Abstract
RECQL5 is a member of the highly conserved RecQ family of DNA helicases involved in DNA repair. RECQL5 interacts with RNA polymerase II (Pol II) and inhibits transcription of protein–coding genes by an unknown mechanism. We show that RECQL5 contacts the Rpb1 jaw domain of Pol II at a site that overlaps with the binding site for the transcription elongation factor TFIIS. Our cryo–electron microscopy structure of elongating Pol II arrested in complex with RECQL5 shows that the RECQL5 helicase domain is positioned to sterically block elongation. The crystal structure of the RECQL5 KIX domain reveals similarities with TFIIS, and binding of RECQL5 to Pol II interferes with the ability of TFIIS to promote transcriptional read–through in vitro. Together, our findings reveal a dual mode of transcriptional repression by RECQL5 that includes structural mimicry of the Pol II–TFIIS interaction.
Keywords: transcription regulation, RNA polymerase II, electron microscopy, X–ray crystallography, RecQ helicase, DNA repair, TFIIS
INTRODUCTION
RecQ helicases constitute a family of highly conserved 3’–5’ DNA helicases that are involved in maintaining genomic stability1. While unicellular organisms such as E. coli have only a single RecQ helicase, multicellular organisms possess several RecQ homologs with non–redundant functions that unwind different kinds of potentially recombinogenic DNA structures1,2. The human genome contains five helicases of the RecQ family, named RecQ1, BLM (RecQ2), WRN (RecQ3), RecQ4 and RECQL5. Mutations in the BLM, WRN and RecQ4 genes are associated with the hereditary disorders Bloom’s syndrome3, Werner’s syndrome4 and Rothmund–Thomson syndrome5, respectively. Patients that carry these syndromes suffer from photosensitivity, have a strong predisposition to develop cancer, and often show signs of premature aging, emphasizing the important roles that RecQ helicases play in DNA repair6. Although no human pathologies have been associated with mutations in RECQL5, the observation that RECQL5−/− mice show increased levels of sister chromatid exchange as well as a predisposition to tumor formation strongly argue for an important role of RECQL5 in the maintenance of genomic stability7,8.
RECQL5 plays a crucial role at the interface of the cellular replication, transcription and recombination machineries. It physically interacts with the DNA sliding clamp PCNA, which is involved in DNA replication and repair9. In addition, the MRN complex, which acts as a DNA double–strand break sensor, has been found to link RECQL5 to the DNA repair machinery10. Furthermore, RECQL5 interacts with RAD51 and can disrupt RAD51 filaments assembled on ssDNA, thereby inhibiting homologous recombination and preventing untimely recombination events8,11.
Recently, RECQL5 was found to interact directly with RNA polymerase II (Pol II) and to inhibit both the initiation and elongation steps of transcription in an in vitro system12–14. RECQL5 is the only one of the human RecQ helicases that can interact with Pol II, hinting at a crucial and specific role for RECQL5 in transcription regulation. The precise interaction surface on Pol II as well as the molecular mechanisms of inhibition remain unclear.
In addition to the canonical DExH helicase and RecQ C–terminal (RQC) domains at the N–terminus, RECQL5 has a long C–terminal extension that is unrelated in sequence to any other RecQ helicase (Fig. 1a). This extension contains two domains that enable RECQL5 to interact with Pol II: a so–called internal RNA polymerase II interacting domain (IRI), and a Set2–Rpb1–interacting (SRI) domain. While the SRI domain binds to the phosphorylated C–terminal domain of Pol II, the IRI interacts with both the phosphorylated and unphosphorylated forms and is therefore thought to bind to the “core” of Pol II.
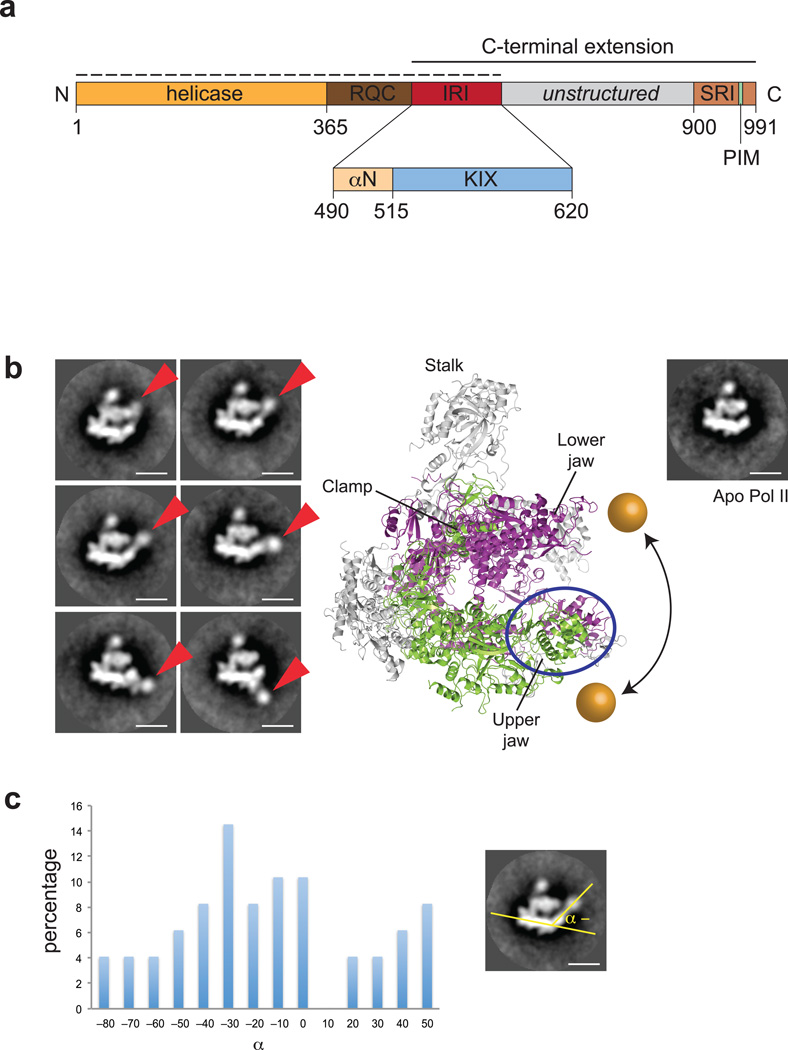
Figure 1. An extended helicase domain fragment of RECQL5 interacts with the upper jaw domain of Pol II.
(a) Domain organization of human RECQL5. RQC, RecQ carboxy–terminal domain; IRI, internal Pol II interaction domain; PIM, PCNA interaction motif; SRI, Set2–Rpb1–interacting domain. Dashed line indicates the construct used for EM studies. Inset shows the organization of the IRI domain, consisting of an N–terminal helix (αN) and the KIX domain (light blue). The crystallized KIX domain includes RECQL5 residues 515–620. (b) Representative 2–D class averages of negatively stained Pol II–RECQL51–620 complex. The RECQL5 helicase domain is indicated by red arrows. The scale bar indicates a length of 100 Å. A crystal structure of yeast Pol II (PDB code 1WCM) and a class average of negatively stained apo Pol II are shown for comparison. Rpb1 is colored in purple, Rpb2 in green and all remaining subunits in grey. The range of movement of the helicase domain is indicated by the golden spheres. (c) Statistical analysis of reference–free 2D class averages reporting on the position of RECQL5 density based on the angle defined schematically on the right.
To gain insight into the molecular mechanism of transcriptional repression by RECQL5, we have defined and structurally characterized the Pol II–RECQL5 IRI interaction surfaces through a combination of biochemical assays, X–ray crystallography and electron microscopy. We show that RECQL5 interacts through its IRI domain with the Rpb1 jaw domain of Pol II, while the RECQL5 helicase domain engages the DNA entering the Pol II downstream cleft. Mutations disrupting the RECQL5–Pol II interaction strongly impair the ability of RECQL5 to inhibit transcription in vitro. In both pull–down assays and a functional transcription read–through assay, RECQL5 competes with the general transcription elongation factor TFIIS for binding to Pol II, suggesting a dual mode of transcriptional repression by RECQL5 that relies, in part, on interfering with the physiological function of TFIIS in promoting transcriptional elongation.
RESULTS
RECQL5 IRI interacts with the Rpb1 jaw domain of Pol II
The N–terminal portion of human RECQL5 contains canonical RecQ helicase and RQC domains that are shared with other RecQ–family helicases; this conserved core is followed by a unique C–terminal extension that consists of three domains: the IRI domain, a ~280–residue long unstructured domain, and the SRI domain (Fig. 1a). To delineate the exact domain boundaries of the RECQL5 IRI domain required for Pol II interaction, we employed a GST pull–down assay to test binding of a series of deletion constructs to human Pol II (hPol II; Supplementary Fig. 1a). While the helicase domain (RECQL51–490) did not bind to Pol II, a protein construct encompassing the minimal IRI domain (RECQL5490–620) was able to interact with Pol II. Secondary structure predictions and homology modeling indicated that the IRI domain is composed of a previously identified three–helix bundle termed KIX domain, and an additional N–terminal helix (termed αN), connected to the KIX domain by a loop segment. Neither the KIX domain alone (RECQL5515–620) nor the N–terminal helix alone (RECQL5490–515) were sufficient for binding to Pol II.
To determine the interaction site of RECQL5 on Pol II, we analyzed the Pol II–RECQL51–620 complex using negative–staining electron microscopy (EM) and reference–free 2D classification (Fig. 1b). In comparison with a class average of apo hPol II or the crystal structure of yeast Pol II (yPol II), a subset of 2D class averages showed clear additional density in the vicinity of the downstream DNA cleft, pivoting around the jaw domain of Pol II and adopting a wide range of different positions relative to Pol II. Statistical analysis showed that while this density has a slight preference for a range of about 40 degrees around its pivot point, it can adopt positions that differ by as much as 130 degrees (Fig. 1c). The additional density is consistent in size with the molecular weight of the RECQL5 helicase domain, and its movement around the upper jaw domain of Pol II suggests that the jaw domain is the likely binding site for the RECQL5 IRI domain.
RECQL5 KIX domain resembles TFIIS domain II
To gain insights into the architecture of the RECQL5 IRI domain, we crystallized a fragment containing the KIX domain and the long N–terminal loop (RECQL5515–620); this construct lacks the N–terminal helix αN that is necessary in addition to the KIX domain for binding to Pol II. The crystal structure, refined to 1.9 Å resolution, shows a domain–swapped dimer of two three–helix bundles connected to each other, whereby helix α1A of molecule A interacts with helices α2B and α3B of molecule B, and vice versa (Fig. 2a). To determine the oligomeric state of RECQL5515–620 in solution, we used small–angle X–ray scattering (SAXS). Analysis of scattering profiles at different concentrations showed that the RECQL5 KIX domain exists as a concentration–dependent dimer (Supplementary Fig. 1b – d). At low concentrations (~280 µM), the KIX domain is predominantly monomeric, but it shifts towards the dimeric state at higher concentrations (~1.4 mM). At the concentrations used for crystallization (~2 mM), the KIX domain is predominately dimeric in solution. To determine whether the dimeric state is a unique feature of the KIX domain or whether it might also be adopted by longer RECQL5 fragments, we used SAXS analysis to determine the oligomeric state of RECQL51–620 (Supplementary Fig. 1b and e). The Porod volume of the scattering data (~115,000 Å3) was consistent with the 68 kDa size of RECQL51–620, suggesting that full–length RECQL5 is monomeric.
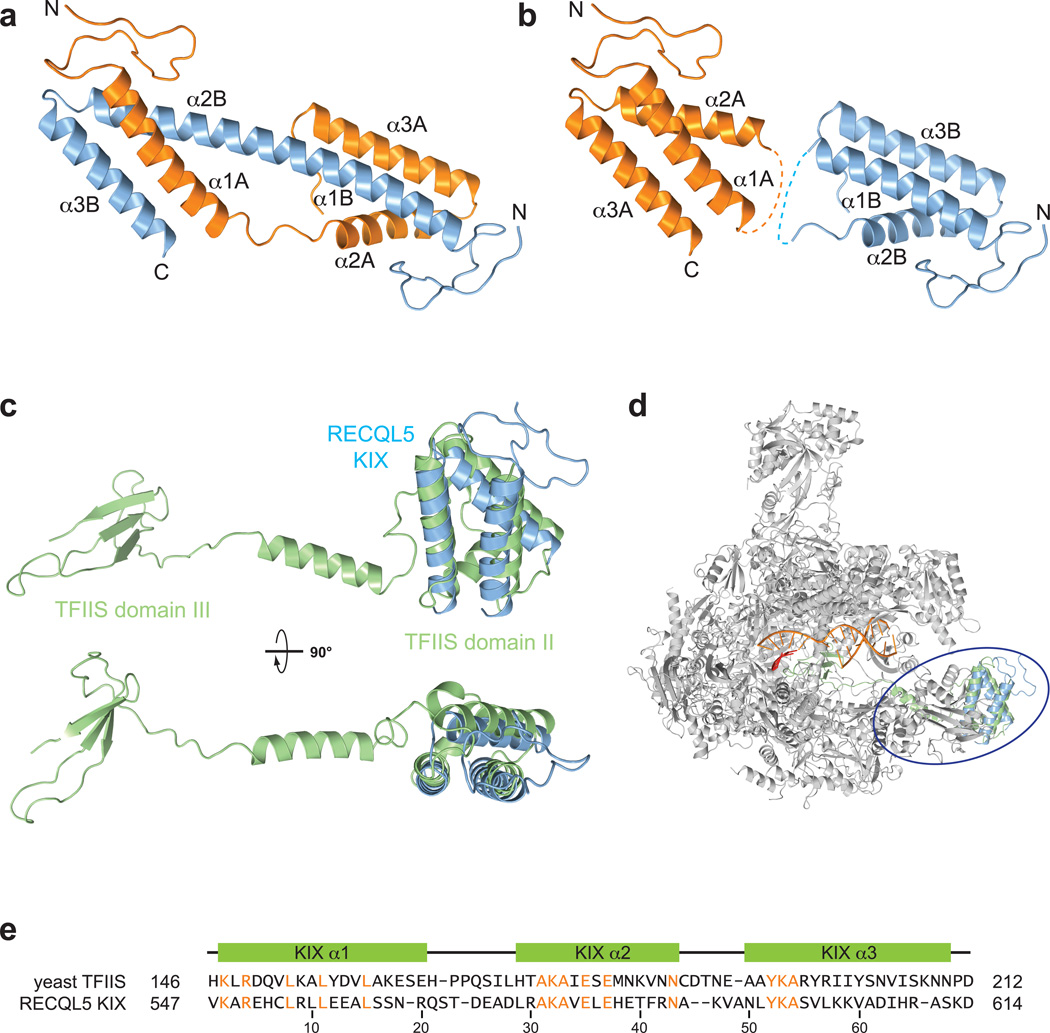
Figure 2. The RECQL5 KIX domain resembles domain II of TFIIS.
(a) Crystal structure of the domain–swapped RECQL5 KIX dimer. Alpha–helical segments are labeled for the two chains. (b) KIX domain monomers derived from the dimeric crystal structure. Dashed lines indicate loops connecting helices α1 and α2 in KIX domain monomers. (c) Superposition of TFIIS (green, PDB code 3PO3) and the RECQL5 KIX domain shown in two orthogonal orientations. (d) Superposition of the RECQL5 KIX domain (blue) onto domain II of TFIIS (green) in complex with Pol II (grey, PDB code 3PO3). (e) Structure–based sequence alignment of domain II of yeast TFIIS and the human RECQL5 KIX domain. Identical residues are highlighted in orange. Secondary structure elements are shown relative to the RECQL5 sequence.
Although our crystal structure shows a domain–swapped dimer, the topology of a monomeric KIX domain can be reconstructed from this model by linking helix α1A of molecule A with helices α2B and α3B of molecule B (Fig. 2b). A search for structural homologs using the DALI server15 revealed that the tertiary structure of monomeric KIX domain resembles domain II of the transcription elongation factor TFIIS, as observed in a co–crystal structure of backtracked yPol II in complex with TFIIS (PDB code 3PO3; Fig. 2c and d). TFIIS is an elongation factor that helps the polymerase to overcome stalling and resume elongation through transcriptional pause sites16,17. Pol II stalling can occur due to nucleotide misincorporation or upon encountering DNA lesions or DNA-binding proteins, and leads to Pol II backtracking and eventual arrest at the stall site18,19. TFIIS resolves the paused state of Pol II by stimulating the cleavage of the backtracked RNA transcript near its 3’-end, allowing Pol II to resume transcription20. Additionally, TFIIS has been implicated to play a role in transcription initiation via its domain I21,22. The three–helix bundles of the RECQL5 KIX domain and domain II of TFIIS align with an rmsd of 2.7 and a Z–score of 4.4 over 65 residues. A structure–based sequence alignment shows a number of conserved residues between the RECQL5 KIX domain and domain II of yeast and metazoan TFIIS orthologs (Fig. 2e, Supplementary Fig. 2a). Domain II of TFIIS binds to the Rpb1 jaw domain of Pol II23, near the RECQL5 binding site identified in the reference–free 2D class averages of the Pol II–RECQL51–620 complex (Fig. 1b).
RECQL5 IRI contacts the TFIIS–binding site on Pol II
To characterize the interaction of the RECQL5 IRI domain and the Pol II Rpb1 jaw biochemically, we designed a codon–optimized construct of the Rpb1 jaw domain (hRpb11168–1302) for recombinant expression in E. coli. The purified Rpb1 jaw domain fragment interacted with both RECQL51–620 and RECQL5490–620 in GST pull–down assays (Fig. 3a). The reciprocal pull–down experiment gave identical results; both GST–fusions of RECQL51–620 and RECQL5490–620 bound to the recombinant Rpb1 fragment. These experiments confirmed that RECQL5 IRI binds to the Rpb1 jaw domain, and led us to construct a structural model of the Rpb1–IRI interaction based on the crystal structure of the Pol II–TFIIS complex (PDB code 3PO3, Fig. 3b). This model suggested that the helices α1 and α3 of the KIX domain contact the Rpb1 jaw domain in the Pol II–RECQL5 complex (Fig. 3b).
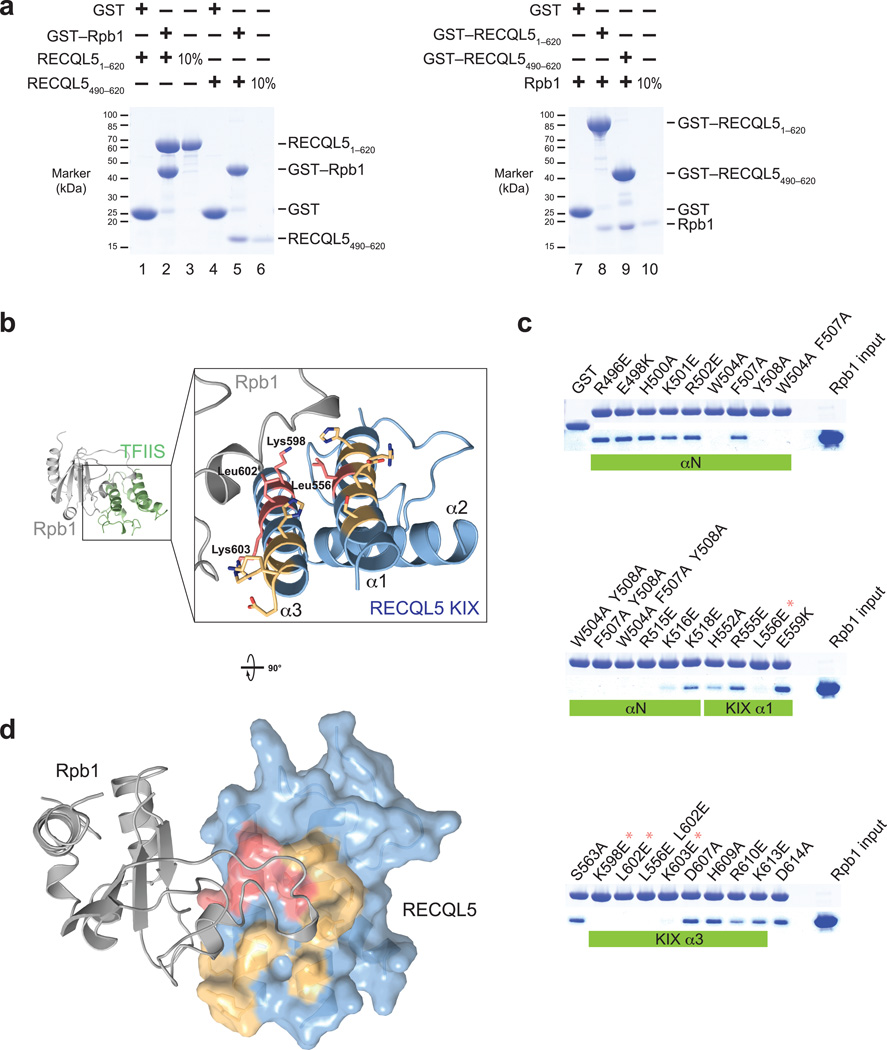
Figure 3. The RECQL5 IRI domain binds to a loop element in the Rpb1 jaw of Pol II.
(a) Pull–down assays of two different RECQL5 fragments and a recombinantly expressed Rpb1 fragment (residues 1168–1302). Left: Recombinant glutathione–S–transferase (GST)–tagged Rpb11168–1302 was immobilized on glutathione sepharose beads and incubated with RECQL51–620 or the RECQL5 IRI domain (residues 490–620). Bound proteins were analyzed by SDS–PAGE and visualized by Coomassie staining. Right: GST–fused RECQL51–620 or RECQL5490–620 were immobilized on glutathione sepharose beads and incubated with untagged recombinant Rpb11168–1302. (b) Model of the RECQL5 KIX–Rpb1 interaction surface based on the TFIIS–Pol II crystal structure, generated by superposition of the KIX domain and TFIIS domain II. Surface residues mutated in (c) are shown in stick representation. RECQL5 residues whose mutations abolish interaction with hRpb1 are highlighted in red, and mutated residues that have no effect on interaction with Rpb1 are shown in yellow. (c) Pull–down assays of Rpb11168–1302 by GST–RECQL5490–620 IRI domain mutants. Large hydrophobic residues important for interaction (W504, F507, Y508) are located in the first alpha–helix of the IRI domain (αN, not present in our crystal structure). Asterisks denote mutations that abolish interaction with Rpb1 and are part of the KIX domain (see b). (d) Model of the RECQL5 KIX–Rpb1 interaction. The KIX domain is depicted as a surface representation and colored as in (b). Mutations that abolish interaction cluster in a single patch on the KIX domain surface.
To further define the binding interface, we tested the ability of a panel of surface mutants of the RECQL5 IRI domain to bind to the recombinant Rpb1 fragment (Fig. 3c). Mutations of aromatic residues in the N–terminal helix αN abolished the interaction with the Rpb1 fragment. In addition, mutations of several charged residues in helices α1 and α3 of the KIX domain led to the loss of binding. Mapping of the mutations onto the surface of RECQL5 KIX showed that residues important for the interaction cluster in a conserved patch located close to the Rpb1 loop element in our docking model (Fig. 3b and d; Supplementary Fig. 3b). Interestingly, this patch coincides with an electropositive patch formed by residues K196, R198, R200, and K209 in yeast TFIIS (Supplementary Fig. 2d); single point mutations in any of these four residues drastically decrease the affinity of the TFIIS–Pol II interaction24. This is consistent with our observation that RECQL5 IRI domain mutants K598E and L602E, which are located at structurally equivalent positions to yeast TFIIS K196 and R200, are impaired in their interaction with Pol II.
Both RECQL5 KIX and domain II of TFIIS form three–helix bundles that are by themselves insufficient to interact stably with Pol II (Supplementary Fig. 1a and Supplementary Fig. 2c). While RECQL5 requires the additional N–terminal helix αN, in TFIIS the additional element required for binding is located at the C–terminus of the three–helix bundle (Supplementary Fig. 2)24. The differing topologies of the Pol II binding modules suggest that the binding sites for the two proteins overlap, but are not identical (Supplementary Fig. 2e).
EM reconstruction of RECQL5 bound to elongating Pol II
To visualize the Pol II–RECQL5 binding interface and to study the effect of RECQL5 binding on the structure and conformation of Pol II, we determined the structure of an elongating Pol II in complex with a fragment of RECQL5 containing the N–terminal helicase, RQC and IRI domains (RECQL51–620). To stabilize the helicase domain with respect to Pol II, we used an artificial transcription bubble containing template and non–template DNA strands as well as product RNA25. The non–template DNA strand contained a single–stranded overhang at its 3’ end to allow the RECQL5 helicase domain to engage to DNA exiting the Pol II cleft. The helicase activity of RECQL5 was abolished by introducing the inactivating single point mutation D157A14. A silver–stained gel and electron micrographs of a uranyl formate–stained Pol II–RECQL51–620 complex showed that the sample was stoichiometric and homogeneous (Supplementary Fig. 5). Statistical analysis of reference–free 2D class averages revealed that in contrast to the Pol II–RECQL5 complex lacking the transcription bubble DNA (Fig. 1c), the RECQL5 helicase domain is found within a narrow range of angles around its pivot point, and positioned close to the DNA downstream cleft (Supplementary Fig. 5c and d). A data set collected from the uranyl formate–stained sample was used to reconstruct an initial model of the complex, which was then further refined to ~13Å resolution using a larger cryo–EM data set (Supplementary Fig. 5). The 3D reconstruction of the elongating complex stalled by RECQL5 displayed clear additional density downstream of the cleft, positioned between the two Pol II jaws (Fig. 4). Local resolution analysis revealed that this domain has a lower resolution than other parts of the complex (Supplementary Fig. 5h), most likely due to the remaining flexibility of the helicase domain with respect to Pol II. However, the density is consistent in size with the two RecA–like domains that form the RECQL5 helicase core (Fig. 4b). In addition to the density between the jaws, extra density is also observed near the Rpb1 jaw at a site that overlaps with the known binding surface for domain II of TFIIS25,26. This density is large enough to accommodate the RQC domain as well as helix αN and the KIX domain of RECQL5. Using the crystal structure of the yPol II–TFIIS complex and our structural alignment between TFIIS and the RECQL5 KIX domain, we docked our crystal structure of RECQL5 KIX into this part of the EM map.
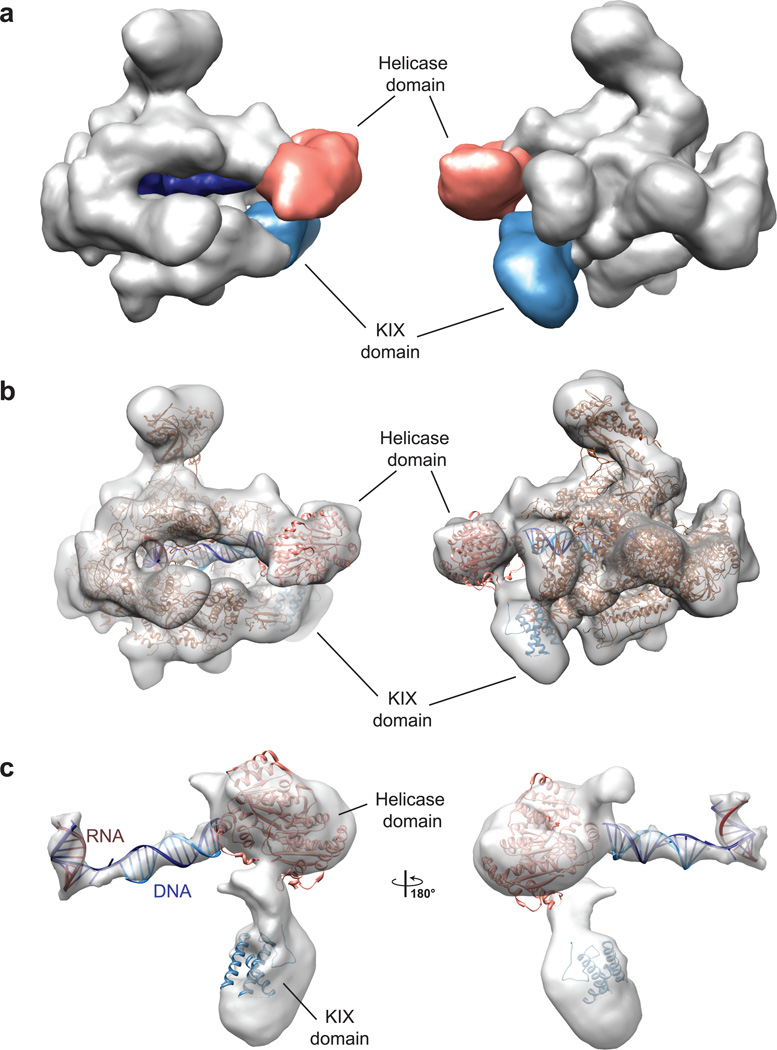
Figure 4. Cryo–EM reconstruction of RECQL5–stalled Pol II elongation complex.
(a) Surface representation of the Pol II–RECQL5 elongation complex shown in two different views. Density not corresponding to Pol II is ascribed to RECQL5 (salmon and light blue) and DNA (dark blue). (b) Cryo–EM map with docked crystal structures. The atomic model of elongating Pol II is based on the crystal structure of yPol II in complex with a transcription bubble (PDB code 1Y1W). The monomeric KIX domain (as shown in Fig. 2) was docked into the additional density observed at the Rpb1 jaw domain, and the helicase domain of RecQ1 (based on PDB code 2WWY) was docked into the density between the Pol II jaws. (c) Close–up view of the additional density with docked crystal structures and an idealized dsDNA and RNA–DNA duplex.
In comparison to apo–Pol II, we also observed additional density in the downstream cleft corresponding to double–stranded DNA, as well as a DNA–RNA duplex in the upstream cleft close to the active site. Interaction of the RECQL5 helicase domain with the single–stranded 3’ overhang of the non–template strand stabilized the DNA beyond the 7 base pairs that were observed in the crystal structure and led us to model the additional double–stranded DNA as ideal B–form DNA for a total of 11 base pairs.
Overall, our EM reconstruction clearly pinpoints the Rpb1 jaw domain as the interaction surface of the RECQL5 IRI domain, and it shows that once RECQL5 is tethered to Pol II through this domain, its helicase domain is located in a position that allows it to engage with DNA entering the Pol II cleft.
IRI domain orients helicase core for repression
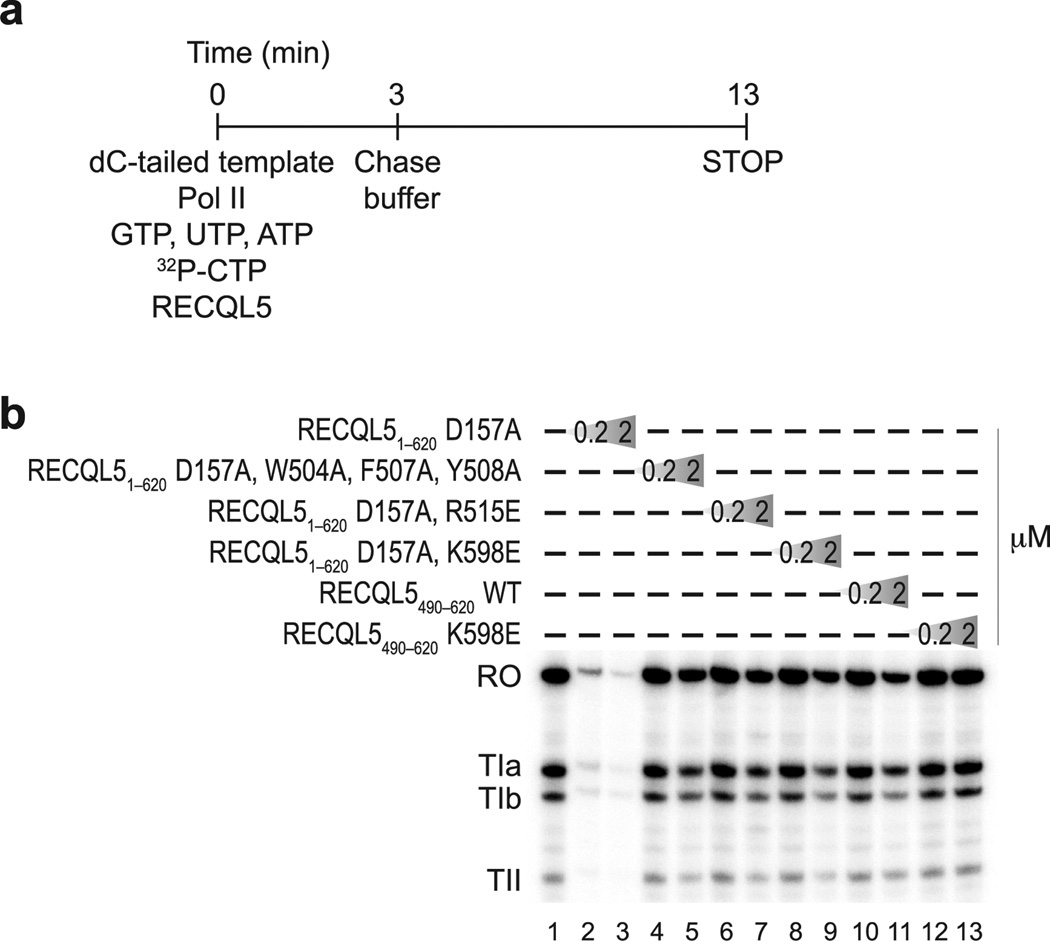
To validate our structural model and confirm the functional significance of IRI domain mutations that result in defects in Pol II binding, we performed an in vitro transcription assay (Fig. 5). We used the well–studied first intron of the human H3.3 gene as a template27, which contains three defined elongation blocks, termed TIa, TIb and TII. While the TIb and TII sites stall Pol II with low efficiency (10% or less), elongation is blocked with ~50% efficiency at the TIa site20. Due to the presence of stall sites in the template, this assay leads to the production of three different stalled transcripts (TIa, TIb, and TII) as well as the run–off transcript. To exclude the possibility that the RECQL5 helicase activity could interfere with transcription, we used the RECQL5 D157A mutant, which can interact with DNA, but is unable to unwind it due to its inability to hydrolyze ATP. In agreement with previous studies14, the assay demonstrated that RECQL51–620 D157A potently repressed transcription, confirming that the helicase activity is not required for repression. Mutant proteins that were unable to interact with Pol II in our pull–down assay were considerably less effective in repressing transcription. The RECQL5 IRI domain alone could repress transcription only when present in high molar excess, while the RECQL5 IRI K598E mutant had no repressive effect at all, further corroborating the importance of this residue for interaction with Pol II. Overall, the results indicated that specific intermolecular contacts between the RECQL5 IRI domain and Pol II, predicted by our structural model, are necessary for transcriptional repression. However, repression required both the helicase core (helicase and RQC domains) and the IRI domain, even though only the IRI domain physically interacts with Pol II. In conjunction with the structure of the Pol II–RECQL5 complex, this suggests that the function of the IRI domain is to recruit the RECQL5 helicase core to Pol II and orient it such that it can engage the DNA template ahead of transcribing Pol II.
Figure 5. Both RECQL5 helicase and IRI domains are required for repression of transcription.
(a) Schematic overview of the in vitro transcription assay. The dC–tailed histone H3.3 intron DNA was used as template and transcribed by purified Pol II (0.8 pmol) in the presence of recombinant wild–type and mutant RECQL5 proteins at two different concentrations (0.2 µM and 2 µM, corresponding to a 5–fold and 50–fold molar excess over Pol II, respectively). (b) Transcription assay performed as outlined in (a) in the presence of recombinant RECQL5 fragments and their mutants, as indicated. Transcripts were resolved by electrophoresis on a denaturing polyacrylamide gel and visualized by phosphorimaging. The transcription template contains three stall sites, giving rise to stalled transcripts TIa, TIb, and TII in addition to the run–off transcript (RO).
RECQL5 interferes with TFIIS function in elongation
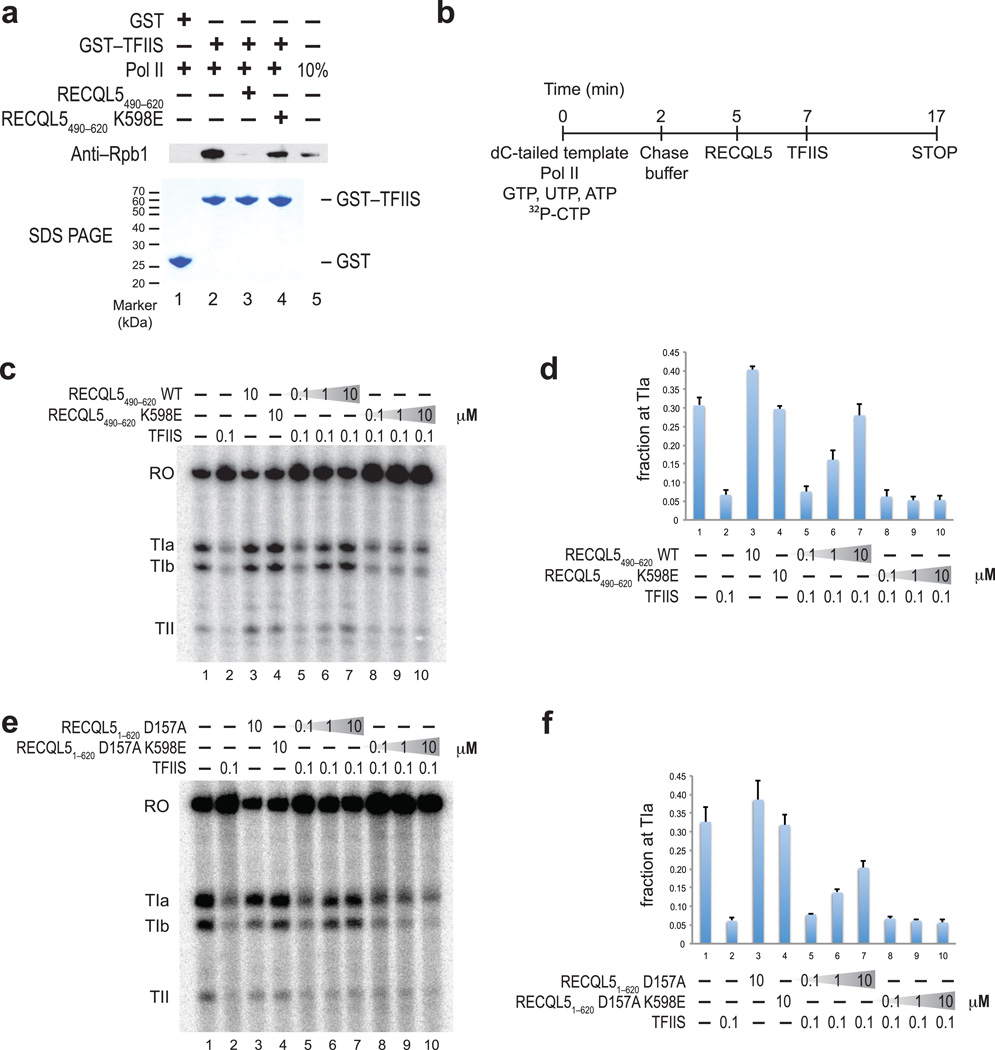
The structural similarity of the RECQL5 IRI domain and TFIIS suggested that the binding of these two proteins to Pol II is mutually exclusive. To test this hypothesis, we employed a GST–pulldown assay (Fig. 6a). While GST–TFIIS could bind to Pol II in the absence of RECQL5, binding was abrogated when Pol II was preincubated with RECQL5 IRI. In contrast, in the presence of an IRI mutant that cannot bind to Pol II (RECQL5 K598E), Pol II was still able to associate with immobilized GST–TFIIS. Using an electrophoretic mobility shift assay, we measured the equilibrium dissociation constant of the Pol II–RECQL51–620 complex (Supplementary Fig. 6). RECQL51–620 bound to Pol II with a Kd of ~40 nM, which is comparable to the dissociation constant of 80 nM previously reported for the TFIIS–Pol II complex using the same methodology24.
Figure 6. RECQL5 IRI domain competes with TFIIS for binding to Pol II and inhibits TFIIS–mediated read–through of intrinsic elongation blocks.
(a) Pull-down assay probing Pol II binding to GST–TFIIS in the presence of either wild–type or K598E mutant RECQL5 IRI domains. Bound proteins were analyzed by SDS–PAGE, followed by staining with Coomassie Blue or Western blotting using an anti–Rbp1 antibody. (b) Schematic overview of the pulse–chase TFIIS read–through transcription assay using histone H3.3 intron DNA template. Following addition of chase buffer, RECQL5 variants (0.1 µM, 1.0 µM and 10 µM final concentration, corresponding to 5–, 50– and 500–fold molar excess over Pol II) and TFIIS (0.1 µM, 5–fold molar excess over Pol II) were added sequentially to test the effect of RECQL5 on the ability of TFIIS to promote read–through of intrinsic blocks to elongation. (c–f) Pulse–chase TFIIS read–through assay in the presence of RECQL51–620 and RECQL5 IRI performed as indicated in (b). Transcripts were resolved on a denaturing polyacrylamide electrophoresis gel and visualized by phosphorimaging. TFIIS stimulates read–through of three distinct blocks to elongation (TIa, TIb, and TII) to produce a run–off transcript (RO). (d, f) Quantitation of the relative amount of transcripts stalled at the TIa site. Error bars denote standard error of the mean (n=3).
The direct competition between RECQL5 IRI and TFIIS suggested that RECQL5 might also inhibit transcriptional elongation by blocking access of TFIIS to Pol II. To test this hypothesis, we employed a pulse–chase transcription assay to study the effect of RECQL51–620 D157A and RECQL5 IRI on TFIIS–mediated read–through of intrinsic stall sites in the histone H3.3 template DNA (Fig. 6b–f). Transcripts initially stalled at the TIa, TIb and TII sites can be elongated to the run–off transcript in the presence of TFIIS20,28. As expected, in the absence of any additional factors, Pol II was stalled at the TIa, TIb and TII sites and the stalling was overcome in the presence of TFIIS. When the RECQL5 IRI domain was titrated in, we observed increased stalling at the TIa site, indicating that the ability of TFIIS to promote read–through was impaired in the presence of the RECQL5 IRI domain. In contrast, increasing concentrations of the K598E mutant protein had no effect, indicating that inhibition is specific and requires the interaction between Pol II and the IRI domain. We also performed this assay in the context of a fragment of RECQL5 encompassing both the helicase and IRI domains. In comparison with the IRI domain in isolation, the competitive effect of RECQL51–620 D157A on transcriptional read–through was slightly less pronounced. This is likely due to RECQL51–620 D157A being sequestered by excess DNA template present in the transcription reactions, which decreases the likelihood of interaction with Pol II. Taken together, these results confirm that the RECQL5 IRI domain interferes, through direct competition, with the ability of TFIIS to promote read–through of intrinsic blocks to elongation.
DISCUSSION
In this study, we have investigated the structural basis of transcription regulation by the DNA helicase RECQL5. We demonstrate that RECQL5 interacts through its IRI domain with the Rpb1 jaw domain of Pol II at a site that overlaps with the known binding site for the elongation factor TFIIS. Our crystal structure of the RECQL5 KIX domain reveals unexpected structural similarity with domain II of TFIIS. Furthermore, we show that binding of RECQL5 and TFIIS to Pol II is mutually exclusive and that binding of RECQL5 to Pol II interferes with the ability of TFIIS to promote elongation through transcriptional stall sites in vitro. Our cryo–EM reconstruction of elongating Pol II in complex with RECQL5 shows that the KIX domain positions the RECQL5 helicase core to engage template DNA that enters the active site of Pol II. In agreement with this model, specific structure–based mutations in the IRI domain impair the ability of RECQL5 to repress transcription in vitro. Taken together, these results provide novel structural and mechanistic insights into RECQL5 function in transcriptional control and suggest that RECQL5 inhibits transcription through two concerted mechanisms.
Dual mechanism of RECQL5–mediated transcription inhibition
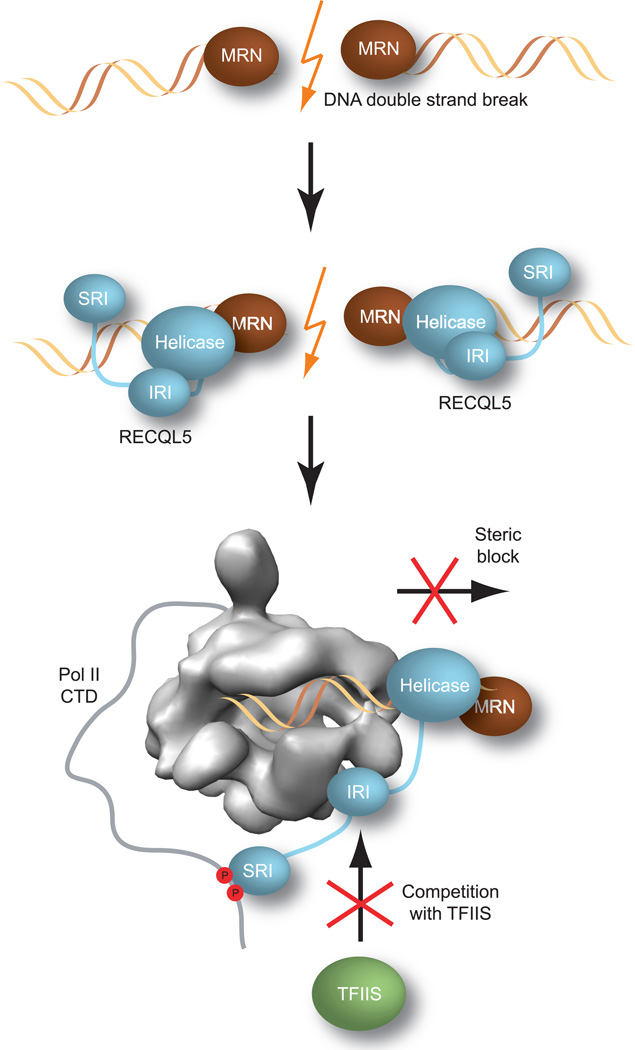
Based on our results, we propose the following mechanisms for transcription regulation by RECQL5 (Fig. 7). (i) Upon recruitment to sites of DNA damage by DNA repair factors such as the MRN complex, RECQL5 engages Pol II through its IRI and SRI domains, and its DNA-bound helicase domain sterically impedes Pol II’s progression along the DNA, thereby blocking transcription towards the repair site. It is also possible that in this context, the ATP–dependent helicase activity of RECQL5 may actively assist in removing Pol II from the DNA. (ii) Concomitantly, the binding of the RECQL5 IRI domain to the Pol II Rpb1 jaw domain occludes the TFIIS binding site on Pol II, thus preventing association with TFIIS that would otherwise stimulate Pol II to overcome stalling and resume transcription.
Figure 7. Dual mechanism of RECQL5–mediated transcriptional repression in DNA repair.
RECQL5 is recruited by the MRN complex to DNA double–strand breaks, where it inhibits transcription through two concerted mechanisms. The DNA–bound helicase domain of RECQL5 acts as a steric block to prevent Pol II from proceeding towards the site of DNA damage. Concurrently, the IRI domain of RECQL5 inhibits binding of TFIIS to Pol II, thereby preventing repeated cycles of TFIIS–mediated backtracking and elongation at the stall site.
Although both TFIIS and RECQL5 bind to Pol II with comparable affinities, and TFIIS is much more abundant than RECQL5 in the cell nucleus, co–localization of Pol II and RECQL5 on the same DNA would strongly favor binding of RECQL5 due to a local concentration effect. The Pol II–TFIIS complex is assumed to be transient, and ChIP assays have indeed shown that elongating Pol II is largely TFIIS–free under regular growth conditions29, which would again facilitate binding of RECQL5. Furthermore, RECQL5 contains the SRI domain, which specifically interacts with a Ser2–Ser5–phosphorylated CTD of Pol II, a phosphorylation state associated with the elongation stage of transcription30,31. The SRI domain might therefore prime RECQL5 for engaging specifically with elongating Pol II by providing an additional tethering site. Thus, the coordinated actions of the helicase, IRI and SRI domains would allow RECQL5 to act as a barrier to elongation that cannot be easily dislodged from the DNA template by the action of TFIIS.
RECQL5 as a transcriptional inhibitor in genomic stability
While RECQL5 has been implicated in DNA repair and the maintenance of genomic stability, its precise functions remain elusive. The helicase activity has been proposed to antagonize the recombinogenic effect of transcription during DNA replication32. RECQL5 has also been shown to be constitutively associated with the MRN complex through the Mre11 and Nbs1 subunits10. Additionally, a recent study has suggested that RECQL5 is involved in the removal of endogenous DNA damage33. It is tempting to speculate that in all these processes, the recruitment of RECQL5 to genomic sites undergoing DNA replication or repair could serve to prevent actively transcribing Pol II from entering these regions and interfering with the molecular machineries responsible for DNA replication or repair. However, it remains to be seen whether transcriptional inhibition is the main role of RECQL5 in all these processes or a separate and secondary function.
Although the IRI domain is necessary for repression, it cannot act in isolation. In turn, the RECQL5 helicase core is required for repression, but does not interact with Pol II by itself. These observations, together with our cryo–EM structure of the Pol II–RECQL5 complex, suggest that the repressive function of RECQL5 stems from its concerted interactions with Pol II and the template DNA and point to a model in which the IRI–Pol II interaction serves to position the RECQL5 helicase domain in a way that permits its association with DNA entering the Pol II active site. As a result, RECQL5 localized near a DNA damage site would be poised to capture and stall approaching Pol II elongation complexes.
Inhibition of TFIIS as a general mechanism of repression
Our studies have uncovered a novel mechanism of transcriptional repression by competition with an elongation factor, suggesting that regulation of Pol II elongation may be an important control mechanism in the context of genome maintenance. Despite the critical role of TFIIS in elongation, one can envision instances in which its stimulatory function is not desirable, for example when a transcribing Pol II encounters DNA regions undergoing repair, replication, or recombination. Competitive inhibition of TFIIS binding to Pol II could therefore offer a safeguard mechanism for preventing Pol II from clashing with replication forks or the DNA repair machinery. Based on a HHPred search34, domains with striking structural homology to TFIIS are present in several proteins that regulate DNA maintenance and repair: PHD finger protein 3 (PHF3), SPOC domain–containing protein 1 (SPOCD1) and death–induced obliterator 1 (DIO–1) in humans, and BYE1 in yeast. While BYE1 has been identified as a negative regulator of transcription elongation35, the precise functions of PHF3, SPOCD1 and DIO–1 have not been characterized. PHF3 is phosphorylated upon DNA damage but its role in DNA repair remains unclear36. SPOCD1 is a member of the SPEN domain–containing family of transcriptional repressors that are involved in developmental signaling37. DIO–1 induces apoptosis and has been implicated as a putative transcription factor38. The TFIIS–like domains in these proteins might allow them to bind Pol II and regulate transcription by occupying the binding site of TFIIS, similar to the mechanism of action we observe for RECQL5. Transcriptional repression by blocking TFIIS function might therefore be a general mode of elongation regulation in response to different cellular signals that require pausing of the transcription machinery.
ONLINE METHODS
Protein expression and purification
All protein constructs were amplified by PCR and inserted into the pGEX–6P1 expression vector (GE Healthcare) using BamHI and XhoI restriction sites. Proteins were expressed in Escherichia coli BL21–CodonPlus (DE3)–RIL cells (Stratagene) and lysed with a cell disrupter (Avestin). For purification of RECQL5515–620, clarified lysate was loaded onto a Glutathione Sepharose 4 Fast Flow column (GE Healthcare) equilibrated in buffer containing 20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM DTT. The GST–tag was cut with PreScission protease during dialysis. The sample was further purified using HiTrap SP HP and Superdex 75 columns (GE Healthcare).
RECQL51–620 was purified using GST, HiTrap Heparin HP, and Superdex 200 columns (GE Healthcare) in buffer containing 20 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM DTT.
Point mutants of the RECQL5 IRI domain (residues 490–620) were generated using the Quikchange Site–directed Mutagenesis Kit (Stratagene). GST–tagged wild–type IRI as well as mutant proteins and the GST–tag by itself were purified using glutathione sepharose and Superdex 200 columns.
Human TFIIS constructs were purified using a glutathione sepharose column in buffer containing 20 mM Tris, pH 8.0, 100 mM NaCl, 5% glycerol, 10 µm ZnCl2, 1 mM DTT. GST–fusion proteins were further purified on a Superdex 200 column. For purification of tag–free TFIIS, the GST–tag was cleaved with PreScission protease and the sample purified on HiTrap SP HP and Superdex 200 columns.
Human Rpb11168–1302 was codon–optimized for expression in E. coli (GeneArt) and purified on a glutathione sepharose column in buffer containing 20 mM Hepes pH 7.5, 100 mM NaCl, 5% glycerol, 1 mM DTT. GST–fusion proteins were further purified on a Superdex 200 column. For purification of tag–free Rpb1, the GST–tag was cleaved with PreScission protease. The sample was further purified using a Q column, a subtractive glutathione sepharose step, and a Superdex 75 column.
Preparation of apo hPol II and elongating Pol II in complex with RECQL5
Human Pol II was prepared as described previously39. To prepare the sample containing Pol II engaged to a transcription bubble and stalled by RECQL5, an artificial transcription bubble was created by aligning synthetic template DNA (5’–ctcaagtacttacgcctggtcattacta–3’), nontemplate DNA (5’–tagtaaactagtattgaaagtacttgagcttagacagcatgtc–3’), and RNA (5’–uauaugcauaaagaccaggc–3’). Pol II was sequentially incubated with a 10–fold molar excess of the transcription bubble and a 10–fold molar excess of RECQL51–620 D157A during the last purification step when Pol II was immobilized on a protein G affinity column. After elution, the sample was diluted to a concentration of ~120 nM, and cross–linked with 0.02% glutaraldehyde for 10 min.
Electron microscopy sample preparation and data collection
For negative stain analysis, continuous carbon grids were glow–discharged using a Solarus plasma cleaner (Gatan). To allow complex formation between Pol II and RECQL51–620, a 2–fold molar excess of RECQL51–620 was incubated with Pol II in buffer containing 10 mM Tris, pH 7.9, 10 mM Hepes, pH 8.0, 4 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.05% NP–40, 0.1% trehalose, 1 mM AMPPNP, for 30 min on ice.
For the sample of crosslinked RECQL5 bound to elongating Pol II, the sample was diluted in transcription buffer (20 mM Hepes pH 8.0, 4 mM MgCl2, 50 mM KCl, 0.05% NP–40, 1 mM TCEP) before 4 µl solution (containing ~15 nM Pol II) were incubated on the EM grid for 20 sec. After blotting with filter paper, the grid was subsequently laid on five 75 µl–drops of 1% (w/v) uranyl formate solution. For cryo–EM analysis, 4 µl sample (containing ~60 nM Pol II) were placed onto C–flat grids (Protochips Inc.) that had a thin carbon film floated on top. Samples were incubated in the chamber of an FEI Vitrobot at 6 °C for 20 sec on grids that had been glow–discharged for 40 sec in an Edwards carbon evaporator before blotting for 2 sec at an offset of 0 mm. Data were collected on a Tecnai 20 transmission electron microscope (FEI) operating at 120 keV at a magnification of 80,000x for negative stain and 100,000x for cryo–EM data collection under low–dose conditions (20e−/Å2) using the Leginon data collection software40. Images were recorded on a Gatan 4k x 4k camera with a final pixel size of 3.01 Å and 2.31 Å, respectively.
Electron microscopy data processing
Particles were picked automatically using DoG picker41. Reference–free 2D class averages were obtained using iterative multivariate statistical analysis (MSA) and multi–reference alignment (MRA)42. For the Pol II–RECQL51–620 negative stain data set (uncrosslinked), 59,168 particles were extracted using a box size of 288 × 288 pixels, decimated by a factor of 2, normalized and phase flipped. Pixels that deviated by more than 4.5 sigma were additionally normalized using XMIPP43. For the uranyl formate–stained data set of crosslinked elongating Pol II in complex with RECQL5, aggregates and particles that were too close to each other were eliminated based on reference–free 2D class averages, yielding a final data set of 55,509 particles. Iterative projection matching was performed using libraries from the SPARX and EMAN2 image processing packages44,45. During the first round of projection matching, three identical models of a cryo–negative stained hPol II reconstruction46, low–pass filtered to 60 Å, were used as starting models and allowed us to further sort the particles. Particles included in the reconstruction of model 1 yielded a model at ~20 Å resolution that showed clear additional density close to the downstream cleft, while the other two models remained at low resolution during refinement (28 Å and 42 Å, respectively). The particles selected for model 1 were extracted for further refinement. Angular increments for projection matching started at 25 degrees and were reduced stepwise to 6 degrees. The model was refined to ~17Å resolution according to the 0.5 cut–off in the FSC curve.
For the cryo–EM data set of crosslinked elongating Pol II in complex with RECQL5, low–pass filtered models of model 1 and model 2 obtained from the negative–stain data set were used as initial models for projection matching to sort the data set, yielding a final stack containing 121,416 particles. The model was refined to ~13Å resolution according to the 0.5 FSC cut–off, and was filtered with bsoft to local resolution47. Crystal structures were docked into the density manually with Chimera48.
Crystallization and data collection
Crystals of the RECQL5 KIX domain were grown at 20 °C in 2 µl hanging drops containing equal volumes of the protein solution at 25 mg/ml and a reservoir solution consisting of 0.2 M NaCl, 0.1 M sodium/potassium phosphate pH 6.2, and 50% PEG 200. Crystals were transferred into Al’s oil (Hampton Research) and flash cooled in liquid nitrogen. Crystals were of the orthorhombic space group P212121 and contained two molecules in the asymmetric unit. The structure was solved using single–wavelength anomalous dispersion using X–ray diffraction data from crystals soaked in mother liquor supplemented with 0.5 M potassium iodide for 30 sec before cryoprotection and flash cooling. Native and anomalous X–ray diffraction data were collected at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory (LBNL), beamline 8.3.1. Diffraction data were processed using XDS49.
Crystal structure determination and refinement
The positions of iodine sites were determined using SHELXD50. Phases were calculated using AutoSharp51 and improved by solvent flipping using Solomon (as implemented in AutoSharp). Iterative model building and refinement were carried out with the programs Coot52 and Phenix53. The final model of the RECQL5 KIX domain contains residues 523–614 for chain A and residues 524–620 for chain B. The model was refined to 1.9 Å and has an Rcryst of 20.14% and an Rfree of 23.36%. Assessment with MolProbity54 showed no Ramachandran outliers and 99.5% of all residues within favored regions of the Ramachandran plot. Figures were generated using PyMOL (www.pymol.org).
Transcription assay
The dC–tailed histone H3.3 intron template was prepared by digestion of the pGEMTerm plasmid (obtained from Caroline Kane, University of California, Berkeley) as described previously20,27. Reactions (15 µl) in transcription buffer (25 mM Hepes pH 7.9, 100 mM KCl, 5 mM MgCl2, 5% glycerol, 6 mM spermidine, 1 mM DTT, 1U RNasin Plus, 800 µM each of GTP, UTP and ATP) contained 20 µg/ml dC–pGEMTerm, 0.8 pmol hPol II, 4 or 40 pmol RecQ5, respectively, or an equivalent amount of buffer. After the addition of 1 µl [α–32P]–CTP [0.01 µM], reactions were incubated for 3 min at 30 °C. Following the addition of 3.5 µl chase buffer (25 mM Hepes pH 7.9, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 800 µM each of GTP, UTP and ATP, 4.8 mM CTP), incubation at 30 °C was continued. After incubation for 10 min, samples were digested with proteinase K for 15 min at 30 °C before the addition of formamide loading buffer (20 mM EDTA, 5% glycerol, 0.02% SDS, bromophenol blue in formamide). RNA transcripts were resolved on 6% polyacrylamide, 7M urea gels, and visualized by phosphorimaging (Storm Scanner, GE Life Sciences).
TFIIS read–through assay
The dC–tailed histone H3.3 intron template was prepared as described above. Reactions (10 µl) in transcription buffer (25 mM Hepes pH 7.9, 100 mM KCl, 5 mM MgCl2, 5% glycerol, 6 mM spermidine, 1 mM DTT, 1U RNasin Plus, 800 µM each of GTP, UTP and ATP, 20 µg/ml dC–pGEMTerm, 0.4 pmol hPol II, 0.25 µl [α–32P]–CTP [0.04 µM]) were incubated for 2 min at 30 °C. Each aliquot was then diluted into 3.5 µl chase buffer and incubated for 3 min at 30 °C. After the addition of 5 µl RECQL5 (2 pmol, 20 pmol, or 200 pmol, a molar excess over Pol II of 1:5, 1:50, 1:500, respectively), or an equivalent volume of buffer, the reactions were incubated for 2 min at 30 °C before the addition of 1.5 µl TFIIS1–301 (2 pmol, a 5x molar excess over Pol II) or an equal volume of buffer. Incubation at 30 °C was continued and samples were quenched after 10 min with formamide loading buffer. For RECQL5 constructs that contained the helicase domain, samples were digested with proteinase K for 15 min at 30 °C before the addition of loading buffer. RNA transcripts were resolved on 6% polyacrylamide, 7M urea gels, and visualized by phosphorimaging (Storm Scanner, GE Life Sciences). Relative amounts of transcription products were quantitated by densitometry using ImageQuant software (GE Life Sciences).
SAXS analysis
SAXS data were collected at the SIBYLS 12.3.1 beamline at the Advanced Light Source, LBNL55,56. Scattering measurements were performed on 20 µl samples at 15 °C loaded into a helium–purged sample chamber, 1.5 m from the Mar165 detector. Sequential exposures (0.5, 0.5, 2, 5, and 0.5 s) were taken at 12 keV. The proteins showed slight radiation–induced aggregation, but the first and second 0.5 s exposures overlaid. Therefore, the initial 0.5 s exposures were used. Data were analyzed using the ATSAS suite57 and FOXS/MES58.
Electrophoretic mobility shift assay
Fluorescently labeled RECQL51–620 was produced using TNT Quick Coupled Transcription/Translation Systems (Promega) and FluoroTect GreenLys (Promega) according to the manufacturer’s protocol. Fluorescently labeled RECQL51–620 was diluted 1:200 and incubated with increasing concentrations of Pol II (0, 4, 8, 16, 24, 32, 40, 80, 200, 400 nM) in a total volume of 10 ul on ice for 30 min before being analyzed on NativePAGE Novex 3–12% Bis–Tris gels (Invitrogen). Protein complexes were visualized using a Typhoon scanner and quantitated using ImageQuant software (GE Life Sciences). Raw data were fit using Prism 5.0 (GraphPad Software) with the equation for one site binding, fitting both total and nonspecific binding (specific=(Bmax*X/(Kd+X); nonspecific=NS*X + background).
Supplementary Material
Table 1.
Data collection, phasing and refinement statistics for crystallographic analysis.
| Native | Iodide soak | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | a=50.7, b=61.2, c=81.7 | a=51.1, b=62.6, c=82.8 |
| α, β, γ (°) | α=β=γ=90° | α=β=γ=90° |
|
peak |
||
| Wavelength | 1.11589 | 1.65315 |
| Resolution (Å) | 100.0–1.90 (1.95–1.90) | 100.0–2.60 (2.67–2.60) |
| Rsym (%) | 4.5 (51.8) | 8.0 (75.0) |
| I / σI | 22.8 (3.6) | 22.2 (2.5) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 7.0 (7.1) | 13.5 (7.4) |
| Refinement | ||
| Resolution (Å) | 49.0–1.9 | |
| No. reflections | 20,613 | |
| Rwork / Rfree | 20.1 / 23.4 | |
| No. atoms | 3041 | |
| Protein | 1,515 | |
| PEG | 34 | |
| Water | 94 | |
| B factors | ||
| Protein | 53.9 | |
| Ligand/ion | 52.6 | |
| Water | 42.1 | |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.04 | |
One crystal was used for each data set. Values in parentheses are for highest–resolution shell.
Acknowledgements
We thank G. Lander and P. Grob for advice on EM data collection and processing, and F. Bleichert for critical reading of the manuscript. We thank C. Kane (University of California, Berkeley, Berkeley, CA 94720, USA) for providing the pGEMTerm plasmid, advice on the transcriptional read–through experiment, and critical reading of the manuscript. We are grateful to J. Holton (beamline 8.3.1, Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA) for assistance with synchrotron data collection. We thank D. King (Howard Hughes Medical Institute, University of California, Berkeley, Berkeley, CA 94720, USA) for synthesis of the CTD peptide used for purification of Pol II. S.K. was supported by a fellowship from the Boehringer Ingelheim Fonds. The work was supported by National Institute of General Medical Sciences grant GM63072 (E.N.). S.T. and the SIBYLS beamline (12.3.1) at the Advanced Light Source are supported by NIH P01 CA092584 and DOE Integrated Diffraction Analysis Technologies (IDAT) under Contract Number DE–AC02–05CH11231. E.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
ACCESSION CODES
The atomic coordinates and structure factors of RECQL5 KIX have been deposited in the Protein Data Bank, www.pdb.org (PDB code 4bk0). The EM map of elongating Pol II in complex with RECQL5 has been deposited in the Electron Microscopy Data Bank under accession code EMD–2367.
Author contributions
S.K. conceived the study, designed and performed experiments, and analyzed the data. M.J. and S.K. collected and processed X–ray diffraction data. J.F. and S.K. purified human Pol II. S.T. collected and processed SAXS data. E.N. oversaw the project and S.K., M.J. and E.N. prepared the manuscript.
REFERENCES
- 1.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 2.Singh DK, Ahn B, Bohr VA. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology. 2009;10:235–252. doi: 10.1007/s10522-008-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis NA, et al. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 4.Yu CE, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 5.Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics. 1999;61:268–276. doi: 10.1006/geno.1999.5959. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, et al. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng L, et al. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwendener S, et al. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem. 2010;285:15739–15745. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izumikawa K, et al. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- 14.Aygun O, et al. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reines D, Conaway JW, Conaway RC. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- 17.Wind M, Reines D. Transcription elongation factor SII. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci U S A. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 20.Christie KR, Awrey DE, Edwards AM, Kane CM. Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, P37. J Biol Chem. 1994;269:936–943. [PubMed] [Google Scholar]
- 21.Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci U S A. 2007;104:16062–16067. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim B, et al. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci U S A. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 24.Awrey DE, et al. Yeast transcript elongation factor (TFIIS), structure and function II: RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem. 1998;273:22595–22605. doi: 10.1074/jbc.273.35.22595. [DOI] [PubMed] [Google Scholar]
- 25.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 26.Wang D, et al. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reines D, Wells D, Chamberlin MJ, Kane CM. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol. 1987;196:299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- 28.Reines D, Chamberlin MJ, Kane CM. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J Biol Chem. 1989;264:10799–10809. [PubMed] [Google Scholar]
- 29.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 30.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 32.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 33.Tadokoro T, et al. Human RECQL5 participates in the removal of endogenous DNA damage. Mol Biol Cell. 2012;23:4273–4285. doi: 10.1091/mbc.E12-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Rossettini A, Hanes SD. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics. 2003;165:1687–1702. doi: 10.1093/genetics/165.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 37.Ariyoshi M, Schwabe JW. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003;17:1909–1920. doi: 10.1101/gad.266203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Domingo D, et al. DIO-1 is a gene involved in onset of apoptosis in vitro, whose misexpression disrupts limb development. Proc Natl Acad Sci U S A. 1999;96:7992–7997. doi: 10.1073/pnas.96.14.7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online methods references
- 39.Kassube SA, et al. Structural Insights into Transcriptional Repression by Noncoding RNAs That Bind to Human Pol II. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 43.Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldwin PR, Penczek PA. The Transform Class in SPARX and EMAN2. J Struct Biol. 2007;157:250–261. doi: 10.1016/j.jsb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Kostek SA, et al. Molecular architecture and conformational flexibility of human RNA polymerase II. Structure. 2006;14:1691–1700. doi: 10.1016/j.str.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 47.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 51.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Classen S, et al. Software for the high-throughput collection of SAXS data using an enhanced Blu-Ice/DCS control system. J Synchrotron Radiat. 2010;17:774–781. doi: 10.1107/S0909049510028566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. Journal of Applied Crystallography. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.