Abstract
The transcription factor Forkhead box M1 (FOXM1) plays important roles in oncogenesis. However, the expression statuses of FOXM1 isoforms and their impact on and molecular basis in oncogenesis are unknown. We sought to determine the identities of FOXM1 isoforms in and the impact of their expression on pancreatic cancer development and progression using human tissues, cell lines and animal models. Overexpression of FOXM1 mRNA and protein was pronounced in human pancreatic tumors and cancer cell lines. We identified five FOXM1 isoforms present in pancreatic cancer: FOXM1a, FOXM1b, and FOXM1c along with two isoforms tentatively designated as FOXM1b1 and FOXM1b2 because they were closely related to FOXM1b. Interestingly, FOXM1c was predominantly expressed in pancreatic tumors and cancer cell lines, whereas FOXM1a expression was generally undetectable in them. Functional analysis revealed that FOXM1b, FOXM1b1, FOXM1b2, and FOXM1c but not FOXM1a promoted pancreatic tumor growth and metastasis. Consistently, FOXM1b, FOXM1b1, FOXM1b2, and FOXM1c activated transcription of their typical downstream genes. Also, Sp1 mechanistically activated the FOXM1 promoter, whereas Krüppel-like factor 4 (KLF4) repressed its activity. Finally, we identified an Sp1- and KLF4-binding site in the FOXM1 promoter and demonstrated that both Sp1 and KLF4 protein bound directly to it. Deletion mutation of this binding site significantly attenuated the transcriptional regulation of the FOXM1 promoter positively by Sp1 and negatively by KLF4. We demonstrated that overexpression of specific FOXM1 isoforms critically regulates pancreatic cancer development and progression by enhancing tumor cell invasion and metastasis. Our findings strongly suggest that targeting specific FOXM1 isoforms effectively attenuates pancreatic cancer development and progression.
Keywords: Progression, angiogenesis, transcription factor, EMT, biomarkers
Introduction
Pancreatic cancer is one of the leading causes of cancer deaths in industrialized countries, and the incidence of this disease appears to be increasing (1). Despite improvements in early diagnosis, surgical techniques, and chemotherapy, the majority of pancreatic cancer patients die of the physiological effects of invasion and metastasis to regional lymph nodes and/or distant organs (2). Unfortunately, little is known about the reasons for the aggressiveness of and dismal prognosis for pancreatic cancer. Therefore, the need for a better understanding of the molecular mechanisms underlying pancreatic cancer progression is urgent (3, 4).
Forkhead box M1 (FOXM1), previously referred to HNF-3, HFH-11, MPP2, Win, and Trident, is a transcription factor in the FOX protein superfamily that is defined by a conserved winged helix DNA-binding domain (5). FOXM1 mRNA is ubiquitously expressed in murine embryonic tissue and murine adult proliferating tissue but its expression is extinguished in differentiated cells (6–8). Recent studies suggested that FOXM1 is required for coupling the S and M phases of the cell cycle in part by regulating the transcription of genes essential for cell-cycle progression, including cyclin D1 and p27 (6–8).
The human FOXM1 gene consists of 10 exons, of which Va (A1) and VIIa (A2) are alternatively spliced. This splicing gives rise to three distinct FOXM1 variants FOXM1a, FOXM1b, and FOXM1c (6–8). FOXM1a harbors Va and VIIa and is transcriptionally inactive owing to disruption of its transactivation domain by VIIa. In comparison, FOXM1b, which contains neither of the two exons, and FOXM1c, which has only Va, are transcriptionally active and can activate their target gene expression via different mechanisms (7). Because FOXM1 is a transcription factor essential for expression of many genes key to regulation of multiple aspects of tumor cell survival, growth, epithelial-to-mesenchymal transition (EMT), angiogenesis, and metastasis, abnormal FOXM1 expression may contribute to human cancer development and progression (9, 10).
Lines of evidence demonstrate that overexpression of FOXM1 occurs frequently in a wide variety of human tumors (11–15). In particular, recent studies suggest that FOXM1 plays important roles in pancreatic cancer development and progression (16–18). Downregulation of FOXM1 expression inhibits pancreatic cancer cell growth, migration, and invasion by decreasing the expression of cyclin B, cyclin D1, Cdk2, matrix metalloproteinase (MMP)-2, MMP-9, and vascular endothelial growth factor (VEGF) (16). Also, FOXM1 may regulate the EMT phenotype of pancreatic cancer cells by activating mesenchymal cell markers (17). Recently, we found that FOXM1/caveolin-1 signaling plays an important role in EMT in, invasion of, and metastasis of pancreatic tumors (18). However, the molecular mechanisms underlying FOXM1 isoform expression and its regulation and the impact of the isoforms on pancreatic cancer development and progression are unknown.
Krüppel-like factor 4 (KLF4) is a zinc-finger transcription factor, and KLF4 mRNA expression is found primarily in postmitotic, terminally differentiated epithelial cells in organs such as the skin and lungs and in the gastrointestinal tract (19). Accumulating clinical, experimental, and mechanistic evidence shows that KLF4 is a potential tumor suppressor in patients with various cancers, including pancreatic cancer (20–22). A recent study by our group showed that loss of expression of KLF4 and overexpression of FOXM1 are evident in human gastric cancer cells and that altered expression and function of FOXM1 contribute to gastric carcinogenesis (13). However, the mechanistic role of FOXM1 in pancreatic carcinogenesis and its causal link with altered KLF4 function are unknown.
In the present study, we sought to determine the roles and effect of regulation of the three FOXM1 isoforms in pancreatic cancer invasion and metastasis. We discovered that KLF4 negatively regulates FOXM1 transcription in pancreatic tumors and cancer cell lines but that Sp1 positively regulates it, that FOXM1c is the predominantly expressed FOXM1 isoform in pancreatic tumors and cancer cells, and that FOXM1c promotes pancreatic tumor invasion and metastasis.
Materials and Methods
Cell lines and culture conditions
The human pancreatic adenocarcinoma cell lines AsPC-1, CaPan-1, CaPan-2, MiaPaca-2, BxPC-3, Hs766T, PANC-1, and PAU8902 and human embryonic kidney 293 (HEK293) cells were purchased from the American Type Culture Collection. The pancreatic cancer cell lines MDA Panc-28 and MDA Panc-48 were gifts from Dr. Paul J. Chiao (The University of Texas MD Anderson Cancer Center). The human metastatic pancreatic adenocarcinoma cell line COLO357 and its fast-growing liver-metastatic variants L3.3 and L3.7 in nude mice were described previously (18, 23–25). All of these cell lines were maintained in plastic flasks as adherent monolayers in Eagle's minimal essential medium supplemented with 10% fetal bovine serum, sodium pyruvate, nonessential amino acids, L-glutamine, and a vitamin solution (Flow Laboratories). Immortalized human pancreatic ductal epithelial (HPDE) cells (provided by Dr. M.S. Tsao, Ontario Cancer Institute) were maintained in a keratinocyte serum-free medium supplemented with epidermal growth factor and bovine pituitary extract (Invitrogen). The cell lines were obtained directly from ATCC that performs cell line characterizations or authentication by the short tandem repeat profiling and passaged in our laboratory for fewer than 6 months after receipt.
Human tissue specimens and immunohistochemical analysis
Expression of FOXM1 and KLF4 was analyzed using human pancreatic cancer specimens and a normal pancreatic tissue microarray (TMA; US Biomax). Use of the tissue specimens was approved by the MD Anderson Institutional Review Board. Standard immunohistochemical staining procedures were performed using anti-FOXM1 (Santa Cruz Biotechnology) and anti-KLF4 (Santa Cruz Biotechnology) antibodies. The staining results were scored by two investigators blinded to the clinical data as described previously (26).
Animals
Pathogen-free female athymic nude mice were purchased from the National Cancer Institute. The animals were maintained in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care International in accordance with the current regulations and standards of the U.S. Department of Agriculture and Department of Health and Human Services.
Orthotopic tumor growth
Tumor cells (1×106) in 0.05 mL of Hank’s balanced salt solution were injected into the subcapsule of the pancreas in nude mice. The tumor-bearing mice were killed when they became moribund or on day 35 after tumor cell inoculation, and their tumors were removed and weighed.
Experimental liver metastases
L3.7 cells (1×105/mouse) or COLO357 and other cells (1×106/mouse) were injected intravenously into mice via the ileocolic vein. The mice were killed on day 35 after tumor cell inoculation or when they appeared to be moribund. Their livers were then removed, and the surface metastases were counted after dissecting the liver into its individual lobes. Every surface metastasis was examined by two investigators unaware of the experimental protocol and scored separately (27). The tissue specimens were then placed in 10% buffered formalin, immersed in an ascending series of alcohols, and embedded in paraffin. The specimens were then cut into sections that were stained with hematoxylin and eosin.
Western blot analysis
Standard Western blotting was performed using whole-cell protein lysates, primary antibodies against FOXM1 and KLF4, and a secondary antibody (anti-rabbit IgG; Santa Cruz Biotechnology). Equal protein-sample loading was monitored using an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (Santa Cruz Biotechnology).
Plasmids and small interfering RNAs
The previously described plasmid pcDNA3.1-FOXM1b and empty control vector plasmid pcDNA3.1 (14) were used as controls. To generate the pcDNA3.1-FOXM1a and pcDNA3.1-FOXM1c plasmids, full-length human FOXM1a and FOXM1c were released via EcoRI and XbaI digestion of the human cytomegalovirus FOXM1a and FOXM1c cDNA expression vectors and subcloned into pcDNA3.1 (Invitrogen). The small interfering RNA (siRNA) sequence targeting FOXM1 (siFOXM1) was CUCUUCUCCCUCAGAUAUAdTdT (13), and the FOXM1a- and FOXM1c-specific siRNA sequence was CCCAGGGUCUCCACAAUUG[dT][dT].
Transfection
Transfection of plasmids and siRNAs into pancreatic cancer cells was performed using the Lipofectamine LTX (Invitrogen) and Lipofectamine 2000 CD (Invitrogen) transfection reagents, respectively. Cells were transfected with plasmids or siRNA at different doses as indicated for 48 hours before the performance of functional assays. Pancreatic cancer cells treated with a transfection reagent alone were included as mock controls.
Construction of FOXM1 promoter reporter plasmids and mutagenesis
A 2.544-kb fragment containing FOXM1 5' sequences from −2469 to +75 relative to the transcription initiation site was subcloned into the pGL3-basic vector (Promega). The final full-length reporter plasmid, which contained multiple Sp1/KLF4-binding sites, was designated pFXM1-2469. Deletion mutation reporters for this plasmid were then generated. All constructs were verified by sequencing the inserts and flanking regions of the plasmids.
Promoter reporter and dual luciferase assay
Pancreatic cancer cells were transfected with the indicated FOXM1 promoter reporters, siRNAs, or specific gene expression plasmids. The FOXM1 promoter activity was normalized by cotransfecting a β-actin/Renilla luciferase reporter containing a full-length Renilla luciferase gene (28). The luciferase activity in the cells was quantified using a dual luciferase assay system (Promega) 24 hours after transfection.
Real-time reverse transcription-polymerase chain reaction
Total RNA extraction from tumor cells was performed using TRIzol reagent (Invitrogen). Next, 2 µg of total RNA was reverse-transcribed using a First Strand cDNA Synthesis Kit (Promega) to synthesize cDNA samples. Real-time reverse transcription-polymerase chain reaction (PCR) analysis of expression of the FOXM1 gene was performed using 2 µL of cDNA and SYBR Green Master Mix (Bio-Rad) as recommended by the manufacturer for with the following primers: FOXM1, 5'-acgtccccaagccaggctc-3' (forward) and 5'-ctactgtagctcaggaataa-3' (reverse); FOXM1a, 5'-tggggaacaggtggtgtttgg-3' (forward) and 5'-gctagcagcactgataaacaaag-3' (reverse); FOXM1b, 5'-ccaggtgtttaagcagcaga-3' (forward) and 5'-tcctcagctagcagcaccttg-3' (reverse); FOXM1c, 5'-caattgcccgagcacttggaatca-3' (forward) and 5'-tcctcagctagcagcaccttg-3' (reverse); and GAPDH, 5'-caccattggcaatgagcggttc-3' (sense strand) and 5'-aggtctttgcggatgtccacgt-3' (antisense strand). GAPDH was used as an internal control. Each PCR product was run in triplicate for the target and internal control genes. The PCR products were loaded onto 2% agarose gels and visualized with ethidium bromide staining under ultraviolet light.
Chromatin immunoprecipitation assay
Tumor cells (2×106) were prepared for chromatin immunoprecipitation (ChIP) assay using a ChIP assay kit (Millipore Corporation) according to the manufacturer’s protocol. The resulting precipitated DNA specimens were analyzed using PCR with the primers 5'-cttcgagcccggaatgccg-3' (forward) and 5'-gggaggggagggggtcccggg-3' (reverse) to amplify a 160-bp region of the FOXM1 promoter. The PCR products were resolved electrophoretically on a 2% agarose gel and visualized using ethidium bromide staining.
Statistical analysis
The significance of the patient specimen data was determined using the Pearson correlation coefficient. The significance of the in vitro and in vivo data was determined using the Student t-test (two-tailed), Mann-Whitney test (two-tailed), or one-way analysis of variance. P values less than 0.05 were considered significant.
Results
FOXM1 protein overexpression in pancreatic tumors and its clinicopathological significance
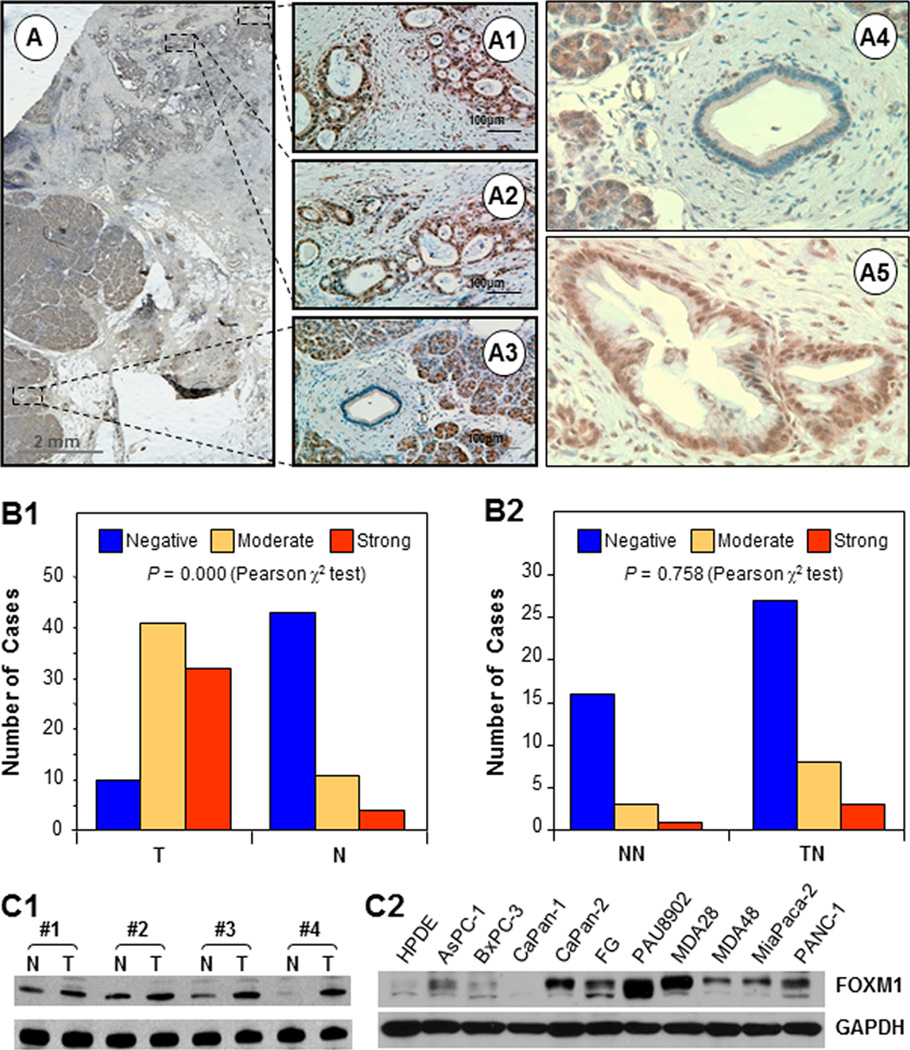
We first detected the expression of FOXM1 protein in a TMA of primary pancreatic tumor and adjacent normal pancreatic tissue specimens using immunohistochemical staining with a specific anti-FOXM1 antibody. We found specific staining for FOXM1 in the nucleus and/or cytoplasm of the tumor cells and negative or weakly positive staining for FOXM1 in the cytoplasm of the tumor-adjacent normal pancreatic cells and more distant normal pancreatic cells (Fig. 1A and B). We did not see a significant difference in FOXM1 expression between the tumor-adjacent normal tissue and normal pancreatic tissue specimens (Fig. 1B). We confirmed that FOXM1 protein expression was increased using Western blot analysis with paired normal pancreatic tissue and pancreatic tumor specimens (Fig. 1C). Consistently, the majority of the pancreatic cancer cell lines expressed FOXM1 protein at levels higher than those in transformed HPDE cells (Fig. 1C).
Figure 1.
FOXM1 protein overexpression in pancreatic tumors and cancer cell lines. Sections were prepared from formalin-fixed, paraffin-embedded specimens of human pancreatic tumors. Immunostaining of the sections was performed using a specific anti-FOXM1 antibody. A, representative photos of FOXM1 expression in tumor cells and adjacent normal cells. B, FOXM1 expression was significantly higher in tumors (T) than in normal tissue (N) (image 1), whereas FOXM1 expression did not differ in tumor-adjacent normal pancreatic tissue (TN) and more distant normal pancreatic tissue (NN) (image 2). C, Western blot analyses of the paired normal pancreatic tissue specimens (N) and pancreatic tumor specimens (T) (image 1) and pancreatic cancer cell lines (image 2).
We then examined the relationship between clinicopathological parameters and FOXM1 expression levels in pancreatic tumors. We observed that increased FOXM1 expression correlated with decreased tumor differentiation and a significant difference between well (grade 1) and poorly differentiated (grade 3) tumors (Supplementary Fig. S1). In addition, FOXM1 expression was positively correlated with disease stage, indicating that FOXM1 expression is upregulated in late-stage pancreatic tumors. This association was significant between stage 1 and stage 4 tumors. Moreover, FOXM1 expression was significantly higher in primary tumor specimens obtained from patients with lymph node or distant metastasis than in those obtained from patients without metastasis. These results strongly demonstrated that FOXM1 expression plays critical roles in pancreatic cancer development and progression and is a valuable biomarker for this disease.
FOXM1 isoform expression in pancreatic tumors and cancer cells
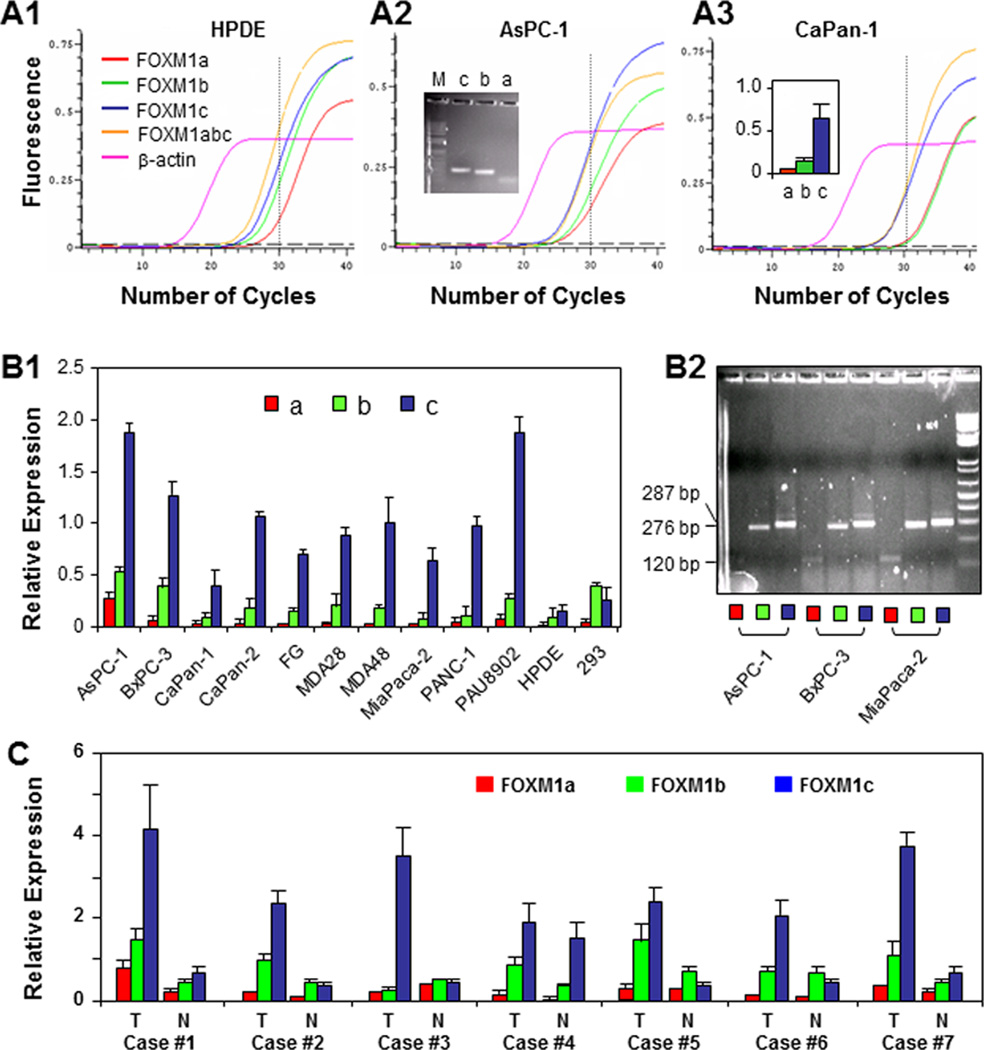
Given that FOXM1 isoform-specific antibodies are currently unavailable, our immunostaining results did not reveal the isoform identities of FOXM1 proteins. We therefore used PCR primers specific for FOXM1a, FOXM1b, and FOXM1c (Supplementary Fig. S2). By using 10 pancreatic cancer cell lines, HPDE cells, and HEK293 cells, real-time PCR analysis revealed that FOXM1c was the predominant isoform expressed in HPDE and pancreatic cancer cells, whereas FOXM1b was the predominant isoform expressed in HEK293 cells (Fig. 2A and B). We also performed real-time PCR analysis of the FOXM1 isoforms in the poorly metastatic human pancreatic cancer cell line COLO357 and the paired highly metastatic human pancreatic cancer cell line L3.7 (Supplementary Fig. S2). The relative level of FOXM1c in L3.7 cells was significantly higher than that in COLO357 cells. Also, the level of FOXM1c expression correlated directly with metastatic ability. We further measured FOXM1 isoform expression in human pancreatic tumor specimens and paired adjacent normal pancreatic tissue specimens using real-time PCR. The relative level of FOXM1c in the pancreatic tumors was significantly higher than that in adjacent normal pancreatic tissue (Fig. 2C). These findings demonstrated that FOXM1c is the predominant FOXM1 isoform expressed in pancreatic tumors.
Figure 2.
Expression of FOXM1a, FOXM1b, and FOXM1c in pancreatic tumors and cancer cell lines. A, qPCR reaction curves created using PCR primers specific for FOXM1a, FOXM1b, and FOXM1c with β-actin used as a control. PCR products generated over 30 cycles were resolved electrophoretically on a 2% agarose gel and visualized using ethidium bromide staining (image 2, insert), and relative levels of FOXM1a, FOXM1b, and FOXM1c expression were measured (image 3, insert). B, relative expression of FOXM1a, FOXM1b, and FOXM1c as determined using qPCR with isoform-specific primers (image 1). The qPCR products over 30 cycles were resolved electrophoretically on a 2% agarose gel and visualized using ethidium bromide staining (image 2). C, relative expression of FOXM1a, FOXM1b, and FOXM1c in seven paired normal pancreatic tissue specimens (N) and pancreatic tumor specimens (T) as determined using qPCR analysis with isoform-specific primers.
Moreover, we found expression of FOXM1b1 and FOXM1b2 in pancreatic cancer cells (Supplementary Fig. S3A). FOXM1b, FOXM1b1, and FOXM1b2 expression did not differ significantly in pancreatic tumors and normal pancreatic tissue (Supplementary Fig. S3B), and the three isoforms had similar functions in transcriptional regulation of genes downstream from FOXM1, such as MMP-2 and VEGF (Supplementary Fig. S3C).
Altered FOXM1 expression affected pancreatic cancer cell migration and invasion in vitro and tumor growth and metastasis in vivo
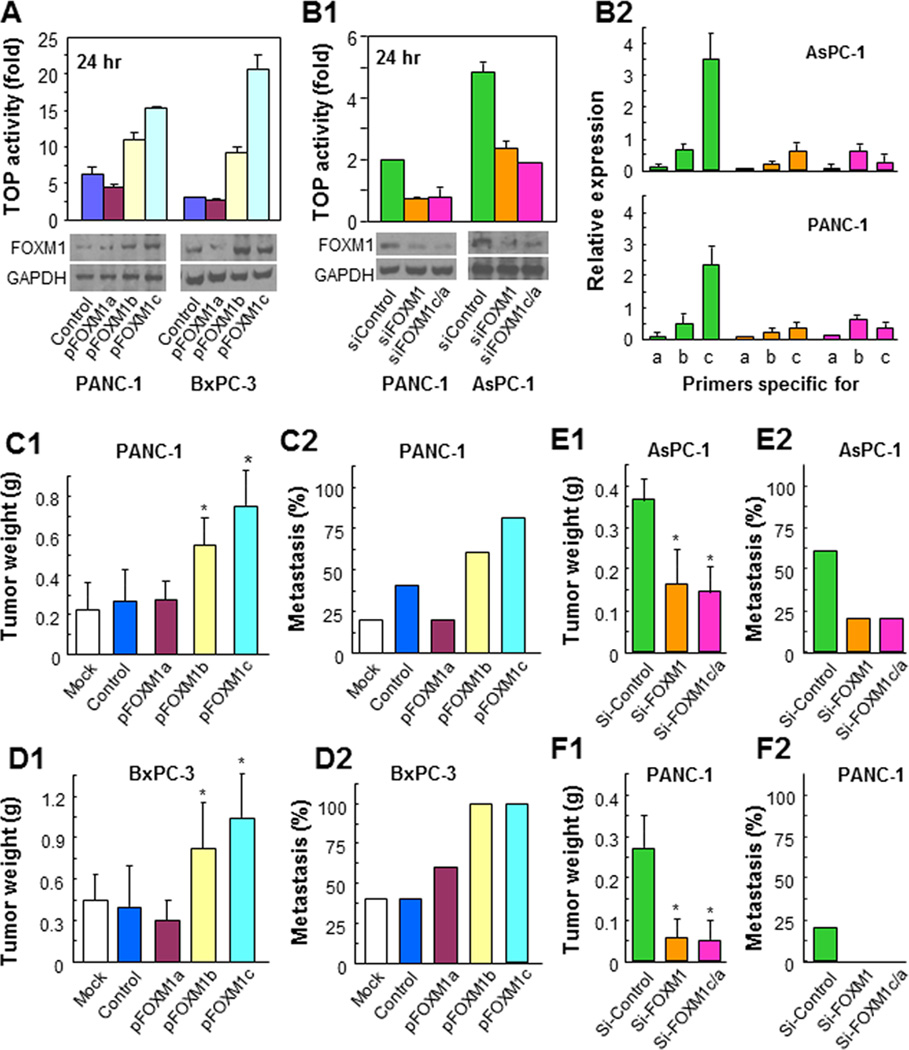
To determine the effect of altered expression of the isoforms of FOXM1 on pancreatic cancer biology, we overexpressed FOXM1a, FOXM1b, and FOXM1c in PANC-1 and BxPC-3 cells using respective expression vectors whereas knocked down overall expression of FOXM1 or FOXM1c/a in PANC-1 and AsPC-1 cells using specific siRNAs. We first confirmed their overexpression (Fig. 3A) and knockdown efficacy (Fig. 3B1) using a TOP reporter activity assay and Western blot analysis. We further confirmed the specific knockdown of isoform expression using quantitative PCR (qPCR) with isoform-specific PCR primers (Fig. 3B2).
Figure 3.
Influence of altered expression of FOXM1 isoforms on pancreatic tumor growth and metastasis. A, PANC-1 and BxPC-3 cells were transfected with the expression vectors for FOXM1a, FOXM1b, and FOXM1c. Successful transfection was confirmed using a TOP reporter activity assay and Western blot analysis. B, PANC-1 and AsPC-1 cells were transfected with si-FOXM1 or with FOXM1a or FOXM1c siRNA; nontargeting siRNA (si-Control) was used as a control. Successful transfection was confirmed using a TOP reporter activity assay and Western blot analysis (image 1) and qPCR analysis with isoform-specific primers (image 2). C, orthotopic growth of PANC-1 cells with increased expression of FOXM1a, FOXM1b, or FOXM1c in pancreas (image 1) and an experimental hepatic metastasis (image 2). D, orthotopic growth of BxPC-3 cells with increased expression of FOXM1a, FOXM1b, or FOXM1c in pancreas (image 1) and an experimental hepatic metastasis (image 2). E, orthotopic growth of AsPC-1 cells with decreased expression of FOXM1 or FOXM1c in pancreas (image 1) and an experimental hepatic metastasis (image 2). F, orthotopic growth of PANC-1 cells with decreased expression of FOXM1 or FOXM1c in pancreas (image 1) and an experimental hepatic metastasis (image 2).
We evaluated the ability of the cells with altered expression of FOXM1 isoforms to migrate and invade in vitro and to grow and metastasize in animal models. Overexpression of FOXM1b and FOXM1c but not FOXM1a consistently promoted pancreatic tumor growth and metastasis, whereas knockdown FOXM1 or more specifically of FOXM1c expression attenuated them (Fig. 3C, 3D, 3E and 3F; Supplementary Fig. S4). Furthermore, overexpression of FOXM1b and FOXM1c but not FOXM1a promoted pancreatic cancer cell migration and invasion (Fig. 4A, 4B, & 4C), whereas knockdown of FOXM1 expression attenuated this migration and invasion (Supplementary Fig. S5). Mechanistically, we found that altered levels of FOXM1c expression impacted on the expression of genes key to cell proliferation and invasion and metastasis (Fig. 4D). Because the levels of both FOXM1a and FOXM1b expression were very low and FOXM1c was predominant, these results strongly suggested that FOXM1c critically regulated pancreatic cancer development and progression.
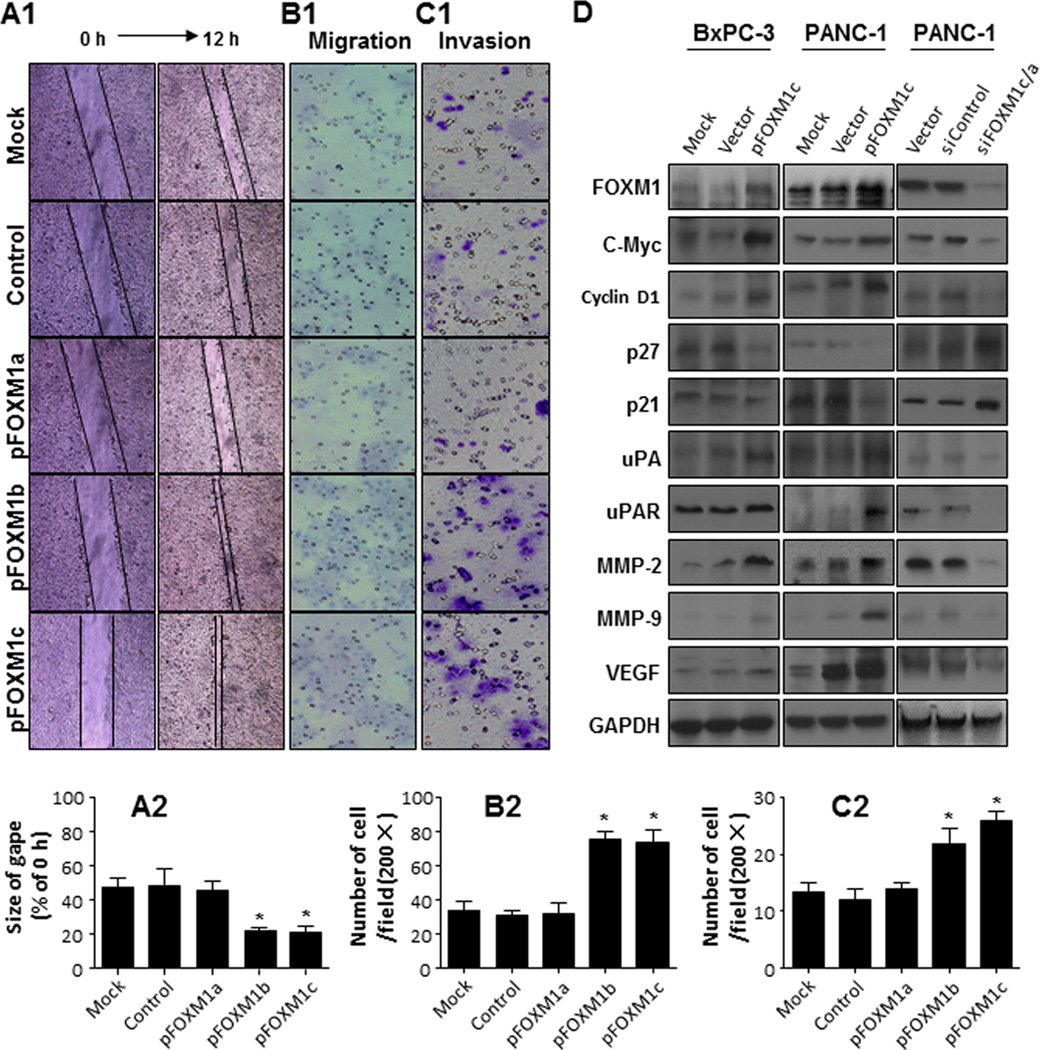
Figure 4.
Promotion of pancreatic cancer cell migration and invasion by overexpression of FOXM1 isoforms. Untransfected COLO357 cells (control) and COLO357 cells transfected with an empty pcDNA3.1 vector (control) or FOXM1a, FOXM1b, or FOXM1c expression vectors were used. A, cell scratch-wound assay. B, cell migration assay. C, cell invasion assay. Assay details are provided in Supplementary Materials and Methods. Note: increased expression of FOXM1b and FOXM1c but not FOXM1a substantially promoted the horizontal and vertical migration and invasion of tumor cells. *P < 0.01 (Student t-test). D. Western blot analyses. FoxM1c upregulated the expression of Cyclin D1, MMP-2, MMP-9, uPA, uPAR, VEGF, and c-myc, but downregulated the expression of p27 and p21.
Transcriptional regulation of FOXM1 expression in pancreatic cancer cells
To determine the molecular basis for altered FOXM1 expression in pancreatic cancer cells, we generated the full-length FOXM1 promoter pFXM1-2469 and deletion mutants of it (Supplementary Fig. S6). One of the deletion mutants, pFXM1-160, exhibited relatively high promoter activity; we therefore identified it as the minimal FOXM1 promoter because another deletion mutant (pFXM1-100) led to decreased promoter activity.
Transcriptional activation of FOXM1 expression by Sp1 and inhibition of FOXM1 transcription by KLF4
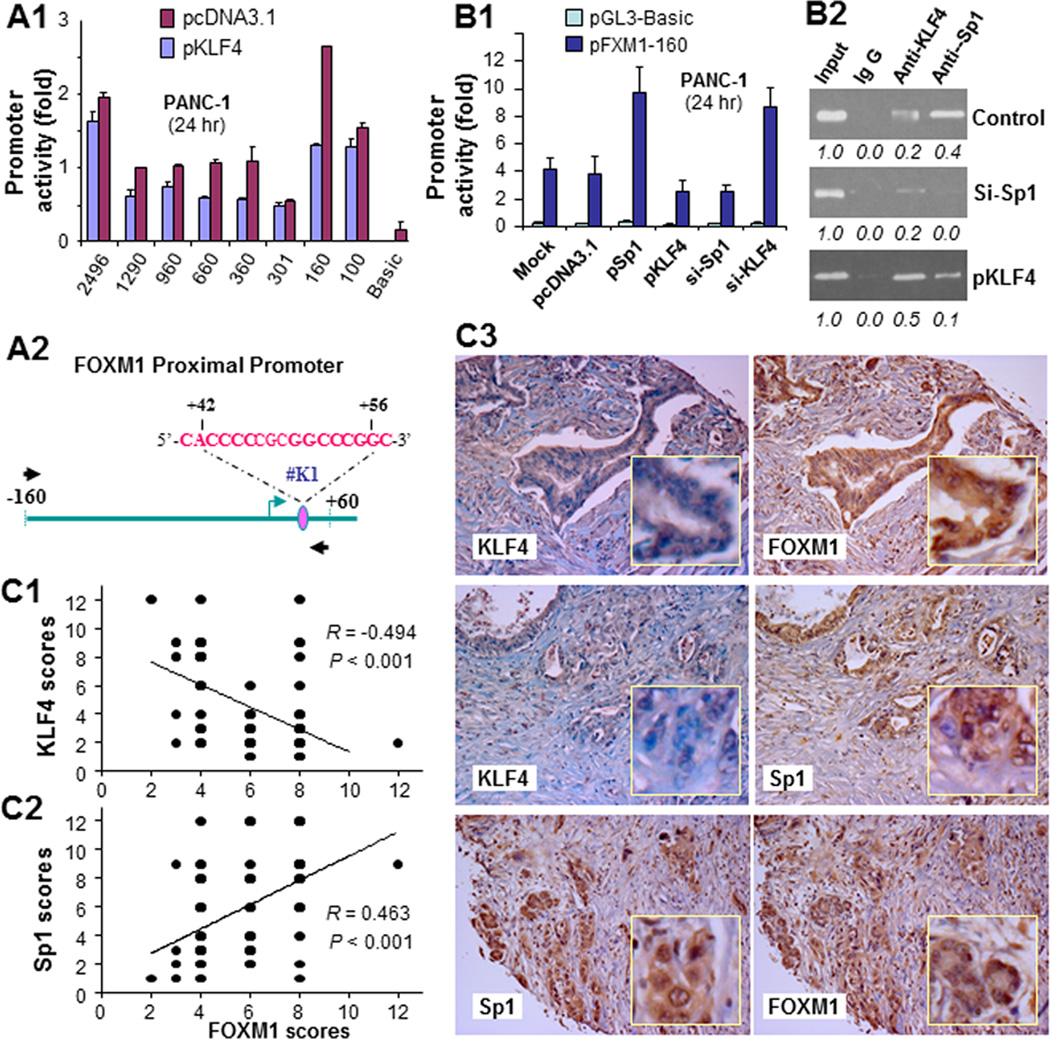
To determine whether KLF4 regulates FOXM1 expression at the transcriptional level, we cotransfected the deletion mutant reporters with or without KLF4 expression vectors into PANC-1 cells. As shown in Fig. 5A, KLF4 inhibited the activity of all mutants but pFXM1-301, which lost a KLF4-binding site downstream from the initiation site. To define the regulatory functions of this putative Sp1/KLF4-binding site, we cotransfected pFXM1-160 with either KLF4 expression vectors or Sp1 or KLF4 siRNA into PANC-1 cells. As shown in Fig. 5B, increased Sp1 expression activated the FOXM1 promoter, whereas knockdown of Sp1 expression attenuated FOXM1 promoter activity. In contrast, overexpression of KLF4 suppressed FOXM1 promoter activity, whereas knockdown of KLF4 expression potentiated FOXM1 promoter activity. Using a ChIP assay, we further found that both KLF4 and Sp1 bound to the putative Sp1/KLF4-binding site (Fig. 5B). These results clearly suggested that Sp1 positively and KLF4 negatively regulated FOXM1 transcription in pancreatic cancer cells.
Figure 5.
Regulation of FOXM1 promoter activity by KLF4 and Sp1. A, the FOXM1 promoter reporters were transfected into PANC-1 cells in triplicate with or without a KLF4 expression vector; pcDNA3.1 was used as a control. The relative FOXM1 promoter activity was measured 24 hours after transfection, and the activity in the treated groups was expressed as the fold of that in their respective control groups (image 1). A schematic of the structure of the minimal FOXM1 promoter reporter construct pFXM1-160 with a putative Sp1/KLF4-binding site is also shown (image 2). B, the pFXM1-160 reporter was transfected into PANC-1 cells in triplicate with or without a KLF4 expression vector (pKLF4), Sp1 expression vector (pSp1), Sp1 siRNA (si-Sp1), or KLF4 siRNA (si-KLF4); nontargeting siRNA (mock) and pcDNA3.1 were used as controls. The relative FOXM1 promoter activity was measured 24 hours after transfection (image 1). The in vivo binding of Sp1 and KLF4 to their putative binding sites and altered binding of them upon increased or decreased expression of KLF4 or Sp1 in PANC-1 cells was examined using a ChIP assay (image 2). C, FOXM1, KLF4, and Sp1 expression in human pancreatic tumors and normal pancreatic tissue was measured using standard immunohistochemical procedures with specific anti-FOXM1, -KLF4, and -Sp1 antibodies. FOXM1 expression correlated inversely with KLF4 expression (image 1) but directly with Sp1 expression (image 2). Representative photos of the expression of FOXM1, KLF4, and Sp1 in pancreatic tumors are shown (image 3). Note: some of the dots on the graphs represent more than one specimen (overlapped scores).
In addition, we analyzed the expression of FOXM1, Sp1, and KLF4 in human pancreatic tumor specimens. We found an inverse correlation between the levels of expression of KLF4 and FOXM1 but a direct correlation between those of Sp1 and FOXM1 (Fig. 5C). These data further supported the notion that Sp1 positively regulated FOXM1 activity but that KLF4 did the opposite, suggesting that loss of KLF4 expression, which is known to occur in pancreatic cancer (29, 30), can cause increased FOXM1 and Sp1 expression and that increased Sp1 expression can lead to further increases in FOXM1 expression (Supplementary Fig. S7).
Discussion
In the present study, we defined the critical roles of FOXM1 isoforms in pancreatic cancer pathogenesis and their underlying mechanisms. For the first time, we demonstrated that human pancreatic cancer cells predominantly express the isoform FOXM1c and that its overexpression correlates directly with increased malignancy of pancreatic cancer. Mechanistically, FOXM1c transcriptionally activated genes that regulate invasion and metastasis of pancreatic cancer cells. Also, Sp1 positively regulated transcription of FOXM1, whereas KLF4 did the opposite. Collectively, our novel clinical and mechanistic findings strongly suggested that dysregulation of the KLF4/Sp1/FOXM1c pathway causes abnormal expression of genes downstream from FOXM1 and critically contributes to the pathogenesis and aggressive biology of pancreatic cancer.
The transcription factor FOXM1 is a key cell-cycle regulator at both the G1/S and G2/M phase (31, 32). Besides its involvement in cell-cycle transitions, FOXM1 plays important roles in tumor angiogenesis, EMT, invasion, and metastasis (9, 10). Lines of evidence demonstrate that FOXM1 is highly expressed in human lung carcinoma (11), cervical cancer (12), gastric cancer (13), glioblastoma (14), and a number of other human tumors (33), suggesting that FOXM1 is involved in the initiation and progression of many different tumors. In the present study, we found that FOXM1 was highly expressed in human pancreatic tumors. We identified FOXM1c as the predominantly expressed isoform of FOXM1 in these tumors, as FOXM1a and FOXM1b were expressed at insignificant levels. The high expression of FOXM1c was significantly correlated with histological differentiation, clinical stage, and lymph node or distant metastasis, demonstrating that it has an important role in pancreatic cancer pathogenesis.
As described above, FOXM1 protein has three isoforms owing to differential splicing of exons Va and VIIa: FOXM1b and FOXM1c function as transcriptional activators, whereas FOXM1a is transcriptionally inactive (34). Several published studies have convincingly demonstrated that FOXM1b is the most important regulator of cellular proliferation in tumorigenesis. For example, FOXM1b induces expression of cyclin A2, JNK1, and ATF2, all of which are critical for G1/S transition and DNA replication (6). FOXM1b also induces transcription of Skp2 and Cks1, which encode for subunits of the Skp/cullin-1/F-box ubiquitin ligase complex, which is required for downregulation of expression of the Cdk2 inhibitors p21Cip1 and p27Kip1 during G1 phase (6). In addition, FOXM1b is a transcriptional activator of various genes critical for EMT, angiogenesis, and metastasis, such as caveolin-1 (18), VEGF (13, 28), and MMP-2 (31). Specifically, FOXM1b may contribute to human cancer progression by upregulating expression of these three genes. Taken together, these published results demonstrated that FOXM1b regulates the expression of proteins required for tumor growth, EMT, angiogenesis, and metastasis and plays important roles in tumorigenesis.
FOXM1c possesses a very strong C-terminal transactivation domain, but full-length FOXM1c is only a weak transactivator under normal physiological conditions because its transactivation domain is completely inhibited by the autoinhibitory N-terminus. However, a recent study demonstrated that cyclin E/Cdk2, cyclin A/Cdk2, and cyclin A/Cdk1 activate FOXM1c (35). In addition, protein kinases such as CK2, PKA, c-Src, and Raf-1 strongly activated FOXM1c by phosphorylating the FOXM1c N-terminus and reversing inhibition of FOXM1c activity by its N-terminus (36). Because cell cyclins and protein kinases in cancer cells are overactivated during tumorigenesis, they may activate FOXM1c and subsequently promote cancer cell growth and metastasis during tumor development and progression. Recently, Wierstra and Alves (37) found that FOXM1c regulates the activity of c-Myc, a key regulator of cell proliferation and differentiation, by directly transactivating the c-Myc promoter via the P1 and P2 TATA boxes. Functionally, studies have demonstrated that enforced expression of FOXM1c in cervical and ovarian cancer cells enhances their proliferation, anchorage-independent growth, and migration and/or invasion ability (29, 38).
In the present study, we found that FOXM1c was the predominantly expressed isoform of FOXM1 in human pancreatic tumors and cancer cells. In addition, the relative level of FOXM1c in the highly metastatic human pancreatic cancer cell line was significantly higher than that in the poorly metastatic human pancreatic cancer cell line. Moreover, we found that FOXM1b and FOXM1c promoted the migration, invasion, and metastasis of pancreatic cancer cells, whereas FOXM1a did not have any such effects. These results demonstrate that both FOXM1b and FOXM1c play important roles in pancreatic cancer cell biology, whereas FOXM1c is more relevant to pancreatic cancer development and progression given that only FOXM1c was overexpressed in pancreatic cancer cells.
Although FOXM1a does not significantly affect the biology of or targeted gene expression in pancreatic cancer cells, it may act as a dominant-negative variant, as it retains normal DNA-binding activity in the absence of a functional transactivation domain (34). This natural splice variant may therefore modulate FOXM1 activity during tumorigenesis, most likely owing to competition with FOXM1b or FOXM1c for binding to FOXM1-binding sites. Therefore, induction of the activity of FOXM1a may be an interesting and effective way to treat human tumors overexpressing FOXM1, such as pancreatic tumors. Interestingly, FOXM1b1 and FOXM1b2 are closely related to FOXM1b and exhibit similar functions, but the significance of their existence in cancer cells remains to be determined. While determining how the interaction and isoform switch among FOXM1a and FOXM1b or FOXM1c contribute to pancreatic carcinogenesis is highly significant, further investigations are equally important and clearly warranted to determine the epigenetic mechanisms underlying differential FOXM1 expression in both normal and tumor cells.
Finally, researchers have proposed that many mechanisms regulate FOXM1 expression (39–42). In the present study, we found that KLF4 negatively regulates the expression of FOXM1, whereas Sp1 does the opposite. Given that KLF4 expression is frequently lost but Sp1 is overexpressed in pancreatic cancer cells (43–46), our data strongly suggest that the combination of reduced KLF4 expression and increased Sp1 expression is a major molecular mechanism of FOXM1 overexpression in these cells. Thus, the novel KLF4/Sp1/FOXM1c pathway may critically contribute to pancreatic cancer development and progression.
In summary, this study provided critical insight into the role of the FOXM1 isoforms in pancreatic cancer pathogenesis and the critical role of the novel KLF4/Sp1/FOXM1c signaling pathway (Supplementary Fig. S7) in pancreatic cancer development and progression. Our study not only describes a detailed molecular mechanism of the distinct expression of the FOXM1 isoforms in pancreatic cancer cells but also uncovers new aberrant FOXM1c signaling pathway as a promising molecular target for novel therapeutic modalities to control pancreatic cancer progression. Thus, this study is fundamentally important, not only in furthering our understanding of the expression and regulation of the important transcription factor FOXM1 but also in identifying an important molecular target for effective therapeutic strategies directed against pancreatic cancer.
Supplementary Material
Acknowledgments
We thank Don Norwood for editorial comments.
Grant Support: The work is supported in part by grant number 30910103911 (to Z. Li) from the National Natural Science Foundation of China, grant number IRT1051 from Innovative Team Plan of the China Ministry of Education, and by grants R01-CA129956, R01-CA148954, R01-CA152309 and R01-CA172233 (to K. Xie) and R01-CA116528 and R01-CA157933 (to S. Huang) from the National Cancer Institute, National Institutes of Health.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Nieto J, Grossbard ML, Kozuch P. Metastatic pancreatic cancer 2008: is the glass less empty. Oncologist. 2008;13:562–576. doi: 10.1634/theoncologist.2007-0181. [DOI] [PubMed] [Google Scholar]
- 3.Pliarchopoulou K, Pectasides D. Pancreatic cancer: current and future treatment strategies. Cancer Treat Rev. 2009;35:431–436. doi: 10.1016/j.ctrv.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 5.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 6.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FOXM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 2011;10:396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychaudhuri P, Park HJ. FOXM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koo CY, Muir KW, Lam EW. FOXM1: from cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 12.Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, et al. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, et al. Critical role and regulation of transcription factor FOXM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FOXM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 15.Priller M, Poschl J, Abrao L, von Bueren AO, Cho YJ, Rutkowski S, et al. Expression of FOXM1 is required for the proliferation of medulloblastoma cells and indicates worse survival of patients. Clin Cancer Res. 2011;17:6791–6801. doi: 10.1158/1078-0432.CCR-11-1214. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–8300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 17.Bao B, Wang Z, Ali S, Kong D, Banerjee S, Ahmad A, et al. Over-expression of FOXM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem. 2011;112:2296–2306. doi: 10.1002/jcb.23150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Huang C, Qiu Z, Wang L, Peng Z, Jia Z, Logsdon CD, et al. A novel FOXM1-Caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 2012;72:655–665. doi: 10.1158/0008-5472.CAN-11-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 22.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan RT, Woods LK, Moore GE, Quinn LA, McGavran L, Gordon SG. Human cell line (COLO 357) of metastatic pancreatic adenocarcinoma. Int J Cancer. 1980;25:591–598. doi: 10.1002/ijc.2910250507. [DOI] [PubMed] [Google Scholar]
- 24.Vezeridis MP, Tzanakakis GN, Meitner PA, Doremus CM, Tibbetts LM, Calabresi P. In vivo selection of a highly metastatic cell line of a human pancreatic carcinoma in the nude mouse. Cancer. 1992;69:2060–2063. doi: 10.1002/1097-0142(19920415)69:8<2060::aid-cncr2820690810>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Wei D, Crum VE, Richardson EL, Xiong HH, Luo Y, et al. A novel model system for studying the double-edged roles of nitric oxide production in pancreatic cancer growth and metastasis. Oncogene. 2003;22:1771–1782. doi: 10.1038/sj.onc.1206386. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 27.Marvin MR, Southall JC, Trokhan S, DeRosa C, Chabot J. Liver metastases are enhanced in homozygous deletionally mutant ICAM-1 or LFA-1 mice. J Surg Res. 1998;80:143–148. doi: 10.1006/jsre.1998.5322. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, et al. FOXM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–8742. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 2008;68:4631–4639. doi: 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei D, Wang L, Kanai M, Jia Z, Le X, Li Q, et al. KLF4alpha up-regulation promotes cell cycle progression and reduces survival time of patients with pancreatic cancer. Gastroenterology. 2010;139:2135–2145. doi: 10.1053/j.gastro.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FOXM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–6219. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 32.Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388:1257–1274. doi: 10.1515/BC.2007.159. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Ahmad A, Li Y, Banerjee S, Kong D, Sarkar FH. Forkhead box M1 transcription factor: a novel target for cancer therapy. Cancer Treat Rev. 2010;36:151–156. doi: 10.1016/j.ctrv.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laoukili J, Stahl M, Medema RH. FOXM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Wierstra I, Alves J. FOXM1c is activated by cyclin E/Cdk2, cyclin A/Cdk2, and cyclin A/Cdk1, but repressed by GSK-3alpha. Biochem Biophys Res Commun. 2006;348:99–108. doi: 10.1016/j.bbrc.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Wierstra I. The transcription factor FOXM1c is activated by protein kinase CK2, protein kinase A (PKA), c-Src and Raf-1. Biochem Biophys Res Commun. 2011;413:230–235. doi: 10.1016/j.bbrc.2011.08.075. [DOI] [PubMed] [Google Scholar]
- 37.Wierstra I, Alves J. FOXM1c transactivates the human c-myc promoter directly via the two TATA boxes P1 and P2. FEBS J. 2006;273:4645–4667. doi: 10.1111/j.1742-4658.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 38.Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, et al. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- 39.Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol. 2008;28:5162–5171. doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gieling RG, Elsharkawy AM, Caamaño JH, Cowie DE, Wright MC, et al. The c-Rel subunit of nuclear factor-kappaB regulates murine liver inflammation, wound-healing, and hepatocyte proliferation. Hepatology. 2010;51:922–931. doi: 10.1002/hep.23385. [DOI] [PubMed] [Google Scholar]
- 41.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. FOXM1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, et al. Plk1-dependent phosphorylation of FOXM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–1082. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C, Xie K. Crosstalk of Sp1 and Stat3 signaling in pancreatic cancer pathogenesis. Cytokine Growth Factor Rev. 2012;23:25–35. doi: 10.1016/j.cytogfr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y, Jia Z, Kong X, Li Q, Chang DZ, Wei D, et al. Combining betulinic acid and mithramycin a effectively suppresses pancreatic cancer by inhibiting proliferation, invasion, and angiogenesis. Cancer Res. 2011;71:5182–5193. doi: 10.1158/0008-5472.CAN-10-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang NY, Woda BA, Banner BF, Whalen GF, Dresser KA, Lu D. Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:1648–1652. doi: 10.1158/1055-9965.EPI-07-2791. [DOI] [PubMed] [Google Scholar]
- 46.Xie K, Wei D, Huang S. Transcriptional anti-angiogenesis therapy of human pancreatic cancer. Cytokine Growth Factor Rev. 2006;17:147–156. doi: 10.1016/j.cytogfr.2006.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.