.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes. Atomic coordinates and structure factors of mTBK1 in complex with BX795 and SU6668 have been deposited in the Protein Data Bank with accession codes: 4JL9 and 4JLC, respectively.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- Cheng G, Baltimore D. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-kappaB activation. Genes & development. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284:14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Fujii K, Nakamura S, Takahashi K, Inagaki F. Systematic characterization by mass spectrometric analysis of phosphorylation sites in IRF-3 regulatory domain activated by IKK-i. J Proteomics. 2010;73:1196–1203. doi: 10.1016/j.jprot.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Godl K, Gruss OJ, Eickhoff J, Wissing J, Blencke S, Weber M, Degen H, Brehmer D, Orfi L, Horvath Z, et al. Proteomic characterization of the angiogenesis inhibitor SU6668 reveals multiple impacts on cellular kinase signaling. Cancer Res. 2005;65:6919–6926. doi: 10.1158/0008-5472.CAN-05-0574. [DOI] [PubMed] [Google Scholar]
- Goncalves A, Burckstummer T, Dixit E, Scheicher R, Gorna MW, Karayel E, Sugar C, Stukalov A, Berg T, Kralovics R, et al. Functional dissection of the TBK1 molecular network. PLoS One. 2011;6:e23971. doi: 10.1371/journal.pone.0023971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- Helgason E, Phung QT, Dueber EC. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: The emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013 doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature structural & molecular biology. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- Kanai F, Yoshida H, Tateishi R, Sato S, Kawabe T, Obi S, Kondo Y, Taniguchi M, Tagawa K, Ikeda M, et al. A phase I/II trial of the oral antiangiogenic agent TSU-68 in patients with advanced hepatocellular carcinoma. Cancer Chemother Pharmacol. 2011;67:315–324. doi: 10.1007/s00280-010-1320-2. [DOI] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Larabi A, Devos JM, Ng SL, Nanao MH, Round A, Maniatis T, Panne D. Crystal Structure and Mechanism of Activation of TANK-Binding Kinase 1. Cell Rep. 2013;3:734–746. doi: 10.1016/j.celrep.2013.01.034. [DOI] [PubMed] [Google Scholar]
- Li J, Li J, Miyahira A, Sun J, Liu Y, Cheng G, Liang H. Crystal structure of the ubiquitin-like domain of human TBK1. Protein Cell. 2012;3:383–391. doi: 10.1007/s13238-012-2929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Helgason E, Phung QT, Quan CL, Iyer RS, Lee MW, Bowman KK, Starovasnik MA, Dueber EC. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc Natl Acad Sci U S A. 2012;109:9378–9383. doi: 10.1073/pnas.1121552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, Brekken R, Wurz R, Tasker A, Polverino T, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41:458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature structural & molecular biology. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhara W, Yoneyama M, Iwamura T, Yoshimura S, Tamura K, Namiki H, Aimoto S, Fujita T. Analyses of virus-induced homomeric and heteromeric protein associations between IRF-3 and coactivator CBP/p300. Journal of biochemistry. 2000;128:301–307. doi: 10.1093/oxfordjournals.jbchem.a022753. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Horiuchi M, Fujii K, Nakamura S, Noda NN, Yoneyama M, Fujita T, Inagaki F. Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change. Genes Cells. 2010;15:901–910. doi: 10.1111/j.1365-2443.2010.01427.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunological reviews. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tu D, Zhu Z, Zhou AY, Yun CH, Lee KE, Toms AV, Li Y, Dunn GP, Chan E, Thai T, et al. Structure and Ubiquitination-Dependent Activation of TANK-Binding Kinase 1. Cell Rep. 2013;3:747–758. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Zhang D, Zhao B, Lu MK, You M, Condorelli G, Wang CY, Guan KL. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc Natl Acad Sci U S A. 2011;108:6474–6479. doi: 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Lo YC, Li Q, Napolitano G, Wu X, Jiang X, Dreano M, Karin M, Wu H. Crystal structure of inhibitor of kappaB kinase beta. Nature. 2011;472:325–330. doi: 10.1038/nature09853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fujita T. Control of IRF-3 activation by phosphorylation. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2002;22:73–76. doi: 10.1089/107999002753452674. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr D Biol Crystallogr. 1993;49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.