Abstract
Glucocorticoid receptor binds to genomic response elements and regulates gene transcription with cell- and gene-specificity. Within a response element, the precise sequence to which the receptor binds has been implicated in directing its structure and activity. We use NMR chemical shift difference mapping to show that non-specific interactions with particular base positions within the binding sequence, such as those of the “spacer”, affect the conformation of distinct regions of the rat glucocorticoid receptor DNA binding domain. These regions include the DNA-binding surface, the “lever arm” and the dimerization interface, suggesting an allosteric pathway that signals between the DNA binding sequence and the associated dimer partner. Disrupting this path by mutating the dimer interface alters sequence-specific conformations, DNA-binding kinetics and transcriptional activity. Our study demonstrates that glucocorticoid receptor dimer partners collaborate to read DNA shape and to direct sequence specific gene activity.
Introduction
Gene expression is tailored to the needs of specific tissues and in response to environmental and developmental changes. Transcriptional regulators coordinate this task by integrating input signals at specific genomic regions1,2 to effect precise transcriptional outputs at target genes. This intricate process relies on combinatorial control, in which distinct combinations of factors assemble into functional regulatory complexes that control the transcriptional activity of associated genes. However, the determinants that define the gene-specific assembly and activity of these regulatory complexes are poorly understood.
Glucocorticoid receptor (GR), a glucocorticoid-activated member of the nuclear receptor superfamily, utilizes combinatorial control to regulate hundreds to thousands of genes in a cell- and gene-specific manner. In part, this specificity arises from context-dependent GR binding regions (GBRs), which can be defined in vivo using genome-wide approaches. Some, but not all, GBRs appear to function in vivo as glucocorticoid response elements (GREs), which confer context-specific glucocorticoid regulation upon nearby genes. While GBR and GRE activities are clearly separable, both rely on the effects of multiple signals, such as hormonal ligands, other regulatory factors, and post-translational modifications. Each of these signals drives distinct conformational changes in the receptor3–8, thereby modulating its transcriptional regulatory activity9–11. For example, two GR ligands, dexamethasone (dex) and RU486, differentially affect the formation of a coactivator interaction surface of the ligand-binding domain8 and induce different transcriptional profiles.
GBRs and GREs are composite elements consisting of binding motifs for non-GR transcriptional regulatory factors and often one or more GR binding sequence (GBSs)12. GBSs are bound with high affinity by purified GR in vitro and mutational studies have confirmed that GBSs within a particular GBR are responsible for GRE activity13, (Thomas-Chollier, M., unpublished). GBSs vary loosely around a 15 base pair (bp) consensus sequence consisting of two hexameric half-sites separated by a 3 bp spacers13. GR binds to a GBS as a homodimer with each dimer partner specifically contacting, at most, three bases within each GBS half-site. Structural studies of free and DNA-bound GR DBD suggest that DNA binding imparts structural changes in the second zinc finger of the DBD, forming the dimerization interface14–16.
Our lab previously demonstrated that DNA binding sequences serve as distinct signals that direct GR structure and activity17,18. Crystallographic studies comparing GR bound to different GBSs revealed alternate protein conformations that are dependent on the precise DNA binding sequence17. The observed alternate conformations were localized to a loop region within the DNA binding domain (DBD) termed the lever arm, which does not itself contact the DNA. Moreover, GBSs that produced different lever arm conformations were invariant at all nucleotide positions that make direct contacts with GR, indicating that non-specific bases affect GR structure. The presence of alternate lever arm conformations suggests that GBS-specific conformational dynamics play a role in GR gene-specific regulation.
These crystallography studies motivated the following questions: (1) how does GR detect sequence differences among GBSs, (2) do GBSs drive distinct “allosteric paths” of conformational changes that extend into and through the lever arm, and (3) how do GBS-dependent differences in GR conformation impact GR activity? To address these questions, we used solution techniques to assess the effects of changing precise nucleotide positions within the GBS and perturbing a functional surface of the GR DBD.
Results
GBS spacer affects GR occupancy, activity and structure
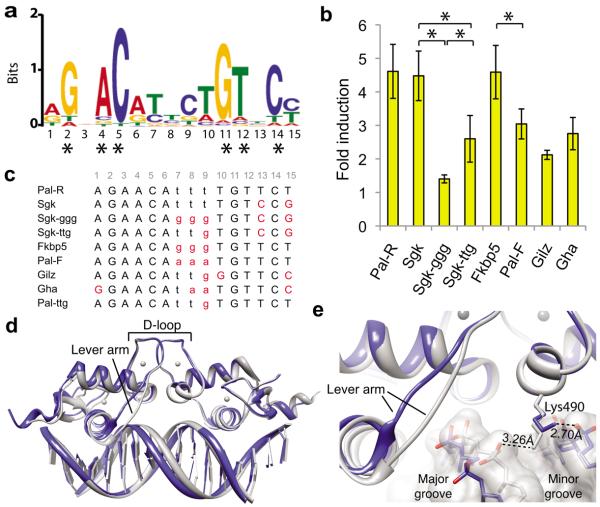
We sought to determine the degree of sequence variability among endogenous GBSs to estimate the potential for DNA sequences to be unique signals that produce distinct GR activities. We identified GR binding regions in U2OS cells exogenously expressing full-length rat GR, using GR chromatin immunoprecipitation (ChIP) followed by deep sequencing of the precipitated DNA fragments. An unbiased search for sequence motifs within 1,000 GR binding regions with the highest number of reads revealed a GBS motif composed of imperfect palindromic hexamers separated by a 3 bp spacer (Fig. 1a), similar to previously identified motifs based on smaller sample sets (e.g.,12). Scanning for this motif among the observed 30,000 GR binding regions revealed that 90% of GBSs are unique, suggesting that there is sufficient diversity for each to be a gene-specific signal. This model would require that non-specific bases contribute to sequence specificity. The GBS positions with the highest information content (>1 bit) correspond to the six bases that are directly contacted by the GR dimer16,17. The remaining nine nucleotide positions each contain less than 1 bit of information, with half-site positions 3 and 13 nearly devoid of sequence preference (0.05 and 0.1 bits, respectively). Notably, however, GR displayed appreciable base-preference at positions that it does not contact directly: pyrimidines at spacer positions 7–9, as well as A and T at positions 6 and 10, respectively, adjacent to the spacer.
Figure 1.
Non-specific GBS bases modulate GR structure and activity. (a) The GR binding motif identified by GR ChIP-sequencing in U2OS-GR cells using MEME in “zero or one motif per site” mode with a 2nd order background Markov Model based on the top 1,000 peaks. Asterisks indicate specific-bases that are directly contacted by GR. (b) Luciferase induction of single GBS reporters with 100 nM dexamethsone (dex) treatment compared to ethanol control in U2OS cells. Error is s.d. of four or more independent experiments. Significant difference in transcriptional response by 2-tailed t-Test (*p < 0.05) is indicated for GBSs that differ only by spacer. (c) List of the GBS sequences used in this study with spacer (lowercase) and bases that differ from the palindromic Pal-R sequence (red). The 15-bp GBSs were centered within identical flanking sequences resulting in a 24-bp double-stranded DNA. (d) Alignment of GR DBD – Pal-F (PDB ID: 3g99, grey) and GR DBD – Fkbp5 (PDB ID: 3g6u, blue) crystal structures. (e) Zoomed view of Lys490 interaction with the GBS spacer of the Pal-F GBS (grey) and the Fkbp5 GBS (blue).
It was previously shown that GBSs differentially modulate GR transcriptional induction using luciferase reporters consisting of a single GBS upstream of a minimal promoter17. To investigate how varying the GBS spacer affects GR transcriptional induction, we compared reporter activity in the presence of 100 nM dex for GBSs that differ only in spacer sequence (Fig. 1b–c). Changing the spacer of Sgk from TTT to GGG resulted in a 69% decrease in transcriptional activation and changing the spacer by only one base (from TTT to TTG) resulted in an intermediate 42% decrease. Alternatively, changing the spacer of Fkbp5 from GGG to AAA (Pal-F), though not TTT (Pal-R) resulted in decreased transcriptional activity. Thus, spacer sequence, within the context of the whole GBS, influences GR activity.
As GR does not directly contact the GBS spacer, we investigated other potential structural mechanisms by which spacer sequence influences GR function. Though prior crystallography studies did not include the Sgk-ggg GBS, we aligned structures of GR DBD bound to the Pal-F and Fkbp5 GBSs, which differ only in spacer (Fig. 1d). These structures revealed that the minor groove of the Pal-F spacer is narrower than that of the Fkbp5 spacer, with average widths of 3.8 Å and 6.4 Å, respectively, as measured by Curves+19. As the Pal-F and Fkbp5 GBSs have spacer sequences of AAA or GGG, respectively, the sequence-specific difference in minor groove width is consistent with previous studies showing that short A-tracts narrow the minor groove20. This led us to hypothesize that DNA shape, defined by the nucleotide sequence of the GBS spacer, serves as a signal that regulates GR activity. Thus, we predict that structural features associated with the DNA spacer impart structural changes that are propagated through the GR DBD. Consistent with this, our examination of GR contacts with the DNA phosphate backbone near the GBS spacer indicated that the orientation of the side chain of Lys490 is dependent on the spacer sequence (Fig. 1e).
D-loop conformation depends on spacer but not half-site
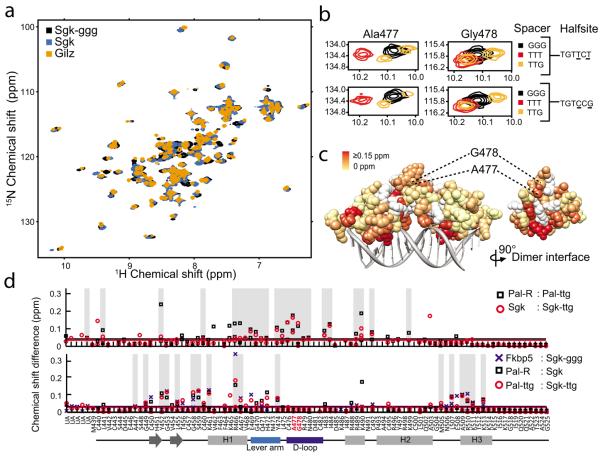
To further investigate how GBS spacer affects GR structure, we monitored GR DBD conformation by 15N-HSQC, which measures the chemical environment of the amide bond of individual amino acid residues. For GR DBD bound to a high-affinity GBS (Gha), we assigned over 90% of the chemical shift peaks to their corresponding residues (Supplementary Fig. 1). We overlaid the HSQC spectra for GR bound to GBSs that differ only at non-specific bases of the spacer or at half-site positions 13 and 15 (hereafter referred to as half-site13:15). In both comparisons, the spectra displayed non-overlapping peaks indicating that each GBS complex is structurally distinct (Fig. 2a), and suggesting that both the sequence of the spacer and half-site13:15 influence GR structure.
Figure 2.
GBS spacer affects the conformation of the D-loop. (a) Comparison of 15N HSQC spectra of GR DBD – GBS complexes that differ at spacer only (Sgk and Sgk-ggg) or at spacer and half-site (Sgk and Gilz, Sgk-ggg and Gilz). (b) Zoomed spectra showing the chemical shift perturbation of D-loop residues Ala477 and Gly478 resulting from changing specific nucleotides in the spacer or half-site13:15. (c) The magnitude of combined 1H and 15N chemical shift difference between GR DBD – Sgk and GR DBD – Sgk-ggg spectra for each assigned residue, colored onto the crystal structure (PDB ID:3g6u). Unassigned residues are colored white. (d) Chemical shift difference (C.S.D.) analysis for pair wise comparison of GR DBD complexes with TTT compared to TTG spacer (top), or TGTTCT compared to TGTCCG half-site (bottom). Peaks unambiguously arising from peak splitting were assigned to their corresponding residues and C.S.D. values for both peaks are plotted. Grey bars indicate residues that have a C.S.D. greater or equal to the mean C.S.D. across pair wise comparisons. UA = unassigned.
We used chemical shift perturbation, which is sensitive to local changes in conformation21–23, to identify regions of GR DBD that are affected by spacer sequence compared to those affected by half-site13:15. GBSs that differ only in the spacer resulted in substantial peak shifts mapping to Ala477 and Gly478 of the GR dimerization loop (D-loop) (Fig. 2b), despite a distance of at least 18 Å between the GR backbone and the nucleotide bases of the spacer. In contrast, half-site13:15 had little effect on Ala477 and Gly478, but instead affected residues at other surfaces of the DBD (Supplementary Fig. 2). Thus, sequence variation within particular regions of the GBS corresponds with conformational changes in distinct subdomains of the DBD.
Extending this analysis across the GR DBD further demonstrated that changing the spacer, but not half-site13:15, induced peak shifts mapping to the D-loop of GR (Fig. 2c–d). Alternatively, half-site13:15, but not the spacer, influenced peaks mapping to outward-facing surfaces of the DBD near the DNA (Fig. 2d and Supplementary Fig. 2). Additionally, the DNA recognition helix (H1) and the lever arm were affected by sequence changes in either the spacer or half-site13:15. This effect of GBS on the lever arm conformation further corroborates the alternate conformations observed by crystallography17.
How might information in the GBS spacer be transmitted across the substantial distance to the D-loop to elicit specific rearrangements? As the lever arm structurally links the DNA recognition helix to the D-loop, the simplest model suggests structural coupling of the D-loop to the DNA recognition helix via the lever arm.
The A477T mutation disrupts D-loop conformation
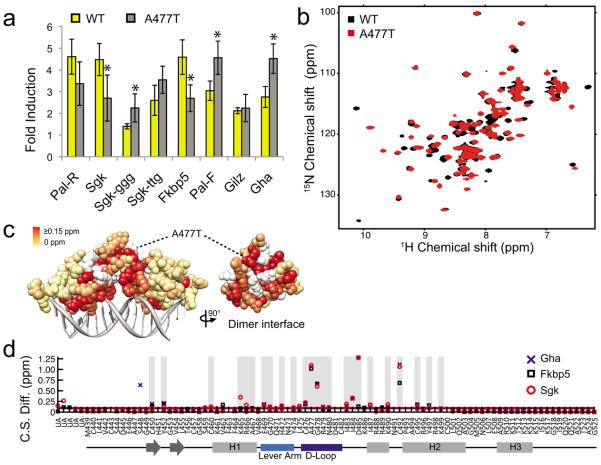
To investigate the functional role of spacer-specific structural changes, we tested whether perturbing the D-loop affected GR activity in a GBS-specific manner. We focused on Ala477 of the D-loop, which makes one of the four dimerization contacts within the GR DBD16#x2014;a backbone hydrogen bond between the carbonyl of Ala477 and the amide of Ile483 from the associated dimer partner. This mutation, A477T, has been shown to alter GR activity in a gene-specific manner24. As reported previously17,25, the extent to which A477T transcriptional induction differed from wild type GR (WT) was GBS-specific. Among eight GBSs tested in reporter assays, the A477T mutation resulted in increased, decreased or unchanged transcriptional induction compared to WT (Fig. 3a). In contrast to WT, A477T activity was indistinguishable for two GBSs that differ in spacer (Sgk and Sgk-ggg). Thus, this mutation in the dimerization interface did not abolish GR activity, but instead resulted in reinterpretation of GBS signals by the mutant GR.
Figure 3.
Disrupting the dimerization interface affects lever arm and DNA recognition helix conformation. (a) Comparison of transcriptional induction of GBS luciferase reporters in U2OS cells expressing either GR wild type (WT) or mutant (A477T) after treatment with 100 nM dex. Error is one s.d. of four or more independent experiments and significant differences in transcriptional response between WT and A477T were determined by 2-tailed t-Test (*p < 0.05). The transcriptional response of A477T is equivalent for Sgk compared to Sgk-ggg or Sgk-ttg, (p-value = 0.44 and 0.19, respectively). (b) 15N-HSQC comparing WT and A477T DBD bound to the Fkbp5 GBS. (c) Magnitude of combined 1H and 15N chemical shift difference between WT and A477T DBD bound to the Fkbp5 GBS, colored onto the WT DBD crystal structure (PDB ID 3g6u). Unassigned WT residues are colored white. (d) Chemical shift difference (C.S. Diff.) between spectra of WT DBD and A477T DBD complexes. Grey bars highlight residues with a chemical shift difference ≥0.05ppm between WT and A477T across all three GBSs. UA = unassigned.
To assess the mechanism by which this mutation differentially affects GBS-specific activity, we characterized the structural and biophysical impacts of A477T. We compared the HSQC spectra of WT and A477T bound to the Fkbp5 GBS (Fig. 3b–c). The A477T DBD spectrum showed many peaks that overlaid well with GR WT, but over 30% of residues were shifted as a result of the A477T mutation. Importantly, these peaks did not overlay with those corresponding to the unbound WT DBD, ensuring that the protein is completely bound to DNA (Supplementary Fig. 3).
We quantified the chemical shift difference as the distance between each peak in the GR WT spectra to the nearest peak in the A477T spectra25. Comparison of WT- and A477T-bound Gha, Fkbp5, and Sgk complexes (Fig. 3d) revealed A477T-specific shifts that mapped to the D-loop and the residues surrounding Ile483, consistent with a disruption of the dimerization interface. Additional chemical shift differences mapped to the N-terminal region of the lever arm and the recognition helix of GR. Thus, the A477T mutation generates local structural changes as well as structural reorganization in regions outside of the dimerization surface. Together with the observation that WT GR produced GBS-specific structural changes in the dimerization interface (Fig. 2b–c), our findings with the A477T mutation indicated that the dimerization and DNA interfaces are structurally coupled.
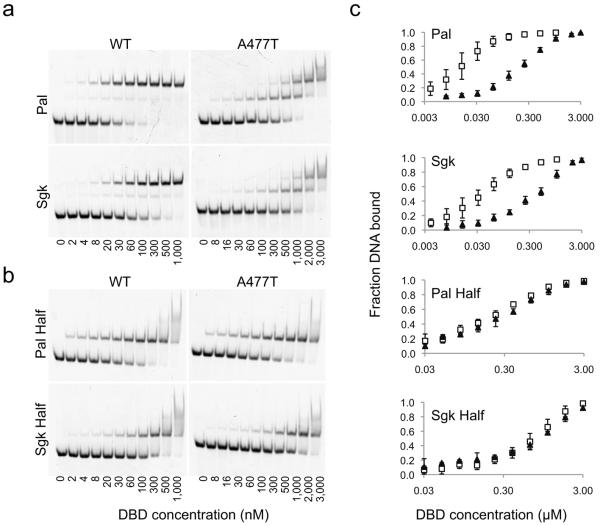
A477T affects cooperativity but not stoichiometry
To pursue the mechanism by which the A477T mutation affects the GBS-specific activity of GR, we assessed DNA binding by an electrophoretic mobility shift assay (EMSA). Comparison of WT DBD and A477T DBD binding to the Pal-R and Sgk GBSs revealed that GR dimer complexes were formed at saturating concentrations of both WT and A477T, though the mutant showed reduced DNA-binding affinity (Fig. 4a). For the WT, the transition from free DNA to dimer complex occurred at lower concentrations of GR, with only a minor population of DNA-bound monomer, indicative of strong positive cooperativity. In contrast, A477T displayed little cooperativity, having nearly saturated the DNA as a monomer prior to dimer formation. To distinguish whether the reduced overall affinity of A477T was due to impaired DNA recognition resulting from the mutation, we compared binding of GR WT and A477T to a GBS half-site (Fig. 4b). We found that half-site binding was equivalent for WT and A477T, indicating that the A477T mutation does not disrupt the DNA-binding ability of the monomer (Fig. 4b–c). Thus, the reduction in overall affinity of the A477T mutant is due to diminished cooperativity.
Figure 4.
A477T impairs dimerization but not monomer DNA-binding. Electrophoretic mobility shift assay (EMSA) monitoring binding of WT or A477T DBD to (a) GBSs conjugated with an Alexa-488 fluorophore at a concentration of 5 nM or to (b) mutated GBSs, where one half-site is changed to the least favorable nucleotide at each position, based on the ChIP-seq binding motif in Fig. 1a. (c) Quantitative comparison of WT DBD (open squares) or A477T DBD (filled triangles) binding by EMSA. Error bars are one s.d. for 2–4 replicates.
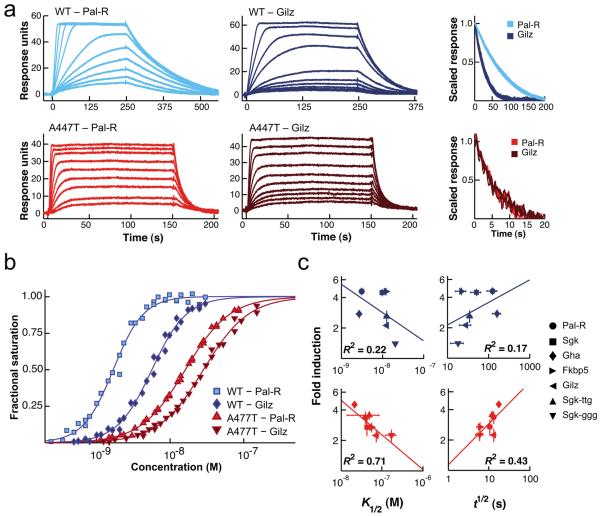
As the EMSA is a non-equilibrium measure, surface plasmon resonance (SPR) was additionally used to monitor the effects of binding sequence on the DNA recognition properties of WT and A477T, under similar conditions as used for the NMR studies. We compared WT DBD or A477T DBD binding to two GBSs whose transcriptional induction in reporter assays was reduced (Pal-R) or unaffected (Gilz) by the A477T mutation (Fig. 5a). Isotherms constructed from maximal binding of WT or A477T DBD to GBS-immobilized surfaces at equilibrium showed that the mutation results in a decrease in binding affinity by a factor of 10 and 5 for Pal-R and Gilz, respectively (Fig. 5b and Supplementary Table 1). Consistent with the EMSA, maximal binding responses were similar for WT and A477T, indicating that the mutant binds to DNA at the same stoichiometry as WT (Fig. 5a). As the concentration dependence was non-Langmuir, the SPR binding isotherms were fit to the Hill equation. Fit Hill coefficients for WT were 1.83 ± 0.28 and 2.13 ± 0.26 for the Gilz and Pal-R GBSs, respectively (Fig. 5b and Supplementary Table 1). The A477T isotherms were well described by Hill coefficients of 1.34 ± 0.16 (Gilz) and 1.41 ± 0.1 (Pal-R) indicating that cooperativity was reduced but not abolished by the A477T mutation. As the Hill coefficient represents an exponential component of the Hill equation, these differences in Hill coefficients between the two GBSs (0.3 for WT and 0.07 for A477T) are substantial for WT and diminished for A477T. This suggests that differential cooperativity contributes to the discrimination between binding sites for WT, but appears to play less of a role for A477T.
Figure 5.
A477T disrupts cooperativity and GBS-specific dissociation. (a) Representative SPR binding traces for WT and A477T DBD binding to immobilized GBSs at 35°C. Comparison of dissociation curves for WT DBD (top right) and A477T DBD (bottom right) (b) Binding isotherms for a GR DBD concentration series (0.700–200 nM for WT, 1.4–400 nM for A477T) binding to immobilized GBS surfaces at 35°C from 3 separate titrations, normalized to maximal binding. Isotherms were fit to the Hill equation: Fractional occupancy = (c/(c+K1/2))nH. (c) Comparison of transcriptional activity (fold induction) versus binding affinity (K1/2) or dissociation half-life (t1/2), for WT DBD (blue) and A477T DBD (red) across seven GBS surfaces at 8°C.
As the transcriptional activity of GR is affected by the fractional occupancy of the active dimer complex on a given response element, we also compared the dissociation kinetics of WT and A477T DBD – GBS complexes (Fig. 5a). To simplify the comparison between WT and A477T across the different binding sites, dissociation traces were fit to a single exponential decay process and the fit parameters were presented as t1/2 values (Supplementary Table 1). For WT, fit t1/2 values of 55 ± 4 s and 23 ± 2 s were determined for Pal-R and Gilz, respectively. In contrast, A477T dissociated from both Pal-R and Gilz GBSs with a t1/2 of ~5 s; a 90% and 80% decrease, respectively, relative to WT. Under these conditions, the dissociation of WT was dependent on the GBS, whereas A477T kinetics appeared undiscriminating of sequence. This suggests that an intact dimerization surface is critical for interpreting GBS-specific signals that modulate GR dissociation.
To assess more broadly the biophysical parameters that might influence GBS-specific transcriptional activity, we extended our analysis to include five additional GBSs. Across all GBSs, A477T had lower affinity, faster dissociation and reduced cooperativity compared to WT (Supplementary Table 1). Interestingly, while Pal-R and Gha shared similar binding parameters, their transcriptional regulatory activities in reporters were not aligned (p-value = 0.004). To assess the relationship between DNA-binding properties and GBS-specific transcription, we compared transcriptional activity to the K1/2 or t1/2 values across this panel of GBSs (Fig. 5c). We found that transcriptional induction of GR WT did not vary as a simple function of DNA affinity. For example, a mutation resulting in reduced binding affinity enhanced transcriptional induction at the Gha GBS (Fig. 3a). Additionally, GBSs with similar GR binding properties such as Gha and Pal-R or Fkbp5 and Gilz differed in transcriptional activity (Fig. 1a), as well as conformation (Supplementary Fig. 4). These results are consistent with those of Bain et al.26, who also describe GBSs with binding affinities and regulatory activities that are not aligned. Unexpectedly, the authors of that work asserted that DNA binding affinity defines transcriptional activity at GBSs. While DNA binding affinity clearly plays a role in the activities of transcriptional regulators, our results as well as those of Bain et al. demonstrated that other factors must also contribute substantially. Having previously proposed that GBS-specific structural changes determine transcriptional activity17,18, we assessed the relationship between binding and activity in the A477T mutant. In comparison to WT, A477T displayed a stronger correlation between transcription and affinity measured by multiple correlation analyses (Supplementary Table 2). Thus, disrupting the dimer interface appears to dampen allosteric signaling, rendering GBS affinity a stronger determinant.
A477T disrupts signaling between dimer partners
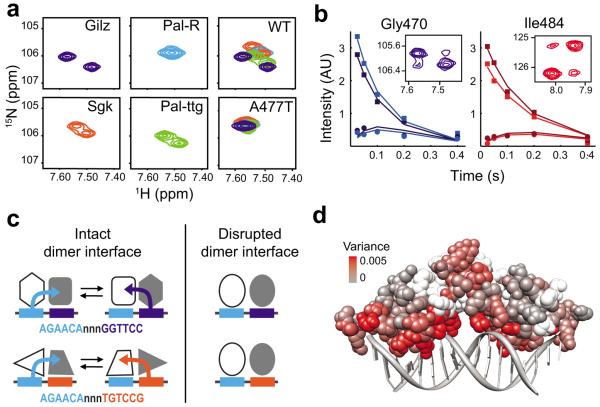
To further dissect the structural mechanism by which A477T alters GBS-specific activity, we used 15N-HSQC to monitor the effects of the A477T mutation on GBS-specific conformations of GR. We focused on the lever arm region because its structure is sensitive to GBS17, and found that, for WT, Gly470 displayed peak-splitting in a GBS-dependent manner (Fig. 6a). Peak-splitting indicates two unique chemical environments for a single residue and can reflect two possibilities: (1) the GR dimer partners have non-equivalent conformations, or (2) GR dimers undergo slow conformational exchange between two distinct states. To distinguish between these, we used ZZ-exchange NMR to detect conformational exchange. This experiment is similar to an HSQC except that a mixing period is introduced between recording the 15N chemical shift and the 1H chemical shift for each amide. Chemical exchange occurring during the mixing period is detected as cross-peaks in the NMR spectrum corresponding to the 15N chemical shift of conformation A and the 1H chemical shift of conformation B—and vice versa (Supplementary Fig. 5a–b). We individually fit the intensity of exchange cross-peaks at different mixing periods27 (Fig. 6b) and determined that the conformational exchange rate of Gly470 (4.36 s−1 and 3.10 s−1) is two orders of magnitude faster than the rate of dissociation from DNA (0.03 s−1) (Supplementary Fig. 5e). This suggests that doublet peaks result from conformational exchange of DNA-bound GR dimer partners rather than dissociation from one half-site and subsequent re-binding binding to the adjacent half-site (Supplementary Fig. 5b).
Figure 6.
Sequence-specific lever arm conformation is dependent on the intact dimerization interface. (a) 15N-HSQC zoomed on the Gly470 peak of the lever arm of WT DBD bound to the asymmetric GBSs (Gilz, Sgk, Pal-ttg) and a palindromic site (Pal-R). Overlay of Gly470 peaks from all for GBSs for WT and A477T DBD (far right). (b) The peak intensity for Gly470 and Ile484 WT DBD residues in ZZ-exchange spectra at five mixing periods. The auto peak and the corresponding exchange peak are shaded equivalently. Insets show the spectra for auto and exchange peaks at a mixing period of 0.2 s. (c) Model showing the role of the dimerization interface in defining GBS-specific conformations. Both half-sites and the spacer determine the conformation of each GR dimer partner by transmitting information from the adjacent GBS half-site across the dimerization interface (represented by the colored arrows). Disruption of the dimerization interface by A477T impairs signal transmission and results in lever arm conformations that are insensitive to GBS sequence. (d) Comparison of WT and A477T chemical shift variance among seven GBS complexes colored by amino acid onto the GR DBD crystal structure.
Because the Gilz GBS consists of two non-identical half-sites, the simplest view is that Gly470 conformations may be determined solely by the sequence of the GBS half-site to which each GR dimer partner is bound. This would predict that GBSs with one identical half-site and that differ at the other half-site will have one overlaid Gly470 peak and one non-overlaid peak (compare Pal-R and Sgk). We found, however, that GBS complexes that are identical at one half-site had two Gly470 chemical shifts that were unique (Gilz, Sgk, Pal-ttg) (Fig. 6a). Thus, the lever arm conformation for each GR dimer depends not only on the sequence of the half-site to which it was directly bound, but also on the sequence bound by the adjacent dimer partner. Taken together with comparisons of Gly470 chemical shifts among different GBS complexes, our data suggest that each GR dimer partner integrates sequence-specific signals from both GBS half-sites as proposed in the model (Fig. 6c).
In contrast to WT, the Gly470 peaks were overlaid for A477T bound to different GBSs (Fig. 6a). Therefore, disrupting the dimer interface at Ala477 abolished the sequence-specific conformations of Gly470 within the lever arm. We expanded this comparison to investigate the extent to which each residue samples distinct conformations when bound to different GBSs by determining the chemical shift variance across a panel of seven GBSs (Fig. 6d and Supplementary Fig. 6). For GR WT, residues with considerable chemical shift variance included those in the recognition helix and lever arm, with the highest variance at Arg466, which makes a direct contact with the GBS. Consistent with the impact of A477T on Gly470, we found that A477T displayed reduced chemical shift variance compared to WT throughout the recognition helix and lever arm. Thus, this global chemical shift perturbation analysis supports our conclusion that the GBS-specific conformations of GR depend on an intact dimer interface.
Building upon NMR chemical shift mapping showing that GBS modulates the GR dimer interface, we conclude that the intact dimerization surface allows for allosteric communication between dimer partners and integration of sequence signals from the GBS as a whole. Consistent with this view, disruption of this communication by the A477T mutation reduced the sequence-specific effects of the GBS on GR conformation and simplified the relationship between GR DNA binding affinity and regulatory activity. Thus, we propose that signals are transmitted from the DNA-binding interface through the lever arm, the dimerization interface, and into domains of GR outside of the DBD that confer transcriptional regulatory activity28,29.
Discussion
Genomic response elements are composed of combinations of sequence motifs that specify binding of distinct transcriptional regulators to execute gene-specific control of transcription. Even combinations of bases within a single binding motif can affect gene-specific activity12. Crystallographic study of the GR DBD revealed that the conformation of the “lever arm”, a region between the DNA recognition helix and the dimer interface, differs at distinct GBSs12. Using NMR analysis of WT and mutant GR DBD – GBS complexes we defined both the origin and the consequence of the lever arm conformational transitions—a path of affected residues including parts of the DNA interface, the lever arm, and the dimerization interface that facilitates allosteric communication between GR dimer partners. This path enables integration of sequence-specific signals from both GBS half-sites, exponentially increasing the informational complexity of the GBS.
We considered the possibility that chemical shift differences in the DNA and dimer interface result from GBS-dependent reorientation of rigid GR dimer partners relative to the DNA or each other, e.g., from differential DNA bending. We found that the magnitude of chemical shift differences did not correlate with the proximity of GR DBD residues to the DNA (data not shown), contrary to the rigid-but-reoriented model. We assessed GBS-dependent DNA bending by FRET analysis. GR binding produced very small increases GBS FRET efficiency (Supplementary Fig. 7), suggesting minimal DNA bending. Furthermore, A477T behaved equivalent to WT among the five GBSs tested. Therefore, the chemical shift differences between GBS complexes and between WT and A477T represent GBS-specific differences in GR structure, not DNA bending.
Protein allostery is critical to concepts of combinatorial control. Structural studies of isolated nuclear receptor ligand binding domains (LBDs) suggest how signaling information residing in small molecule ligands is transmitted to a coregulator recognition surface8,30–34. Here we show that GBSs drive structural changes at the DNA-binding interface that are coupled with changes in the GR dimerization interface and dimer partner, as well as correlated with distinct biophysical and transcriptional outcomes. Such structural transitions likely extend into distinct domains of intact GR to specify regulatory complex assembly and activity. These findings provide the clearest mechanistic perspective to date on functional studies showing that binding sequences modulate receptor interactions with coregulators17,35,36, that ligands modulate interactions with DNA37 and that both DNA and ligands direct interactions with coregulators38. Thus, the DBD residues identified here, together with LBD residues that interpret ligand signals39 and affect gene-specific regulation40–42 could begin to define a molecular “map” that, in the cellular context, integrates GBSs, ligands, chromatin, coregulators and post-transcriptional modifications to determine the composition and function of gene-specific transcriptional regulatory complexes.
Other than specific base contacts, what DNA signals might trigger changes in GR structure? Our data suggest that GR “measures” the spacer minor groove width as an indirect readout of spacer sequence. In the Pal complex, the Lys490 side chain reaches across the spacer minor groove to contact the phosphate backbone at the complement of spacer position 7. In the Fkbp5 complex, which has a wider spacer minor groove, Lys490 contacts the phosphate backbone of the proximal strand at position 11 (Fig. 1e). Indirect recognition of narrow minor grooves by insertion of positively charged side chains is a general feature of DNA recognition20 and a contributor to specificity among transcription factors43,44. For GR, we observe a distinct mechanism, where minor groove width imposes structural constraints on lysine-mediated backbone contacts to DNA. Thus, flexible regions of the protein may adopt conformations that accommodate differences in DNA shape, as has been demonstrated for other transcriptional regulators45.
In addition to identifying regions of GR that can adopt distinct conformational states among different GBS complexes, we found that dimer partners undergo dynamic exchange between two discrete conformations while bound to a particular GBS (Fig. 6a–b and Supplementary Fig. 5). This is consistent with the structural asymmetry between dimer partners observed by crystallography17. Lever arm Gly470 and dimer interface Ile484 residues displayed dynamics with similar timescales, consistent with structural coupling between these two regions of the DBD. We speculate that GR – GBS complexes may differentially access conformational states that interact preferentially with particular transcriptional coregulators, thus providing “assembly instructions” for different regulatory complexes. GBS-specific dissociation kinetics may, in part, impact GR activity through altering the turnover of GR – DNA complexes. Indeed, interactions with response elements are highly dynamic, on the timescale of seconds46, and regulatory complexes are actively and continuously disassembled47. How GR DNA-binding kinetics are regulated and how they impact transcriptional activity remain open questions.
Structural characterization of related nuclear receptors have shown that DNA-binding mediates conformational changes in the dimerization surface, providing a mechanism for cooperative dimerization48. We showed that GR cooperativity is affected by the precise binding sequence and impaired by the A477T mutation. Differential cooperativity is well-established as a mechanism for achieving gene-specific activity and suppressing transcriptional noise49,50. We propose that sequence-dependent conformational changes in the dimer interface modulate gene-specific cooperativity, in turn regulating the level of transcriptional activation by GR. While this manuscript was in revision, Hudson et al.51 reported that GR binds with negative cooperativity at “nGREs” where GR represses transcription. Thus, GBS-mediated allosteric regulation of cooperativity may enable GR to exhibit exceptional specificity of gene regulation ranging from transcriptional activation to repression.
Online Methods
Protein expression and purification
Expression vector pET28a containing rat GR WT DBD, residues 440–525, has been described previously17. Vector for A477T DBD was derived by PCR site-directed mutagenesis. BL21 Gold E. coli cells were grown in 50mL LB media to optical density of ~0.5–1.0, then pelleted and resuspended in 1L minimal media containing 2 g/L 15NHCl4 as the only nitrogen source. Cultures were grown to an optical density of ~0.7 and expression was induced with 0.5 mM IPTG for ~16 hours at 18°C or 8 hours at 30°C (both produced equivalent spectra). Cells were pelleted and resuspended at 40 mL/L cells in lysis buffer containing 50 mM Tris pH 7.5, 500 mM NaCl and 15 mM Imidazole, then frozen in liquid nitrogen, and stored at −80°C. Cells were lysed using an EmulsiFlex C5 homogenizer. Lysate was centrifuged for 45 min at 40,000 rpm at 4°C and run over a Nickel Sepharose (GE Healthcare) column and eluted with a linear gradient to 350 mM Imidazole. Pooled fractions were dialyzed into 20mM Tris pH 7.5, 50 mM NaCl, 2.5 mM CaCl2, and 0.5 mM β-mercaptoethanol and cleaved at 4°C overnight with 50–100 U Thrombin. Protein was further purified over a Resource S ion exchange column with a linear gradient of 50 mM-300 mM NaCl, 20 mM Tris pH 7.5, and 0.5 mM β-mercaptoethanol. Protein was concentrated using Amicon Ultra 5K MWCO (Millipore) and run over a 16/60 Superdex75 gel filtration column in NMR buffer (20 mM Sodium Phosphate pH 6.7, 100 mM NaCl, 1 mM DTT).
Protein-DNA complex formation
Single-stranded GBS oligos (IDT) were purified by 10/10 MonoQ as described previously17, and resuspended at 2 mM. Oligos were annealed at 1 mM in boiling water and cooled slowly to room temperature. To ensure solubility of GR DBD – DNA complexes, dilute protein was combined with dsDNA 1× diluted in cold NMR buffer. DNA was in ~33% excess to DBD, to ensure saturated binding. Dilute GR DBD – DNA complexes were concentrated slowly at 4°C using a 3K MWCO centrifugal filter (Amicon) to ~300 μM dimer complex and filtered with Ultrafree PVDF 0.22 μm columns (Millipore).
Protein NMR assignment
Triple-labeled GR DBD was prepared as described above except that 50 mL LB cultures were resuspended in 1 L unlabeled minimal media, grown to optical density of ~0.7, then pelleted and resuspended in 1 L of minimal media containing 2 g of 15NHCl4, 2g 13C-glucose in 90–100% D2O. Expression was induced at 30°C for 12 h. Assignments in the absence of DNA were with 1.7 mM GR DBD. The following experiments were run at 25°C on a Bruker 500 MHz spectrometer: 15N-HSQC, HNCO, HNCACO, HNCA, HNCOCA, HNCOCACB52,53, and CC(CO)NH-TOCSY. A 15N-edited NOESY54 was ran on a Bruker 800 MHz spectrometer. For assignment of DNA-bound GR, purified DBD and the Gha GBS were combined at a ratio of 2:1 monomer to GBS and concentrated to 500 μM complex (1mM GR DBD). NMR assignments were generated from 15N-edited NOESY54, TROSY-HNCO, TROSY-HNCOCA, TROSY-HNCA, TROSY-HN(CO)CA, TROSY-HNCACB, and TROSY HN(CA)CO55 experiments conducted on 600, 800 and 900 MHz spectrometers at 25°C and 35°C because some peaks gave stronger signal at 35°C. Assignments were aided by 15N-HSQC of 15N specific amino acid labeling of Ile, Leu, Val, Phe, Tyr, Lys, and reverse-labeled Arg using DL39 cells for expression. All NMR data were processed in NMRPipe56, and analyzed using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). WT DBD – GBS assignments were transferred to A477T DBD complexes, in general, according to the minimal combined 1H and 15N chemical shift difference for each assigned GR WT peak to the nearest A477T peak25. Assignment transfer was aided by 15N reverse-labeling of Arg or Lys A477T residues.
Chemical shift difference and ZZ-exchange NMR
15N-HSQC spectra were acquired on a Bruker 800 MHz spectrometer at 35°C. Peak assignments were transferred from the WT DBD - Gha complex to additional GBS complexes by measuring the minimal chemical shift difference from each assigned WT DBD - Gha peak using the formula: chemical shift difference, Δδ = [(Δ1H ppm)2 + (Δ15N ppm / 5)2]1/2 for each assigned GR WT peak to the nearest A477T peak25. Similarly, peak assignments for each A477T - GBS complex were transferred by minimal chemical shift difference using the A477T DBD - Gha complex as a reference.
For ZZ-exchange, 15N-HSQC spectra were acquired on a Bruker 800 MHz spectrometer at 35°C using a pulse sequence modified to include a mixing time of 0, 0.025, 0.05, 0.1, 0.2 and 0.4 s. The intensities of auto and exchange peaks were plotted against mixing period and curves were fit individually for Gly470 and Ile484 residues according to the formulas described by Farrow et al. 57. The kex and relaxation rate for each conformation were fit separately.
Transcriptional Reporter Assays
GBS reporter plasmids were either those generated by Meijsing et al.17 or were constructed equivalently. Reporter assays were performed as previously described17, except that we used hGR WT and hGR A477T (generated by PCR site-directed mutagenesis). Briefly, U2OS cells were seeded in DMEM + 5% FBS in 24-well plates at ~20,000 cells per well one day prior to transfection. Cells were transfected with 20ng/well GR plasmid, 20ng/well GBS-luciferase plasmid, 200 pg/well pRL Renilla, 120 ng/well empty p6R plasmid, 1 μL PLUS Reagent and 0.7 μL/well Lipofectamine Reagent (Invitrogen) for 4 hours in no-serum DMEM media. Cells were washed and recovered in DMEM + 5% FBS for 3 hrs, then treated with 100 nM dexamethasone or ethanol for 12 hrs. Luciferase induction was measured using Dual-Luciferase Reporter kit (Promega) in 96-well format using a Tecan plate reader. Data is normalized to Renilla for each well, then to ethanol-treated control, and empty pGL3 vector control.
Surface Plasmon Resonance
SPR analysis of WT and A477T DBD interaction with GBSs was carried out on a Biacore T100. Matrix-free surfaces were prepared by injection of Neutravidin (Invitrogen) across a planar saccharide monolayer with covalently coupled biotin (BP chips, Xantec Bioanalytics) at 25°C in 20 mM HEPES (pH 7.2), 150 mM NaCl. Double-stranded GBS oligos with a single 5' biotin-TEG label (IDT) were subsequently captured at immobilization levels ranging from 20 to 65 RU. WT and A477T DBDs were dialyzed overnight in NMR buffer. Following dialysis, 0.1 mg/mL BSA (Sigma) was added to both the assay buffer and the protein samples. Fifteen point concentration series were prepared by serial dilutions spanning 0.700–200 nM for WT and 1.4–400 nM for A477T. Association and dissociation times were selected to ensure equilibrium and complete dissociation. All data were processed and analyzed in Matlab. Isotherms were fit to the Hill equation: Fractional occupancy = (c/(c+K1/2))nH. Errors in K1/2 and nH were determined using a bootstrap method with replacement: after scaling of n equilibrium responses for each GBS, a random set of n data points was selected with the possibility of selecting the same data point multiple times. After 500 iterations, the 100 best-fit parameters (s.s.e) were used to find mean values and standard deviations. t1/2 parameters were fit from the following equations: R = Ro e^(-t•koff) and t1/2 = ln(2)/koff, where R = response units.
Electrophoretic Mobility Shift Assay (EMSA)
Unlabeled GR WT DBD and A477T DBD were purified and annealed as described above. Double-stranded GBS with an Alexa488 fluorophore conjugated to one of the 3' ends (IDT) were incubated for 30 min at 5 nM final concentration with GR DBD titrations in Binding Buffer (20 mm Tris pH 8, 50 mM NaCl, 1mM EDTA, 10ug/mL dIdC, 5 mM MgCl2, 200 μl/mL BSA, 5% glycerol, and 1 mM DTT) on ice. Native 8% polyacrylamide gels were run at 200 V in 0.5 × TBE at 4°C. Alexa488 signal was imaged on a Typhoon scanner (GE Healthcare) and quantified using ImageQuant. Fraction GR DBD bound was determined as 1 - [DNAfree].
GR ChIP-sequencing
Details of GR ChIP-sequencing methods and data will be described in a separate publication. Briefly, U2OS cell lines stably expressing GR WT or A477T were treated with 100 nM dex for 90 min, then cross-linked with 1% formaldehyde and quenched with 125 mM glycine. Cells were lysed for 30 min at 4°C, and nuclei pelleted at 600 × g for 5 min and resuspended in RIPA buffer (10 mM Tris pH8.0, 1 mM EDTA, 150 mM NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100). Chromatin was fragmented by Diagenode Biorupter sonication and incubated for 16 hrs at 4°C with pre-washed anti-GR antibodies bound to Protein G Dynabeads in RIPA buffer containing protease inhibitor cocktail and 2 μg/μL BSA. Complexes were washed extensively with RIPA buffer containing 500 mM NaCl followed by LiCl buffer and cross-links were reversed. DNA was column-purified (Zymogen Clean and Concentrator Kit). ChIP-sequencing libraries were prepared by end-repair, dATP-addition and ligation of sequencing adaptors containing in-house barcodes. Libraries were amplified by 17 cycles of PCR and purified by PAGE. Libraries were sequenced using the Genome Analyzer II (Illumina) with 2 × 75 bp paired-end reads, and aligned with Bowtie58. Motif analysis was performed using MEME59.
DNA oligos for NMR, EMSA, SPR and FRET
| GBS | 5' → 3' |
| Fkbp5 | gtacAGAACAgggTGTTCTtcgac |
| Gha | gtacGGAACAtaaTGTTCCtcgac |
| Gilz | gtacAGAACAttgGGTTCCtcgac |
| Pal-F | gtacAGAACAaaaTGTTCTtcgac |
| Pal-R | gtacAGAACAtttTGTTCTtcgac |
| Pal-ttg | gtacAGAACAttgTGTTCTtcgac |
| Sgk | gtacAGAACAtttTGTCCGtcgac |
| Sgk-ggg | gtacAGAACAgggTGTCCGtcgac |
| Sgk-ttg | gtacAGAACAttgTGTCCGtcgac |
Supplementary Material
Acknowledgements
We thank M. Kelly (UCSF) and J. Pelton (UC Berkeley) for NMR support; J. Cheney and S. Floor for providing scripts for NMR analysis; and S. Cooper and S. Meijsing for developing GR ChIP-seq protocols. We thank R. Fletterick, A. Johnson, G. Narlikar and members of the Yamamoto Lab for thoughtful discussions; and M. Knuesel and S. Meijsing for critical reading of the manuscript. This work was supported by US National Institutes of Health (NIH) grant CA020535 (K.R.Y), the Biophysics Graduate Group training grant NIHT32GM008284 (K.M.K), NIH grants GM08537 and 5K99CA149088 (M.A.P.), the Leukemia and Lymphoma Society Fellowship (M.A.P), the Larry S. Hillblom Foundation Fellowship (L.C.W), the Genentech and Sandler Foundation Graduate Fellowship (L.C.W), and the Cancer Research Coordinating Committee Fellowship (L.C.W).
Footnotes
Author Contributions: L.C.W. designed, performed and analyzed experiments. J.D.G. and L.C.W. designed and performed experiments for NMR sequential assignment. K.M.K. and L.C.W. designed, performed, and analyzed SPR and FRET experiments. B.J.S. and L.C.W. designed and performed ChIP-seq experiments and B.J.S. analyzed the data. M.A.P. and K.R.Y. contributed to the direction of project and L.C.W. and K.R.Y. wrote the manuscript.
References
- 1.Yamamoto K, Darimont B, Wagner R, Iniguez-Lluhi J. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harbor Symposia on Quantitative Biology. 1998;63:587–598. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- 2.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Development. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 3.Ricketson D, Hostick U, Fang L, Yamamoto K, Darimont B. A conformational switch in the ligand-binding domain regulates the dependence of the glucocorticoid receptor on Hsp90. J Mol Biol. 2007;368:729–741. doi: 10.1016/j.jmb.2007.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garza AMS, Khan SH, Kumar R. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol Cell Biol. 2010;30:220–230. doi: 10.1128/MCB.00552-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bledsoe RK, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 6.Frego L. Conformational changes of the glucocorticoid receptor ligand binding domain induced by ligand and cofactor binding, and the location of cofactor binding sites determined by hydrogen/deuterium exchange mass spectrometry. Protein Science. 2006;15:722–730. doi: 10.1110/ps.051781406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoch GA, et al. Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations. J Mol Biol. 2010;395:568–577. doi: 10.1016/j.jmb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Kauppi B, et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem. 2003;278:22748–22754. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- 9.Shah N, Scanlan TS. Design and evaluation of novel nonsteroidal dissociating glucocorticoid receptor ligands. Bioorganic & Medicinal Chemistry Letters. 2004;14:5199–5203. doi: 10.1016/j.bmcl.2004.07.052. [DOI] [PubMed] [Google Scholar]
- 10.Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 11.Rogatsky I, Waase CL, Garabedian MJ. Phosphorylation and inhibition of rat glucocorticoid receptor transcriptional activation by glycogen synthase kinase-3 (GSK-3). Species-specific differences between human and rat glucocorticoid receptor signaling as revealed through GSK-3 phosphorylation. J Biol Chem. 1998;273:14315–14321. doi: 10.1074/jbc.273.23.14315. [DOI] [PubMed] [Google Scholar]
- 12.So AY-L, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-Specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Baer J, Yamamoto KR. Analysis of the DNA-binding affinity, sequence specificity and context dependence of the glucocorticoid receptor zinc finger region. J Mol Biol. 1994;239:664–688. doi: 10.1006/jmbi.1994.1405. [DOI] [PubMed] [Google Scholar]
- 14.Baumann H, et al. Refined solution structure of the glucocorticoid receptor DNA-binding domain. Biochemistry. 1993;32:13463–13471. doi: 10.1021/bi00212a011. [DOI] [PubMed] [Google Scholar]
- 15.Härd T, et al. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990;249:157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- 16.Luisi BF, et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 17.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 19.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves. Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci USA. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci USA. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci USA. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heck S, et al. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO. 1994;13:4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Floor SN, Borja MS, Gross JD. Interdomain dynamics and coactivation of the mRNA decapping enzyme Dcp2 are mediated by a gatekeeper tryptophan. Proc Natl Acad Sci USA. 2012;109:2872–2877. doi: 10.1073/pnas.1113620109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bain DL, et al. Glucocorticoid receptor-DNA interactions: binding energetics are the primary determinant of sequence-specific transcriptional activity. J Mol Biol. 2012;422:18–32. doi: 10.1016/j.jmb.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 28.Iñiguez-Lluhí JA, Lou DY, Yamamoto KR. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 29.Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes & Development. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-a. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 31.Egea PF, et al. Crystal structure of the human RXRα ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lusher SJ, et al. Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators. J Biol Chem. 2011;286:35079–35086. doi: 10.1074/jbc.M111.273029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 34.Brzozowski AM, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–757. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 35.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 36.Engel KB, Yamamoto KR. The glucocorticoid receptor and the coregulator Brm selectively modulate each other's occupancy and activity in a gene-specific manner. Mol Cell Biol. 2011;31:3267–3276. doi: 10.1128/MCB.05351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J-C, et al. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes & Development. 2006;20:689–699. doi: 10.1101/gad.1400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, et al. DNA binding alters coactivator interaction surfaces of the intact VDR–RXR complex. Nat Struct Mol Biol. 2011;18:556–563. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 40.Rogatsky I. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA. 2003;100:13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao Y-G, Xu Y, Xu HE, Simons SS. Mutations of glucocorticoid receptor differentially affect AF2 domain activity in a steroid-selective manner to alter the potency and efficacy of gene induction and repression. Biochemistry. 2008;47:7648–7662. doi: 10.1021/bi800472w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee G-S, Simons SS., Jr. Ligand binding domain mutations of the glucocorticoid receptor selectively modify the effects with, but not binding of, cofactors. Biochemistry. 2011;50:356–366. doi: 10.1021/bi101792d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi R, et al. Functional specificity of a hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slattery M, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scully KM, et al. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 46.McNally JG, Müller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 47.Stavreva DA, Müller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmbeck SM, Dyson HJ, Wright PE. DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J Mol Biol. 1998;284:533–539. doi: 10.1006/jmbi.1998.2207. [DOI] [PubMed] [Google Scholar]
- 49.Ackers GK, Johnson AD, Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci USA. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robblee JP, Miura MT, Bain DL. Glucocorticoid receptor–promoter interactions: energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation. Biochemistry. 2012;51:4463–4472. doi: 10.1021/bi3003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013;20:53–58. doi: 10.1038/nsmb.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress in Nuclear Magnetic Resonance Spectroscopy. 1999;34:93–158. [Google Scholar]
- 53.Salzmann M, Wider G, Pervushin K, Senn H, Wüthrich K. TROSY-type Triple-Resonance Experiments for Sequential NMR Assignments of Large Proteins. J Am Chem Soc. 1999;121:844–848. [Google Scholar]
- 54.Talluri S, Wagner G. An optimized 3D NOESY-HSQC. J Magn Reson B. 1996;112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- 55.Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 57.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 58.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.