Abstract
OBJECTIVES
To assess the applicability of the Prostate Cancer Prevention Trial High Grade (Gleason grade ≥ 7) Risk Calculator (PCPTHG) in ten international cohorts, representing a range of populations.
METHODS
25,512 biopsies from 10 cohorts (6 European, 1 UK, and 3 US) were included; 4 implemented 6-core biopsies and the remaining had 10- or higher schemes; 8 were screening cohorts and 2 were clinical. PCPTHG risks were calculated using prostate-specific antigen (PSA), digital rectal examination, age, African origin and history of prior biopsy and evaluated in terms of calibration plots, areas underneath the receiver operating characteristic curve (AUC), and net benefit curves.
RESULTS
The median AUC of the PCPTHG for high grade disease detection in the 10- and higher-core cohorts was 73.5% (range 63.9% to 76.7%) compared to a median of 78.1 (range = 72.0 to 87.6) among the four 6-core cohorts. Only the 10-core Cleveland Clinic cohort showed clear evidence of under-prediction by the PCPTHG, and this was restricted to risk ranges less than 15%. The PCPTHG demonstrated higher clinical net benefit in higher- compared to six-core biopsy cohorts, and among the former, there were no notable differences observed between clinical and screening cohorts, nor between European and US cohorts.
CONCLUSIONS
The PCPTHG requires minimal patient information and can be applied across a range of populations. PCPTHG risk thresholds ranging from 5 to 20%, depending on patient risk averseness, are recommended for clinical prostate biopsy decision-making.
Keywords: Calibration, Discrimination, Net Benefit, High Grade Prostate Cancer, Risk, Prostate Cancer Prevention Trial
INTRODUCTION
The Prostate Cancer Prevention Trial (PCPT) was a North American double-blind randomized study of the chemoprevention effects of finasteride versus placebo on prostate cancer development. To enter the trial men had to be greater than 54 years of age, have a prostate-specific antigen (PSA) level less than or equal to 3 ng/mL and a normal digital rectal exam (DRE) result. Then during seven years on the trial men were annually screened and referred to interim biopsy (6-core) whenever their PSA exceeded 4 ng/mL or their DRE was abnormal. At the end of the trial, all men were requested to undergo prostate biopsy regardless of their current PSA and DRE examination, or whether they had previously undergone prostate biopsy that was negative for prostate cancer. Both the stringent inclusion criteria and follow-up protocol contribute to the uniqueness of the PCPT population; this population was more generally older, healthier and with more Caucasian representation than the rest of the US. The PCPT Risk Calculator for predicting high grade (Gleason grade ≥ 7) prostate cancer (PCPTHG) was developed from 5,519 men from the placebo arm who all underwent 6-core prostate biopsy, 257 (4.7%) of whom were diagnosed with high grade disease [1]. The calculator includes the risk factors PSA, DRE, African origin, age and history of a prior biopsy. The PCPTHG was designed for high grade risk assessment based on a typical urological visit based on PSA and DRE; it therefore does not incorporate prostate volume, which is used in many other contemporary prostate cancer nomograms. Recently, an option to the calculator for missing DRE was also installed to the online calculator, so a DRE is also not required.
The PCPTHG was posted online in 2006 and is assessed by patients and clinicians worldwide as a counseling aid for the decision to undergo prostate biopsy. To our knowledge it is the only high grade prostate cancer risk calculator with analytical formulas published. Hence to date it has been externally evaluated in three studies [2–4]. These single validation studies have compared performance of the PCPTHG to nomograms that were developed on the same cohort of patients, thus yielding biased conclusions that the PCPTHG does not externally validate and requires extra parameters such as %freePSA or prostate volume. But, as we have previously illustrated, performance of a risk prediction tool is a property of both the tool and the population to which it is applied [5,6] and a virtue of the PCPTHG is that it is based on the standard risk factors and thus more widely applicable. Therefore, this report presents the first external validation study of the PCPTHG on multiple cohorts in order to identify potential populations where it may or may not be applicable.
METHODS
Data were included from ten European and US cohorts belonging to the Prostate Biopsy Collaborative Group (PBCG), where criteria for biopsy referral and sampling scheme have been previously described [5]. These included five screening cohorts from the European Randomized study of Screening for Prostate Cancer (ERSPC), all following a primarily 6-core biopsy sampling scheme similar to the PCPT except for Tarn (10–12 core); three additional screening cohorts, San Antonio Biomarkers Of Risk of cancer study (SABOR), Texas, US, ProtecT, United Kingdom, and Tyrol, Austria, each following a primarily 10-core biopsy scheme; and two US clinical cohorts, Cleveland Clinic, Ohio, and the Durham VA, North Carolina, both following a mixed but primarily 10–14-core biopsy scheme. All cohorts except for ERSPC Goeteborg and Rotterdam Rounds 1 included some patients who had been previously screened. All biopsies after a diagnosis of prostate cancer and 221 biopsies positive for prostate cancer but with missing Gleason grade (across five of the cohorts) were excluded from the analysis.
Clinical characteristics of each cohort were summarized in terms of descriptive statistics, including median, ranges and percents. For each biopsy in the dataset, the PCPTHG risk of high grade cancer was computed, requiring PSA (ng/mL), DRE (normal vs. suspicious), African origin (yes vs. no), age and history of a prior negative prostate biopsy (yes vs. no). An iterative multiple imputation procedure was used to impute missing values of any of the risk factors when the percentage of missing data for a risk factor in a cohort was less than 100% [7]. For cohorts with African origin or DRE not recorded for any participants, single imputation of “not of African origin” or “negative DRE”, respectively, were implemented.
Calibration plots were computed by comparing observed proportions of cancer to mean PCPTHG risks by PCPTHG deciles observed in the cohort; pointwise 95% confidence bands were computed using the binomial formula with spline smoothing. Discrimination was calculated and displayed via receiver operating characteristics curves (ROC). Areas underneath the ROC curve (AUC) were calculated for PCPTHG risks using the Wilcoxon test statistic and compared to the AUC of PSA by non-parametric U statistics. Net benefit curves were computed by calculating for each possible PCPTHG risk (p) ranging from 0 to 100%, Sensitivity × π − (1 − Specificity) × (1−π) × [p/(1−p)], and were assessed for their gain over the corresponding net benefit curves of referring no patients to biopsy (horizontal line at 0) or simply referring all patients to biopsy, π − (1−π) × [p/(1−p)], where Sensitivity and Specificity are defined at the cutpoint p and π denotes the cohort prevalence of high grade disease (the fraction of all participants, with and without cancer, that have high grade disease)[8].
RESULTS
High grade prostate cancer rates ranged from a low of 4% in Goeteberg Rounds 2–6 to a high of 22% in the Durham VA cohort, which also had the highest reported percentage of men of African origin (45%), one of the risk factors included in the PCPTHG (Table 1). Across the 25,512 biopsies from the ten cohorts combined, the AUC of the PCPTHG was 74.6%, a modest 3 point increase over the AUC for PSA (71.5%, p < 0.0001). Use of PCPTHG risk thresholds of 5%, 10% and 20% as definitions of a positive test for referral to biopsy would have resulted in 84.4%, 41.7% and 15.0%, respectively, of all high-grade negative biopsies testing positive (percent unnecessary biopsies), and 4.7%, 24.0%, and 51.5% missed high-grade prostate cancer cases, respectively. Statistics according to the individual cohorts are shown in Table 1.
Table 1.
Clinical characteristics of each cohort: age and PSA report median (range), the rest report number n (%), and areas underneath the receiver operating characteristic curves for the PCPTHG and PSA with p-value for comparison of the two.
| Cohort (screening vs. clinical, primary number of cores) | ERSPC Goeteborg Round 1 (screening, 6 cores) | ERSPC Goeteborg Rounds 2–6 (screening, 6 cores) | ERSPC Rotterdam Round 1 (screening, 6 cores) | ERSPC Rotterdam Rounds 2–3 (screening, 6 cores) | ERSPC Tarn (screening, 10–12 cores) | SABOR (screening, 10 cores) | Cleveland Clinic (clinical, 10– 14 cores) | ProtecT (screening, 10 cores) | Tyrol (screening, 10 cores) | Durham VA (clinical, 10–14 cores) |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of patients | 740 | 1241 | 2889 | 1494 | 295 | 389 | 2631 | 7324 | 4029 | 1846 |
| Number of biopsies | 740 | 1241 | 2889 | 1494 | 295 | 389 | 3286 | 7324 | 5449 | 2405 |
| Age | ||||||||||
| Median (range) | 61 (51,70) | 63 (53, 71) | 66 (55, 75) | 67 (59, 75) | 64 (55, 71) | 63 (50,75) | 64 (50,75) | 63 (50,72) | 62 (50,75) | 64(50,75) |
| PSA | ||||||||||
| Median Range | 4.7 (0.5, 226.0) | 3.6 (2.0, 88.8) | 5.0 (0.0, 245.0) | 3.5 (0.4, 99.5) | 4.4 (1.6, 131.0) | 3.4 (0.2,919.2) | 5.8 (0.2,491.7) | 4.4 (3.0, 847.0) | 4.1 (0.1,3210.0) | 5.2 (0.1,1250.3) |
| DRE result | ||||||||||
| Normal | 614 (83%) | 1117 (90%) | 2135 (74%) | 1182 (79%) | 177 (60%) | 279 (72%) | 3083(94%) | 0 | 4958(91%) | 887(37%) |
| Abnormal | 126 (17%) | 124 (10%) | 754 (26%) | 312 (21%) | 91 (31%) | 110 (28%) | 203 (6%) | 0 | 491 (9%) | 265 (11%) |
| Unknown | 0 | 0 | 0 | 0 | 27 (9%) | 0 | 0 | 7324(100%) | 0 | 1253(52%) |
| African origin | ||||||||||
| No | 0 | 0 | 0 | 0 | 0 | 346 (89%) | 2818(86%) | 6933(95%) | 0 | 1212(50%) |
| Yes | 0 | 0 | 0 | 0 | 0 | 43 (11%) | 422 (13%) | 34 (0%) | 0 | 1071(45%) |
| Unknown | 740 (100%) | 1241 (100%) | 2889 (100%) | 1494 (100%) | 295 (100%) | 0 | 46 (1%) | 357 (5%) | 5449(100%) | 122 (5%) |
| Prior biopsy | ||||||||||
| Yes | 0 | 0 | 0 | 0 | 0 | 95(24%) | 1091(33%) | 0 | 1524(28%) | 565(23%) |
| No | 740(100%) | 1241(100%) | 2889(100%) | 1494(100%) | 295(100%) | 294(76%) | 2195(67%) | 7324(100%) | 3925(72%) | 1840(77%) |
| Cancer | 192 (26%) | 322 (26%) | 794 (27%) | 388 (26%) | 93 (32%) | 130 (33%) | 1292(39%) | 2570(35%) | 1367 (25%) | 1134(47%) |
| High-grade cancer(% biopsies) | 40 (5%) | 53 (4%) | 286 (10%) | 91 (6%) | 51 (17%) | 35 (9%) | 623 (19%) | 867 (12%) | 456 (8%) | 528 (22%) |
| AUC of PCPTHG % (AUC PSA, p- value to PSA) | 87.6 (82.4, 0.01) | 72.0 (59.6, <.001) | 82.2 (77.5, <.001) | 74.1 (69.8, 0.046) | 76.7 (64.1,<.001) | 69.5 (68.0,0.60) | 63.9 (59.3,<.001) | 75.4 (75.1,0.35) | 73.2 (69.2,<.001) | 73.9 (69.6,<.001) |
| Number of unnecessary biopsies for thresholds 5%, 10%, 20% (percent of negative biopsies) | 632, 275, 123 (90.3, 39.3, 17.6) | 1054, 222, 35 (88.7, 18.7, 2.9) | 2512, 1575, 646 (96.5, 60.5, 24.8) | 1246, 448, 111 (88.8, 31.9, 7.9) | 233, 134, 38 (95.5, 54.9, 15.6) | 219, 116, 34 (61.9, 32.8, 9.6) | 2334, 1517, 579 (87.6, 57.0, 21.7) | 5849, 2083, 448 (90.6, 32.3, 6.9) | 3197, 1705, 649 (64.0, 34.1, 13.0) | 1691, 1306, 699 (90.1, 69.6, 37.2) |
| Number of missed high-grade cancers for thresholds 5%, 10%, 20% (percent of positive biopsies) | 0, 3, 8 (0, 7.5, 20.0) | 2, 25, 41 (3.8, 47.2, 77.4) | 0, 26, 72 (0, 9.1, 25.2) | 5, 28, 55 (5.5, 30.8, 60.4) | 0, 4, 29 (0, 7.8, 56.9) | 5, 14, 25 (14.3, 40.0, 71.4) | 39, 162, 377 (6.3, 26.0, 60.5) | 27, 266, 526 (3.1, 30.7, 60.7) | 56, 154, 266 (12.3, 33.8, 58.3) | 7, 45, 162 (1.3, 8.5, 30.7) |
ERSPC: European Randomized Study of Screening for Prostate Cancer; SABOR: San Antonio Biomarkers of Risk of Prostate Cancer Study; VA: Veteran’s Administration; PSA: prostate-specific antigen; DRE: digital rectal examination; AUC:area underneath the receiver operating characteristic curve; PCPTHG: Prostate Cancer Prevention Trial Risk Calculator for High Grade Prostate Cancer.
Evaluation of the PCPTHG for10–12 core biopsy schemes – comparison to 6-core
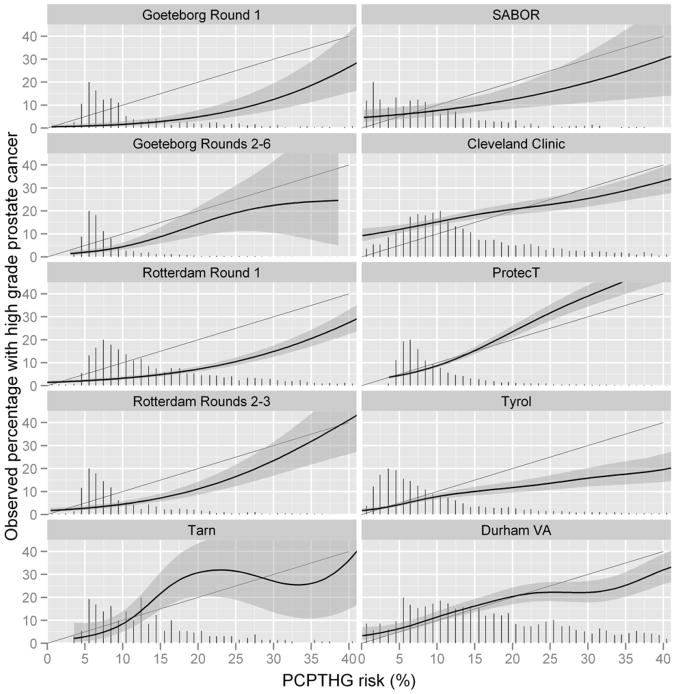
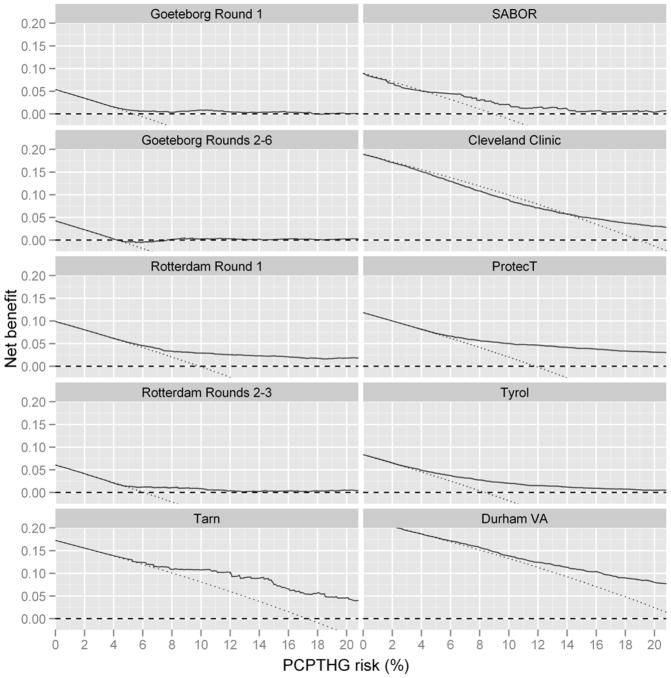
The last six cohorts of Table 1 and Figures 1 and 2 (going down the columns) implemented 10-and higher-core schemes. The median AUC of the PCPTHG for high grade disease detection in the 10- and higher-core cohorts was 73.5% (range 63.9% to 76.7%). Both the median and range were lower than that for the 4 ERSPC cohorts that had 6-core biopsy schemes (median = 78.1, range = 72.0 to 87.6). In two of the six 10- and higher-core cohorts, the PCPTHG was not statistically significantly better than PSA for high grade cancer discrimination (p-values > 0.05); in all four 6-core cohorts the PCPTHG performed statistically significantly better than PSA (p-value < 0.05). Of all cohorts included in the analysis, only the 10-core Cleveland Clinic cohort showed clear evidence of under-prediction, and this was restricted to risk ranges less than 15% (Figure 1). The PCPTHG primarily over-predicted high grade prostate cancer in all 6-core ERSPC screening studies. Clinical net benefit was not lower for the six higher-core biopsy scheme cohorts compared to the 6-core biopsy cohorts, in fact it was often higher (Figure 3). In three of the four 6-core ESRPSC screening cohorts, there was no clinical benefit to using the PCPTHG across all risk thresholds.
Figure 1.
Calibration plots for the PCPTHG showing average PCPTHG risks for men grouped by their PCPTHG risk value (x-axis) compared to the actual percentage of these groups which were diagnosed with prostate cancer (y-axis). Shaded areas represent approximate 95% confidence intervals. Perfect calibration would fall on the diagonal line where predicted risks equal observed rates of prostate cancer and adequate calibration is indicated where shaded regions overlap the black diagonal lines. Vertical bars at the bottom are scaled histograms depicting relative frequencies of participants obtaining specified PCPTHG risks.
Figure 2.
Net benefit curves for the PCPTHG (solid black line) versus the rules of biopsying all men (dashed line) and no men (dotted horizontal line at 0). A risk tool has clinical benefit when a specific risk threshold (x-axis) is used for referral to biopsy when its net benefit curve is higher than the curves corresponding to biopsying all men or no men.
Comparison of the PCPTHG in healthy/ screening to clinically-referred populations
Restricting attention to cohorts with 10- and higher biopsy core schemes, the four screening cohorts had PCPTHG AUCs 76.7% (Tarn), 69.5% (SABOR), 75.4% (ProtecT), and 73.2% (Tyrol), respectively, which overlapped with the AUCs observed in the clinical cohorts, 63.9% (Cleveland Clinic) and 73.9% (Durham VA). Of note is the large 10 point difference between the Cleveland Clinic and Durham VA AUCs (Table 1). There were no obvious differences between calibration or in clinical net benefit in the higher-core screening cohorts compared to the higher-core clinical cohorts (Figure 2).
Comparison of the PCPTHG for US versus European populations
Restricting attention to cohorts with 10- and higher biopsy core schemes, this comparison involves the 3 US cohorts: SABOR (AUC = 69.5%), Cleveland Clinic (63.9%) and Durham VA (73.9%) versus the 3 European cohorts: Tarn (76.7%), ProtecT (75.4%) and Tyrol (73.2%). The range of AUCs for the European cohorts is in fact shifted higher than that for the US cohorts. The sample size of Tarn is too low to make inference concerning calibration and for low levels of high grade risk (< 10%) the PCPTHG appears as good or better calibrated in the two remaining European higher-core cohorts (ProtecT and Tyrol) compared to the US cohorts (Figure 1). The higher-core European screening cohorts Tarn, ProtecT and Tyrol show comparable clinical net benefit to the US higher-core cohorts, with the exception of the US Cleveland Clinic cohort, where the PCPTHG had lower clinical net benefit.
DISCUSSION
This study did not show decreased performance of the PCPTHG for contemporary cohorts that use a higher number of cores compared to cohorts that had implemented 6-core biopsy schemes (used in the PCPT), in cohorts comprising clinical patients rather than healthy patients undergoing screening, or in European versus U.S. cohorts. Two primary advantages of the PCPTHG are that it requires only easily obtainable patient parameters that are part of a routine clinical exam (not including prostate volume), and that it is available on the internet. Performance characteristics of the PCPTHG were graded on three scales: discrimination, calibration and net benefit. On some populations and judged by some criteria, the PCPTHG was no better than other screening methodologies, for example, in SABOR and ProtecT, the AUC of the PCPTHG did not differ statistically significantly from PSA (Table 1). These two cohorts implemented contemporary 10- and higher-core biopsy schemes. Extended core sampling has been shown to increase both prostate cancer and high-grade disease detection [9–11]. Nevertheless on no population and according to no scale, was the PCPTHG worse than simpler screening measures such as PSA, and this combined with the PCPTHG’s simplicity and availability implies that it can be implemented as a complementary aid to the physician and patient in their decision to go forward or not with prostate biopsy, without the expectation that it could cause harm to the patient.
There are several limitations to the current study. The primary limitation is that comparison of cohorts that evolved under different protocols as a means of assessing whether specific factors, such as 6- versus higher-core biopsy schemes, affects performance characteristics of a risk tool is no substitution for a single protocol analysis where individual factors, such as actual number of biopsy cores taken, are recorded for each patient. Cohorts were classified according to the primary number of cores used. Nevertheless, given this limitation, we believe a multiple external validation of a risk tool gives a more balanced assessment of the operating characteristics of a risk tool than a single evaluation study, and can be more informative as to when and where the risk tool works in practice.
Other limitations are that all men underwent prostate biopsy, and thus had one or more risk factors for prostate cancer. It was not possible to account for subtle differences in biopsy technique that might have had significant impact on high grade cancer detection rates, such as choice of specific location to obtain cores independent of the number of cores. Furthermore, central pathology review was not achievable, so it is possible that variation in aggressiveness in declaring biopsy specimens to have high grade cancer might have occurred. The PCPTHG was designed to predict high-grade disease defined as Gleason 7 and higher, but contemporary risk prediction typically focuses on clinically significant cancer, which may not include Gleason 7. African origin, a key risk factor in the PCPTHG, was entirely missing for 6 of the cohorts. Since these cohorts were all European, it could be assumed that their African origin proportion was negligible. DRE was not recorded for the ProtecT cohort so assumed to be normal for all participants in that cohort. This can alternatively be considered a bonus evaluation of the robustness of the online PCPTHG, since it now allows use without DRE performed and then defaults to normal. This feature followed a prior study on SABOR that revealed DRE to be highly unstable, reverting to normal the year after an abnormal result in nearly 75% of incidences [12].
There are currently many online nomograms and risk calculators available for prostate cancer, but since validation is both a property of the risk tool and cohort under question and external validations are performed on single cohorts, it can be confusing figuring which calculator is optimal [13]. Though novel biomarkers, such as %freePSA, and additional parameters, such as prostate volume, could improve upon existing calculators, the cost of including a more difficult to obtain risk factor has to be weighed against a more widely-applicable risk calculator. The rate of complications from prostate biopsy range from 2 to 4%, and individual patients and doctors will vary in their assessment of how high a risk of high grade disease needs to be to prompt them to biopsy [14]. Therefore, we recommend that PCPTHG risks in the range of 5% to 20% be used depending on how much the individual weights the harm of a missed high grade cancer to the harm of an unnecessary biopsy.
Acknowledgments
FUNDING ACKNOWLEDGEMENT
Statistics supported by a National Cancer Institute Cancer Cancer Center Support Grant for the Cancer Therapy and Research Center at the University of Texas Health Science Center at San Antonio [P30-CA054174]. Grants to support the work of the ERSPC include: European Union Grants SOC 95 35109, SOC 96 201869 05F022, SOC 97 201329, SOC 98 32241, the 6th Framework Program of the EU: PMark:LSHC-CT-2004-503011; The Dutch Cancer Society (KWF 94-869, 98-1657, 2002-277, 2006-3518); The Netherlands Organisation for Health Research and Development (ZonMW-002822820, 22000106, 50-50110-98-311); The Prostate Cancer Research Foundation of Rotterdam (SWOP); Institut National du Cancer convention 12-2008 Convention : 07/3D1616/SPC-111-13/NG-NC Appel à projets 2007 : Santé publique et Cancers Ligue Nationale Contre le cancer PROJET N° PRE08/AV – CONV. DRC 2009/119 (Tarn); Beckman-Coulter-Hybritech Inc; Abbott Pharmaceuticals, Sweden; Af Jochnick’s foundation; Catarina and Sven Hagstroms family foundation; Gunvor and Ivan Svensson’s foundation; Johanniterorden, King Gustav V Jubilée Clinic Cancer Research Foundation; Sahlgrenska University Hospital ; Schering Plough, Sweden, Swedish Cancer Society (Contract numbers 09 0107, 080315 and 083455); Wallac Oy, Turkku, Finland. The Tyrol study is supported by the International Agency for Research on Cancer, Lyon and the Tyrolean Prostate Cancer Early Detection Group. The SABOR project is supported by the San Antonio Center of Biomarkers of Risk for Prostate Cancer CA086402. The ProtecT study is funded by the UK NIHR Health Technology Assessment Programme (projects 96/20/06, 96/20/99).
Footnotes
CONFLICT OF INTEREST: The authors declare that they have no conflict of interest.
References
- 1.Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, Lucia MS, Feng Z, Parnes HL, Coltman CA., Jr Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–534. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen CT, Yu C, Moussa A, Kattan MW, Jones JS. Performance of Prostate Cancer Prevention Trial Risk Calculator in a contemporary cohort screened for prostate cancer and diagnosed by extended prostate biopsy. J of Urol. 2010;183:529–533. doi: 10.1016/j.juro.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Ngo TC, Turnbill BB, Lavori PW, Presti JC., Jr The prostate cancer risk calculator from the Prostate Cancer Prevention Trial underestimates risk of high grade cancer in contemporary referral patients. J Urol. 2011;185:483–488. doi: 10.1016/j.juro.2010.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nam RK, Kattan MW, Chin JL, Trachtenberg J, Singal R, Rendon R, Klotz LH, Sugar L, Sherman C, Izawa J, Bell D, Stanimirovic A, Venkateswaran V, Diamandis EP, Yu C, Loblaw A, Narod SA. Prospective multi-institutional study evaluating the performance of prostate cancer risk calculators. J Clin Oncol. 2011;29:2959–2964. doi: 10.1200/JCO.2010.32.6371. [DOI] [PubMed] [Google Scholar]
- 5.Vickers AJ, Cronin AM, Roobol MJ, Hugosson J, Jones JS, Kattan MW, Klein E, Hamdy F, Neal D, Donovan J, Parekh DJ, Ankerst D, Bartsch G, Klocker H, Horninger W, Benchikh A, Salama G, Villers A, Freedland SJ, Moreira DM, Schroeder FH, Lilja H. The relationship between prostate-specific antigen and prostate cancer risk: the Prostate Biopsy Collaborative Group. Clinical Cancer Research. 2010;16:4374–4381. doi: 10.1158/1078-0432.CCR-10-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ankerst DP, Boeck A, Freedland SJ, Thompson IM, Cronin AM, Roobol MJ, Hugosson J, Jones JS, Kattan MW, Klein EA, Hamdy F, Neal D, Donovan J, Parekh DJ, Klocker H, Horninger W, Benchikh A, Salama G, Villers A, Moreira DM, Schroeder FH, Lilja H, Vickers AJ. Evaluating the PCPT Risk Calculator in ten international biopsy cohorts: results from the prostate biopsy collaborative group. World Journal of Urology. 2012 doi: 10.1007/s00345-011-0818-5. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Buuren S. Multiple imputation of discrete and continuous data by fully conditional specification. Statistical Methods in Medical Research. 2007;16:219–242. doi: 10.1177/0962280206074463. [DOI] [PubMed] [Google Scholar]
- 8.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Medical Decision Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takenaka A, Hara R, Hyodo Y, Ishimura T, Sakai Y, Fujioka H, Fujii T, Jo Y, Fujisawa M. Transperineal extended biopsy improves the clinically significant prostate cancer detection rate: a comparative study of 6 and 12 biopsy cores. Int J Urol. 2006;13:10–14. doi: 10.1111/j.1442-2042.2006.01221.x. [DOI] [PubMed] [Google Scholar]
- 10.O’Connell MJ, Smith CS, Fitzpatrick PE, Keane CO, Fitzpatrick JM, Behan M, Fenlon HF, Murray JG. Transrectal ultrasound-guided biopsy of the prostate gland: value of 12 versus 6 cores. Abdom Imaging. 2004;29:132–136. doi: 10.1007/s00261-003-0089-8. [DOI] [PubMed] [Google Scholar]
- 11.Eskicorapci SY, Baydar DE, Akbal C, Sofikerim M, Guenay M, Ekici S, Ozen H. An extended 10-core transrectal ultrasonography guided prostate biopsy protocol improves the detection of prostate cancer. 2004;45:444–448. doi: 10.1016/j.eururo.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Ankerst DP, Miyamoto R, Nair PV, Pollock BH, Thompson IM, Parekh DJ. Yearly prostate specific antigen and digital rectal examination fluctuations in a screened population. Journal of Urology. 2009;181:2071–2075. doi: 10.1016/j.juro.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickers AJ, Cronin AM. Everything you always wanted to know about evaluating prediction models (but were too afraid to ask) Urology. 2010;76:1298–1301. doi: 10.1016/j.urology.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson IM, Ankerst DP. The benefits of risk assessment tools for prostate cancer. European Urology. 2012 doi: 10.1016/j.eururo.2011.12.012. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]