Abstract
Recent studies have suggested that changes in serum phosphate levels influence pathological states associated with aging such as cancer, bone metabolism, and cardiovascular function, even in individuals with normal renal function. The causes are only beginning to be elucidated but are likely a combination of endocrine, paracrine, autocrine, and cell autonomous effects. We have used an integrated quantitative biology approach, combining transcriptomics and proteomics to define a multi-phase, extracellular phosphate-induced, signaling network in pre-osteoblasts as well as primary human and mouse mesenchymal stromal cells. We identified a rapid mitogenic response stimulated by elevated phosphate that results in the induction of immediate early genes including c-fos. The mechanism of activation requires FGF receptor signaling followed by stimulation of N-ras and activation of AP-1 and serum response elements. A distinct long-term response also requires FGF receptor signaling and results in N-ras activation and expression of genes and secretion of proteins involved in matrix regulation, calcification, and angiogenesis. The late response is synergistically enhanced by addition of FGF23 peptide. The intermediate phase results in increased oxidative phosphorylation and ATP production and is necessary for the late response providing a functional link between the phases. Collectively, the results define elevated phosphate, as a mitogen and define specific mechanisms by which phosphate stimulates proliferation and matrix regulation. Our approach provides a comprehensive understanding of the cellular response to elevated extracellular phosphate, functionally connecting temporally coordinated signaling, transcriptional, and metabolic events with changes in long-term cell behavior.
Keywords: Inorganic phosphate, FGF receptor signaling, immediate early genes, angiogenesis, AP-1
INTRODUCTION
It is becoming increasingly apparent that diet can have profound effects on functional genomics and represents an area of research that has yet to be exploited for potential health benefits. Inorganic phosphate (Pi) represents a common dietary element that may directly alter cell phenotype in just such a manner. Pi is critical to the human body on many levels. At the cellular level it is required as a component of energy metabolism, kinase signaling and in the formation and function of DNA and lipids. At the systemic level it is critical for normal skeletal and dentin formation. Recent in vivo studies in rodents and humans have suggested positive correlations between high serum Pi and cancer (Camalier et al., 2010; Jin et al., 2009) and cardiovascular disease (Dhingra et al., 2007; Ferro et al., 2009; Mathew et al., 2008; Onufrak et al., 2008; Tonelli et al., 2005), and a negative correlation with bone quality (Huttunen et al., 2006; Huttunen et al., 2007; Koshihara et al., 2005a; Koshihara et al., 2005b). In addition to diet a number of disease states cause changes in serum and cellular phosphate levels including; end-stage renal disease, hyperparathyroidism, multiple myeloma, oncogenic osteomalacia, and chronic alcoholism among others. The underlying mechanisms by which elevated serum Pi influence cell behavior likely involve autocrine, paracrine, and endocrine signaling as well as cell autonomous effects.
Cell culture studies have linked elevated Pi to changes in cell phenotype including; osteoblast mineralization (Beck, 2003), chondrocytes differentiation (Fujita et al., 2001; Julien et al., 2007; Mansfield et al., 2001), cementoblast formation (Foster et al., 2006), odontoblast differentiation (Lundquist et al., 2002), osteoclast differentiation (Kanatani et al., 2003; Mozar et al., 2007; Takeyama et al., 2001; Yates et al., 1991) as well as pathological calcification of osteoarthritic cartilage (Cecil et al., 2005) and vascular smooth muscle (Giachelli, 2003; Jono et al., 2000), and altered kinetics of transport in the kidney (Kido et al., 1999). Pi has been identified as a limiting nutrient in the proliferation of Swiss 3T3 cells (Hilborn, 1976; Holley and Kiernan, 1974; Weber and Edlin, 1971), to alter cell growth properties (Chang et al., 2006; Conrads et al., 2005; Cunningham and Pardee, 1969; Engstrom and Zetterberg, 1983; Roussanne et al., 2001) and is required in the regulation and stimulation of transformation (Rubin and Chu, 1984; Rubin and Sanui, 1977). The cellular and molecular mechanism(s) by which elevated Pi alters cell behavior remains to be fully elucidated, however, data suggest a complex temporally controlled series of specific events likely as specific as many of the more traditional signaling molecules.
Certain effects of elevated Pi on cell behavior have been demonstrated to be cell autonomous. It was noted almost four decades ago that contact inhibited 3T3 cells respond to serum stimulation with a rapid increase in Pi transport (Barsh et al., 1977; Cunningham and Pardee, 1969; de Asua et al., 1974). Pi transport is regulated by a family of sodium dependent phosphate transporters (Collins et al., 2004; Tenenhouse, 2005; Werner et al., 1998). Type II transporters (current nomenclature Slc34a1-3) are thought to be responsible mainly for absorption in the intestine and resorption in the kidney (reviewed in (Tenenhouse, 2007)) although recent data suggests the possibility of a more diverse function (Lundquist et al., 2007). Type III transporters (current nomenclature Slc20a1-2) are expressed more ubiquitously but evidence suggest important roles in calcifying tissues (reviewed in (Collins et al., 2004)). Recent studies have demonstrated the requirement of at least one specific Pi co-transporter, Slc20a1 (Pit-1, Glvr-1), for Pi-induced changes in cell behavior suggesting direct cell autonomous effects of elevated extracellular Pi (Kimata et al., 2010; Li et al., 2006; Suzuki et al., 2006; Yoshiko et al., 2007). Slc20a1 has also recently been linked to proliferation of tumor cells (Beck et al., 2009) and transformation of NIH3T3 cells (Byskov et al., 2012) emphasizing the role of Pi transport in the response.
In this study, transcriptomic, proteomic, and computational analyses were used to facilitate an understanding of the coordinated temporal and spatial response of cells to elevated Pi levels including prediction and validation of stimulated signaling pathways, transcriptional regulation, changes in cell processes, and changes in cell phenotype. This approach has allowed us to begin to comprehensively understand the cellular response functionally connecting temporally coordinated membrane, signaling, transcriptional, metabolic events with changes in long-term cell behavior in response to elevated Pi. The demonstration of a universal Pi-induced signaling pathway in cell types from different developmental origins suggest a common and conserved mechanism by which cells respond to changes in extracellular Pi. Recent studies have suggested that serum Pi levels may influence initiation and/or progression of pathological states associated with cardiovascular disease, bone metabolism, kidney function, and cancer. Results presented herein provide insight into the cell autonomous mechanism(s) involved and identify potential novel therapeutic targets to modulate the response.
MATERIALS AND METHODS
Cell culture
The murine calvaria-derived osteoblast MC3T3-E1 cells (Beck et al., 1998; Sudo et al., 1983) were grown in αMEM (Irvine Scientific, Santa Ana CA) and supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA), 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM L-glutamine (Invitrogen Corp., Carlsbad, CA). αMEM contains 1mM Pi and added Pi was in the form of NaPO4 (pH7.4). Unless otherwise noted, all short time point experiments (<6hr) were performed on cells which had been serum starved overnight (0.2% FBS) and all long term experiments (>6 hours) were performed on cells cultured in medium containing 10% FBS. Inhibitors; FGFR (PD173074 and PD166866), EGFR (Cyclopropanecarboxylic acid), IGFR (N-(2-Methoxy-5-chlorophenyl)-N′-(2-methylquinolin-4-yl)-urea), VEGFR (KRN951), Rotenone, GDPβS (Guanosine 5′-O-(2-Thiodiphosphate)) and U0126 were purchased from CalBiochem (Gibbstown, NJ) and PDGFR (6,7-Dimethoxy-3-phenylquinoxaline (5μM)) from Sigma (St. Louis, MO). FGF2 was purchased from Sigma. hFGF23 (Prospec-Tany, East Brunswick, NJ: cyt-374) was combined with mKlotho (R and D Systems: Minneapolis, MN 1819-KL) and heparin (Sigma) at a 1:1:10 ratio. FGF23 was added at 500ng/ml simultaneously with the addition of Pi.
Human bone marrow stromal cells
Human bone marrow was obtained from patients undergoing iliac crest bone graft to repair traumatic mandible fracture at Emory University, Atlanta GA, 30322. All studies were approved by the Emory Institutional Review Board. Samples from six patients were analyzed, 4 males and 2 females ranging in age from 27–53. Bone marrow was diluted in DMEM (1:20) with 10% FBS, penicillin, and streptomycin as described in (Beck et al., 2012). After 10–14 days the adherent cells were sub-cultured twice before being treated with Pi.
Mouse bone marrow stromal cells
Bone marrow was obtained from iliac crest and femur by centrifugation after sacrifice. Bone marrow cells were resuspended in DMEM+10%FBS and sub-cultured for multiple passages to generate sufficient cell numbers similar to hBMSCs. Studies were approved by and conducted in accordance with the Emory Institutional Animal Care and Use Committee.
Northern blotting and plasmids used for probes
Northern blots were performed as described previously (Camalier et al., 2010) using a 1% formaldehyde-agarose gel. 32P-labelled probes were prepared using a random prime labeling kit (Roche Diagnostics Corp., Indianapolis, IN). Between successive probes, blots were stripped by treatment with boiling 0.1% SDS. Radiochemicals were obtained from Perkin Elmer Life Sciences Inc. (Boston, MA).
Plasmids used for Northern probes
The murine osteopontin plasmid, mop3, was provided by Marian Young and described in (Fisher et al., 1995). The actin, cox-2 (ptgs2), Nrf2 (nfe2l2), osteocalcin, and Egr1 probes have been described previously (Beck et al., 2003; Beck et al., 1998; Camalier et al., 2010). The c-fos plasmid was provided by R. Bravo (Bravo et al., 1986). The cDNA for Slc20a1 was purchased from OpenBiosystems (Huntsville, AL). Primers used for northern probes were generated from Pi treated MC3T3-E1 cell cDNA and are listed in Table S5.
Quantitative real time PCR
RNA was isolated using TRizol (Invitrogen). cDNA was generated using First strand synthesis kit (Invitrogen) according to manufacturer’s instruction. qRT-PCR was performed using EvaGreen qPCR master mix (Biotium, Hayward CA) on an Applied Biosystems-7000 thermocycler. Primers were designed by website qPrimerDepot (http://mouseprimerdepot.nci.nih.gov/), purchased from IDT (Coralville, IA), and sequences detailed in Table S5. Analysis of hSOST RNA was performed using TAQMAN primers (Life Technologies; Carlsbad, CA) according to manufacturer’s suggestion (Hs00228830). All qRT-PCR results were calculated using the ΔΔCT method.
Transcriptomics
MC3T3-E1 cells in growth medium were treated with 10 mM Pi for times from 15 minutes through 72 hours as indicated (Fig. S1). All Pi-induced time points were compared with untreated control. Samples were harvested in parallel for proteomics analysis. Oligo microarrays, printset Mm_MEEBO_ATC_VIP4_031405, were printed by the Laboratory of Molecular Technology (Frederick, MD). Total RNA was labeled with either Cy3 or Cy5 Mono-Reactive Dye (Amersham Pharmacia Biotech, Inc., Piscataway, NJ). Probes were purified using a Qiaquick PCR purification kit (Qiagen, Valencia, CA). cDNA was labeled using Pronto dye (Corning, Corning NY) and hybridized in a 50% formamide buffer at 42°C overnight. Arrays were washed and scanned using a GenePix microarray scanner (Axon Instruments, Union City, CA) and data analyzed by GenePix Pro 4.0.
Clustering of transcriptomic results
Clustered transcriptomic results were based on the four peaks of transcriptional activation, calculated as the percent of genes changed by greater than 2 fold. The four clusters corresponded to Early (15, 30, 45, 60, and 90 min), Delayed early (90 min, 2, 4, and 8 h), Intermediate (12, 16, 20, and 24 h) and Late (36, 48, and 72 h). Within each cluster of transcriptional events genes were filtered by requirement of a greater than 2-fold change on at least 3 arrays (2 of 3 for the late group) and a greater than 2-fold average change across the cluster. The output identified generally 400 genes per cluster which are henceforth referred to as Pi responsive. For promoter analyses we further refined the early cluster to include “Immediate” (15 and 30 min).
Proteomic analysis by cleavable Isotope-Coded Affinity Tags (cICAT)
The general methods used have been described previously (Conrads et al., 2004). Briefly, MC3T3-E1 osteoblast cell proteins (~750 μg each) were labeled either with the light (control, cICAT-12C9) or the heavy (phosphate-treated, cICAT-13C9) isotopic versions of the cleavable isotope coded affinity tags (cICAT) reagent using a modified method from that recommended by the manufacturer (Applied Biosystems, Inc. Foster City, CA). The samples were digested with trypsin and labeled peptides isolated using avidin columns. The lyophilized cICAT-labeled peptides were dissolved and injected onto a strong cation exchange liquid chromatography (SCXLC) column (PolyLC Inc., Columbia, MD). A multistep gradient was used to elute the cICAT-labeled peptides from the column. Each SCXLC fraction was lyophilized and reconstituted prior to microcapillary reversed-phase liquid chromatography (μRPLC). Ten cm long μRPLC-electrospray ionization columns were coupled online with an ion-trap MS (LCQ Deca XP, ThermoElectron, San Jose, CA) and the cICAT-labeled peptides were eluted using a linear step gradient. The MS was operated in a data dependent tandem MS (MS/MS) mode in which each full MS scan was followed by three MS/MS scans where the three most abundant peptide molecular ions were dynamically selected for collision-induced dissociation (CID) using a normalized collision energy of 35%. The MS spectrum for the molecular ions was acquired using 2 microscans for the mass range of m/z 475–2000 and the CID spectrum for the fragment ions was acquired using 3 microscans. The heated capillary temperature and electrospray voltage were set at 160 °C and 1.7 kV, respectively.
Peptide identification and quantitation
The CID spectra were searched using SEQUESTagainst the Mus musculus proteome database (27,612 entries) downloaded from the European Bioinformatics Institute (EBI) (http://www.ebi.ac.uk/proteome/index.html). The Archaea-derived database (12,038 entries) utilized in the false-positive bioinformatic analysis was constructed using genomic sequence information from: Aeropyrum pernix, Archaeoglobus fulgidus, Pyrobaculum aerophilum, Sulfolobus tokodaii, and Thermoplasma volcanium. Dynamic modifications for cysteinyl (Cys) residues were set by mass additions of the cleaved cICAT labels (227.13 Da for the light label, 236.16 Da for the heavy label) in a single search. SEQUEST criteria were set as Xcorr ≥ 1.9 for [M+H]1+ ions, ≥2.2 for [M+2H]2+ ions and ≥ 2.9 for [M+3H]3+ ions, and DeltaCn ≥ 0.08 for the identification of fully tryptic peptides within the cICAT-labeled samples. The identified peptides were quantified using XPRESS (ThermoElectron), which calculates the relative abundances of peptides based on the area of their mass chromatograms.
Luciferase promoter reporter assay
The c-fos luciferase plasmid as well as mutant plasmids; delta SIE, mTCF, mCRE and mSRE were provided by Brent Cochran (Kim et al., 1998). The mutant AP-1 (mAP-1) plasmid was created using a site directed mutagenesis kit (Stratagene, La Jolla CA) with primer 5′-TAGGACATCTGCGTTGGCAGGTTT-3′. Cells (1 × 105) were plated and transfected with 500 ng of DNA and Fugene (Roche) according to manufacturer’s recommendations. Medium was changed to serum starve (0.2% FBS) 24 h after transfection and assay performed 24 h later. The same conditions were used for the Elk-1 reporting system (Pathdetect-Stratagene). Firefly luciferase activity was measured according to manufacturer’s recommendations (Promega, Madison, WI) using a Dynex Technologies MLX microtiter plate luminometer. Results were statistically compared by student T test.
Ras activation assay
Cells were serum starved overnight and supplemented with NaPO4 as indicated (in addition to the 1 mM in the medium). Ras activity was measured using the Ras-Binding Domain of Raf-1 to pulldown active ras according to manufacturer’s protocol (Cell BioLabs, San Diego, CA). Blots of whole cell lysate (50 μg) from the input were probed with antibodies to total ERK1/2 (Promega) and phospho-ERK1/2 and N-ras (Santa Cruz), Pan-ras antibody was from Cell BioLabs.
Immunoblotting and Immunoprecipitations
MC3T3-E1 cells were cultured as above and nuclear isolation and immunoblotting performed as described previously (Conrads et al., 2005) with 50μg of lysate. All antibodies were purchased from Santa Cruz Biotechnologies Inc., (Santa Cruz, CA), except ERK1/2 from (Promega), and FRS2α and phospho-tyr-196 FRS2α from (Cell Signaling; Beverly, MA). Immunoprecipitations were performed on cells that were serum starved overnight and treated with Pi for indicated times. IgG (Santa Cruz) was used as a control. Cells were lysed in IP Lysis Buffer with HaltTM Protease & Phosphatase Single-Use Inhibitor Cocktail (Thermo Scientific, Rochford, IL). The lysate (1 mg) was pre-cleared and then incubated with antibody and Protein A/G PLUS-Agarose (Santa Cruz) overnight with rotation. The immunoprecipitates were washed with IP Lysis Buffer twice and analyzed by SDS-PAGE.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described previously (Meng et al., 2006). A consensus AP-1 oligonucleotide (Santa Cruz) was radio-labeled according to manufacturer’s protocol (Promega). DNA binding reactions were performed using 5 μg of nuclear extract, 5X Gel Shift Binding Buffer (Promega), labeled oligonucleotide, and nuclease-free water.
ATP assay
Cells were plated at a density of 5,000 cells per well in a 96 well white-walled, clear-bottom plate in 100μL of medium. ATP was quantified in MC3T3-E1 cells 20 h after treatment with Pi using Cell-Titer Glo kit (Promega) with background subtracted. Results were statistically compared by student T test.
Angiogenesis blots of conditioned medium from Pi treated cells
MC3T3-E1 cells were grown in growth medium (1mM Pi) or in growth medium supplemented with 10mM Pi for 96 hrs. The medium was removed and serum free and phenol free DMEM (Mediatech, Manassas, VA) plus pen/strep and glutamine (1mM Pi) was added for 20 hrs. The resulting conditioned medium from three 10cm plates/condition was concentrated using Amicon Ultra centrifugal filters (Millipore, Billerica, MA). The resulting conditioned medium (300μg) was used with the Proteome Profiler™ Mouse Angiogenesis Array Kit (#ARY015) R&D Systems, Inc. (Minneapolis, MN) according to the manufacturer’s protocol. Western blots were performed on additional samples derived following the same procedure.
Tube formation Assay
Human Embryonic Vascular Endothelial Cells (HUVEC) were purchased from Invitrogen (Catalogue is C-003-5C) and cultured in Medium 200 (m-200PRF-500) with Low Serum Growth Supplement (S-003-10) at 37°C, 5% CO2 incubator according to manufacturer’s suggestions (protocol-MAN0001687). Conditioned medium was generated from control E1 cells or cells treated with 10mM Pi for 7 days (the medium was changed every 2 or 3 days). Cells were plated in serum free, phenol-free DMEM and concentrated as described above. Condition medium (5ul) was added to HUVEC cultures (3×104 cells) in 96 well plates. The cells were photographed after 6 hours. Tube formation results were quantified by Wimasis GmbH (Munich, Germany) software.
Bioinformatic analyses of promoters
We have previously described an in-house software tool, WholePathwayScope (WPS) (Yi et al., 2006) that displays HTP data in user-defined or stored gene groups or pathways and offers statistical evaluation of global functional category (GO term, pathway, etc.) enrichment in the user’s gene lists. We then used the newly developed PPEP method (Yi et al., 2009) in WPS to perform pathway-level comparative analysis on the differential gene lists derived for each of the time points across the 72 h time course (Fig. S1A). The promoter regions within positions −1500 and +200 of transcription initiation sites of all annotated genes in the mouse genome were searched using a perl script for regular expression comparison for defined consensus binding sequences of all annotated transcription factors from the TFD database (www.ifti.org). The obtained target genes with the corresponding binding sequences for transcription factors having sequence frequency smaller than cutoff value of 1 × 10−5 for binding sequence hit probability in the genome were retrieved and incorporated into WPS program database. Transcription factor binding profiles were generated either with complete profiles or profiles claimed as good quality from locally installed BioBase (http://biobase.abcc.ncifcrf.gov; the original URL of BioBase: www.biobase-international.com). MATCH program was used to search for transcription binding sites within positions −1500 and +200 of transcription initiation sites of all annotated genes in the mouse genome. Based on these predicted target genes of transcription factors, comparative analysis of the enrichment levels of transcription factor target genes within the differentiated lists obtained from the time-course experiments were performed using PPEP method (Yi et al., 2009).
Bioinformatic analyses of signaling and function
Data was analyzed using Ingenuity Pathway Analysis software (www.Ingenuity.com). In addition to transcriptomics, proteins with changes in abundance of >1.75 were also used for this analysis (from Table S4). The software is also used to map and unify IDs of all the genes including protein IDs, GenBank IDs from microarray or protein datasets. The statistical significance of the results was validated using permutation analysis. Briefly, the permutation analysis is done with a utility program for permutation of the data and R-package for statistical calculation of correlation coefficients for data in each of the permutated data files. To obtain permutated correlation coefficients for each candidate pathway or GO group to be validated, each gene and its data in the pathway or GO group is shuffled randomly within the data pool of the corresponding dataset to generate a permutated file with the same number of genes as the original file but with permutated data. This process is iterated for 1000 times to generate 1000 permutated files for each intended pathway or GO group.
RESULTS
Transcriptomic analysis
To better understand the temporal cellular response to elevated extracellular Pi levels a detailed time course of transcriptomic and proteomic results were combined. Fifteen time points starting at 15 min and extending to 72 h were chosen for transcriptomic analysis (Fig. S1A). All studies were performed in normal growth medium containing 1mM Pi with additional Pi added for the indicated times. A partial list of some of the more interesting Pi responsive genes is presented in Table S1. The entire data set is presented in Table S2. Array results were validated using Northern blotting and revealed rapid, dramatic, and dynamic changes in gene expression following cell exposure to elevated Pi levels (Fig. S1B). Multiple patterns of gene changes were detected with many of the earliest upregulated genes following the pattern of previously defined immediate early genes (Cochran et al., 1983; Greenberg et al., 1985; Herschman, 1991; Lau and Nathans, 1985). This gene expression pattern is associated with cell growth and suggested that Pi behaves as a mitogen.
Proteomic analysis
Global changes in protein abundances were measured using cICAT labeling (Gygi et al., 1999) at 1, 2, 8 and 24 h after Pi treatment. A total of 1716 (1078 ≥ 2 peptide hits), 1847 (1119 ≥ 2 peptide hits), 1063 (645 ≥ 2 peptide hits) and 1559 (1003 ≥ 2 peptide hits) proteins were quantified at 1, 2, 8, and 24 h after Pi treatment, respectively. A partial list of the most interesting Pi responsive proteins and the changes in abundance is presented in Table S3 with the entire list detailed in Table S4. The increase in protein abundance and RNA levels correlated well for genes such as Egr1, Cyr61 but a number of proteins, including many associated with metabolism, did not show correlation between protein and RNA levels (Fig S2). The overall correlation of transcriptomic data acquired at 15, 30, 45, and 60 min after Pi treatment with the proteomic data acquired at 60 min after Pi treatment was low (R2 < 0.003) over this temporally coordinated time frame (Fig. S3).
Bioinformatic prediction of Pi enhanced promoter elements and transcription factor activation
To predict the complement of transcription factors involved in Pi response, we identified the transcription factor binding elements that are enriched in the promoters of Pi responsive genes. The Pathway Pattern Extraction Pipeline (PPEP) method (Yi et al., 2009) provided by the in-house tool WholePathwayScope (Yi et al., 2006) was used to define and compare the enrichment of transcription factor binding elements within the promoters of Pi-responsive genes across the time course of experiments. Northern blotting suggested the “early” genes (Fig. S1B) could actually be separated into a 15–30 minute subset “immediate” and we therefore used these 5 time points as the basis for analysis of transcriptional regulatory events. The results identified approximately 10 to 20 different statistically enriched transcription factor binding elements that change over the 72 h time period. A list of the most relevant predicted elements is presented in Table 1. The results identified the enrichment of binding elements of a number of known immediate early gene and osteoblast relevant transcription factors. Although these sites are enriched in the promoters of genes induced by Pi, it does not necessarily suggest that they are activated, but in fact could be negative regulators.
Table 1.
Predicted enriched elements in phosphate responsive genes
| Immediate | Early | Delayed | Intermediate | Late |
|---|---|---|---|---|
| Creb/ATF/AP-1 | Egr1 | ATF | E2F/E2F4,6/DP | Myc/max |
| Egr1 (NGFI-A) | Srf | Ets/Elk | Ets/Elk | NF-E2 |
| Srf | NFκB | Runx | Runx | GATA |
| Oct | MyoD | Ets-Erg | Msx2 | UBP1 |
| SREBP | C/EBPβ | LEF1 | ||
| T3R | Oct | C-abl | ||
| E box fact. | LEF1 | Oct | ||
| TFII-I | Myb | Hox1 |
Italicized factors: identified as altered by either transcriptomic or proteomic analyses.
Pi stimulation of immediate early genes requires serum response and AP-1 binding elements
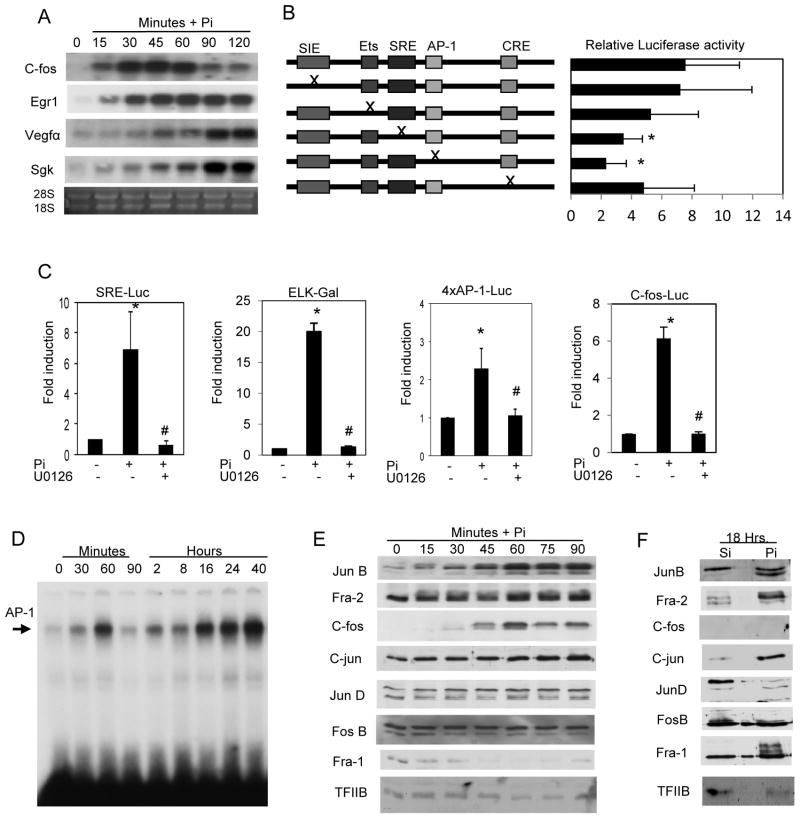
Our transcriptomic analyses defined c-fos as one of the most highly responsive early Pi-regulated genes (Fig. 1A and Table S2). To determine if the changes in RNA level were reflective of an endogenous promoter, a previously defined luciferase reporter construct was used (Kim et al., 1998). Mutant constructs of the c-fos promoter were used to determine the functionally relevant elements in the promoter (Fig. 1B) and identified SRE and AP-1 sites as necessary for the Pi-induced response. To further examine these specific elements luciferase constructs containing a consensus serum response element (SRE) (Suzukawa et al., 2002) or 4xAP-1 element (Rincon and Flavell, 1994) were used. Pi strongly stimulated the promoter activity of these consensus reporter constructs (Fig. 1C). Activation of the Serum Response Factor (SRF) has been demonstrated to require the Ets binding protein Elk-1 (Murai and Treisman, 2002). We used an Elk-Gal reporter construct to ask if Elk-1 was Pi-responsive and in fact it was strongly stimulated by elevated Pi (Fig. 1C). The robust stimulation of ERK1/2 phosphorylation by elevated Pi in various cell types (Beck and Knecht, 2003; Camalier et al., 2010; Chang et al., 2006; Jin et al., 2006; Julien et al., 2007; Orfanidou et al., 2012; Wada et al., 2004; Yamazaki et al., 2010), including drosophila (Bergwitz et al., 2012) suggests the conserved importance of these proteins in Pi signaling and represents both an endpoint to study required upstream events as well as a link to downstream events. We therefore used the upstream inhibitor of ERK1/2, U0126, to identify the requirement of this signaling pathway as a critical intermediate from phosphate sensing to the activation of early transcriptional events (Fig. 1C). No change was detected for the empty luciferase control construct (not shown).
Figure 1. Elevated Pi stimulates immediate early gene expression through AP-1 and Serum response elements.
(A) MC3T3-E1 cells grown in normal growth medium (1mM Pi and 10% FBS) and Pi (10mM) was added for the indicated times. Samples were analyzed by Northern blotting and the resulting membrane was probed as indicated. Egr1; early growth response-1, Vegfα; Vascular endothelial growth factor alpha, Sgk; Serum glucocorticoid kinase. (B) Cells were transiently transfected with a c-fos reporter construct or mutants as indicated, serum starved overnight, and luciferase measured in response to Pi. Results are expressed as fold activation to Pi. (Avg. 10 exp.) *P<0.01 relative to full length construct. (C) MC3T3-E1 cells were transiently transfected with the indicated reporter plasmids. Cells were serum starved overnight and pretreated with the MEK-ERK1/2 inhibitor U0126 (30μM) followed by addition of 10mM Pi for 4 h (control medium contains 1mM Pi). Results are expressed as fold activation relative to control for each construct. *p<0.01 compared to control (1mM), #p<0.01 compared to Pi-stimulated (student T test). (D) Cells in growth medium were treated with Pi for the indicated times. The resulting nuclear lysate was used for EMSA using a specific AP-1 consensus oligo. (E) MC3T3-E1 cells were serum starved overnight and cells harvested at indicated time points after addition of Pi. The resulting nuclear lysate was analyzed by Western blotting and probed for AP-1 proteins as indicated. TFIIB was used as a loading control. (F) MC3T3-E1 cells in growth medium were treated with Pi for 18 h and nuclear lysate analyzed by Western blotting and the membrane probed as indicated. Results are representative of multiple experiments.
Pi increases AP-1 DNA binding and dynamic changes in AP-1 proteins
The predicted binding of AP-1 in response to elevated Pi (Table 1) was validated using electrophoretic mobility shift assays (EMSA). MC3T3-E1 cells were treated with Pi for the indicated times and EMSA’s performed using an AP-1 consensus oligo (Fig. 1D). The DNA binding profile suggests a positive bi-phasic activation of AP-1 corresponding to a bi-phasic ERK1/2 phosphorylation described previously (Beck and Knecht, 2003). The AP-1 transcription factor consists of 7 bzip family members that function as either homo or heterodimers and include four FOS (c-fos, Fra-1, Fra-2, and fosB) and three JUN members (c-jun, junB, and junD) (Curran and Franza, 1988). To determine the individual AP-1 proteins involved, specific AP-1 antibodies were used to supershift the DNA binding assays at both early (75 min) and late (22 h) time points (Fig. S4A,B). Binding specificity was confirmed using excess cold oligo (Fig. S4C). In agreement with the supershift results, Western blotting revealed dynamic changes in AP-1 factors JunB, Fra-2, c-jun, and c-fos early and Fra-1, Fra-2, JunB, and C-jun late in the Pi response (Fig. 1E,F). Results show that early in the response Fra-2, c-fos, fosB, and JunB are bound to AP-1 sites and late in the response the complex is dominated by Fra-1, c-jun and JunB.
Bioinformatic analyses of datasets; cell signaling and function
To determine the signal transduction pathways required for transcriptional regulation during the temporal response to elevated Pi the transcriptomic and proteomic data were analyzed using multiple approaches. Four different databases were analyzed including; Biocarta (B), Gene Ontology/Biological Process (GB), and Ingenuity Pathway analysis of the transcriptomic data (InM), and the proteomics data (InP). The analyses were performed on the filtered genes clustered around the four transcriptionally active time points; 0–60 min, 75 min–2 hrs, 4–20 h, and 24–72 h defined as early, delayed early, mid, and late, respectively. Pathways scored as significantly responsive are listed in Table 2. The results provide an overall picture of the dynamic changes in signaling pathways, individual cell functions, and ultimately changes in cell phenotype. The identification of GFR tyrosine kinase as well as G protein signaling represented novel insights into the cellular response to elevated Pi. Identified pathways such as ERK1/2, Akt (Chang et al., 2006), and Nitric oxide (NO) (Teixeira et al., 2001; Zhong et al., 2010) have been previously demonstrated to respond to Pi in cell types of ranging from keratinocytes to chondrocytes to lung cells highlighting the applicability and validity of our analyses to cell types of different origins.
Table 2.
Predicted phosphate regulated changes in signaling pathways
| Early | |
| Growth Factor Receptor Tyrosine kinase | B, InM, GB |
| Regulation of MAPK pathways | B, InM |
| G-Protein signaling | B, InM |
| IFN signaling | B, InM |
| cAMP-Mediated signaling | InM |
| Insulin signaling pathway | B |
| IL-6 signaling | B |
| Inactivation of Gsk3 by Akt * | B |
| IL-7 signaling * | B |
| Delayed Early | |
| Regulation of MAPK by DUSP | B, GB |
| Growth Factor Receptor Tyrosine kinase | InP, InM |
| Regulation of MAPK pathways | InM |
| NF-κB signaling | B |
| B cell receptor signaling | InP |
| Intermediate | |
| ERK/MAPK signaling | InP |
| Growth Factor Receptor Tyrosine kinase | InP |
| Integrin signaling | InP |
| E2F1 destruction pathway | B |
| NF-κB signaling * | B |
| CD40L signaling pathway * | B |
| Late | |
| NO mediated signaling | GB, InM |
| Growth Factor Receptor Tyrosine kinase | InP, InM |
| PI3K/AKT signaling | InP |
| G protein coupled signaling | InM |
| Ras protein signaling | GB |
| ER signaling | InP |
B; Biocarta, InM; Ingenuity for microarrays, InP; Ingenuity for proteomics, GB; gene ontology/biological processes database.
=down regulated.
Pi stimulated signaling required G protein signaling and activates N-ras
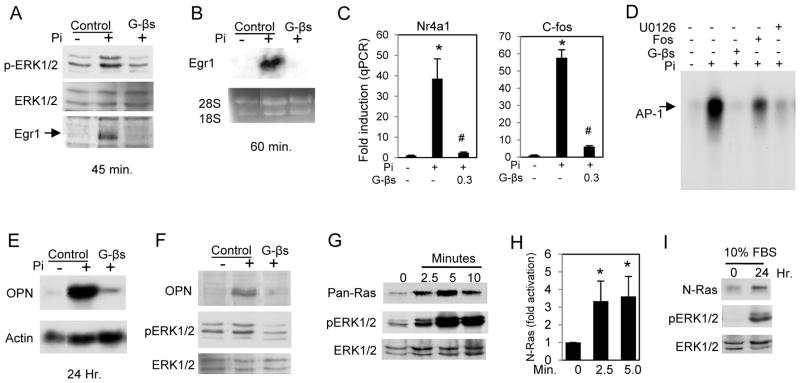
The analyses predicted the involvement of ERK1/2, growth factors, and G protein signaling early in the Pi response. To determine if GTP signaling is required upstream of ERK1/2, cells were pretreated with GDPβS, a non-hydrolyzable GDP analog, followed by addition of Pi for 15 min. Western blotting results demonstrated that inhibition of GTP signaling reduced ERK1/2 phosphorylation (Fig. 2A), suggesting that the GTP required signaling event is upstream of ERK1/2 phosphorylation. To determine the requirement of GTP signaling in Pi-responsive gene expression, GDPβS-treated MC3T3-E1 cells were subjected to RNA analysis. Northern blotting revealed that inhibition of GTP signaling completely blocked expression of the early Pi marker genes Egr1, c-fos, and Nr4a1 (Fig. 2B,C). Using an electrophoretic shift mobility assay we determined that Pi-stimulated activation of AP-1 was blocked by pretreatment of cells with GDPβS, U0126 or foscarnet (Fig. 2D). The requirement of G protein signaling was also determined for the expression of the late Pi marker gene OPN as well as subsequent increase in OPN protein and phosphorylation of ERK1/2 (Fig. 2E,F). Our previous results using keratinocytes showed that elevated Pi levels activated N-Ras signaling (Camalier et al., 2010). To test the hypothesis that N-Ras is also stimulated in mesenchyme derived cells, a pan-Ras activation assay was performed on cells treated with Pi for 2.5, 5, or 10 min (Fig. 2G). The densitometry from four assays using antibodies showed that N-Ras is activated in response to elevated Pi (Fig. 2H). The ras activation assay was performed on cells treated with Pi for 24 h in standard growth medium (10% FBS) (Fig. 2I). The results showed that N-Ras is activated by elevated Pi at both early (min.) and late (after 24 h) time points. Taken together, these results suggest that activation of N-ras is an important early and late signaling event in the cellular response to elevated Pi.
Figure 2. Pi-induced signaling requires GTP and activates N-ras.
(A) MC3T3-E1 cells were serum starved overnight and pretreated with 3mM GDPβS (a non-hydrolyzable GDP analog) followed by addition of Pi for 15 min and harvested for Western blotting and probed with antibodies as indicated. (B) Cells were treated as in (A) for 60 min and samples harvested for Northern blot analysis and probed for Egr1. (C) Cells were treated as in (A) and resulting RNA analyzed by qRT-PCR for c-fos and Nr4a1 expression. (D) Electrophoretic mobility shift assay using an oligo for AP-1. Cells were pretreated with indicated inhibitor for 60 min followed by addition of Pi for 60 min (GDPβS:3mM, U0126:30μM, Fos:1mM). (E) Cells were pretreated with 3mM GDPβS in growth medium followed by Pi for 24 h. The resulting Northern blot was probed as indicated. (F) Cells were treated as in (E) and protein analyzed by Western blotting. (G) Cells were serum starved overnight and supplemented with Pi for 2.5, 5, and 10 min and total-ras activity measured with a pan-ras antibody. Parallel whole cell lysate samples were used for Western blotting and probed with antibodies to phospho and total ERK1/2. Results are representative of multiple experiments. (H) N-ras activity was quantified at 0, 2.5 and 5 min after Pi addition from 4 separate experiments. Results are expressed as arbitrary units obtained from densitometry analysis. (I) Cells cultured in growth medium were treated with Pi for 24 h and the resulting samples used to measure N-ras activity and parallel whole cell lysate samples probed for phospho and total ERK1/2. *p<0.05 compared to control; #p<0.05 compared to Pi-stimulated (student T test).
Pi induced gene expression requires FGF receptor signaling
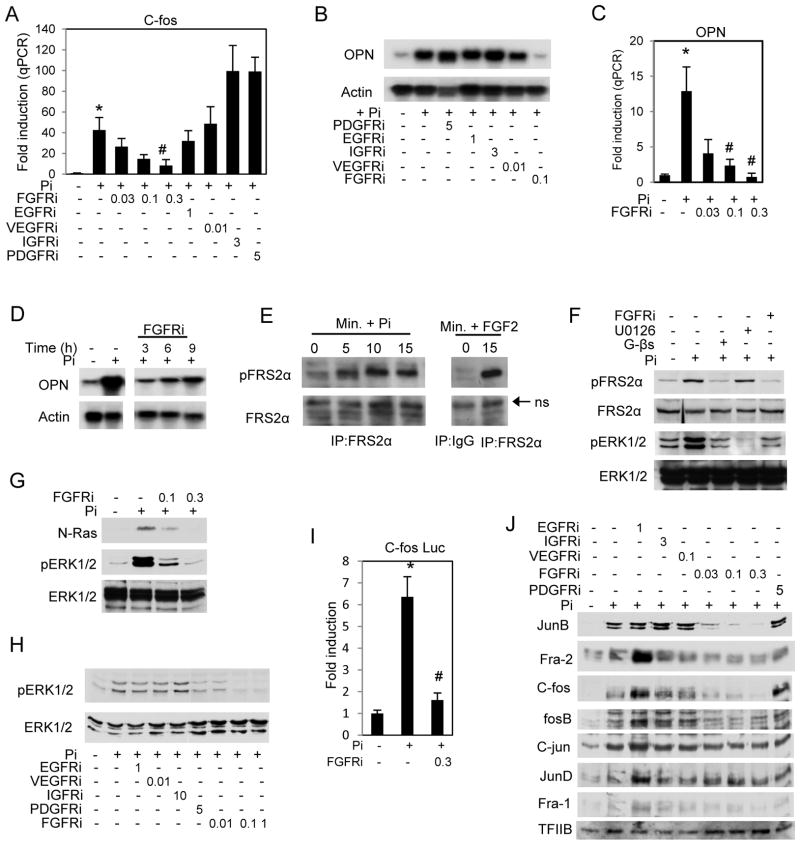
The analyses also predicted the requirement of Pi regulated GFR signaling, which has not previously been linked to the Pi response in osteoblasts. To validate the involvement of GFR signaling in the Pi response, cells were treated with specific inhibitors of common GFRs at concentrations within the previously reported range of specificity (Girnita et al., 2004; Ji et al., 2007; Mulvihill et al., 2008; Nakamura et al., 2006; Panek et al., 1998; Stromberg et al., 2006). Inhibition of FGFR with PD173074 blocked Pi-induced c-fos gene expression early in the response (<1 h) as well as OPN (spp1) at 24 h and this inhibition was dose responsive. (Fig. 3A,B,C). To determine if FGFR signaling was required specifically for the late response the inhibitor was added at 3, 6, and 9 h after addition of Pi (Fig. 3D). Results suggest that there is an FGFR initiated signaling event specifically required for the induction of the late response genes occurring after the early response. These results illustrate the requirement of FGFR as one of the earliest signaling events in Pi response as well as generating a separate late response.
Figure 3. FGF receptor signaling is required for the Pi induced response.
(A) Cells were serum starved overnight (1mM Pi) and pretreated with DMSO (vehicle) or growth factor receptor inhibitors (μM) as indicated for 2 h followed by Pi for 45 min. The resulting RNA was used to analyze c-fos levels by quantitative real time PCR. (B) Cells were cultured in growth medium and pre-treated with inhibitors as indicated followed by Pi for 24 h and resulting RNA was analyzed by Northern blotting and probed for osteopontin (OPN) or actin. (C) Cells cultured in growth medium were pretreated with FGF receptor inhibitor as indicated followed by 10mM Pi for 24 h. RNA was analyzed for OPN levels by qRT-PCR. (D) FGFR inhibitor was added to cells cultured in growth medium at the indicated times after Pi addition and resulting RNA analyzed by Northern blotting. (E) Cells were serum starved overnight, treated with Pi as indicated, and resulting cell lysate was immunoprecipitated with an antibody to FRS2α. (F) Cells were serum starved overnight and pretreated with inhibitors as indicated followed by Pi for 10 min. The resulting samples were analyzed by Western blotting. (G) Cells were serum starved overnight, pretreated with FGFR inhibitor, and Pi added for 2.5 min. A ras activation assay and parallel whole cell lysate blot were probed with antibodies for N-ras, phospo-ERK1/2 and ERK1/2 as indicated. (H) Cells were treated as in (A) with inhibitors as indicated and treated with Pi for 15 min. The resulting protein was used for Western blotting and probed as indicated. (I) Serum starved cells were transfected with c-fos luciferase construct, pretreated with FGFR inhibitor (300nM), and luciferase activity measured after 4 h of Pi. (J) Cells were treated as in (A) and nuclear lysate analyzed by Western blotting as indicated. *p<0.01 compared to control (1mM), #p<0.01 compared to Pi-stimulated (student T test).
We next investigated the requirement of FGFR activation on downstream signaling proteins. Phosphorylation of the adapter/scaffold protein FRS2α is one of the earliest events in the defined FGFR signal transduction pathway (Gotoh, 2008). FRS2α was immunoprecipitated from a time course of Pi treated cells and identified increased phosphorylation within 5 minutes of Pi addition (Fig. 3E). FGF2 was used as a positive control. The requirement of the FGFR, GTP, and ERK1/2 signaling upstream of FRS2α phosphorylation were tested using pharmacological inhibitors. Inhibition of FGFR activation predictably inhibited phosphorylation of FRS2α and inhibition of ERK1/2, which is traditionally downstream of FRSα, had no effect (Fig. 3F). Using the GDPβS we also unexpectedly identified the requirement of GTP signaling upstream of FRS2α (Fig. 3F). The requirement of FGFR signaling for the activation of N-ras was investigated and revealed that FGFR signaling is an upstream event in the Pi-induced signaling pathway (Fig. 3G) as well as phosphorylation of ERK1/2 (Fig. 3H). Using the c-fos promoter luciferase construct revealed that FGFR signaling was required for Pi-induced gene expression as well as changes in the AP-1 proteins. (Fig. 3I,J). The results define a Pi-activated signaling pathway consisting of FGFR, GTP signaling, FRS2α, N-ras, and ERK1/2 leading to changes in AP-1 proteins and ultimately increased gene expression.
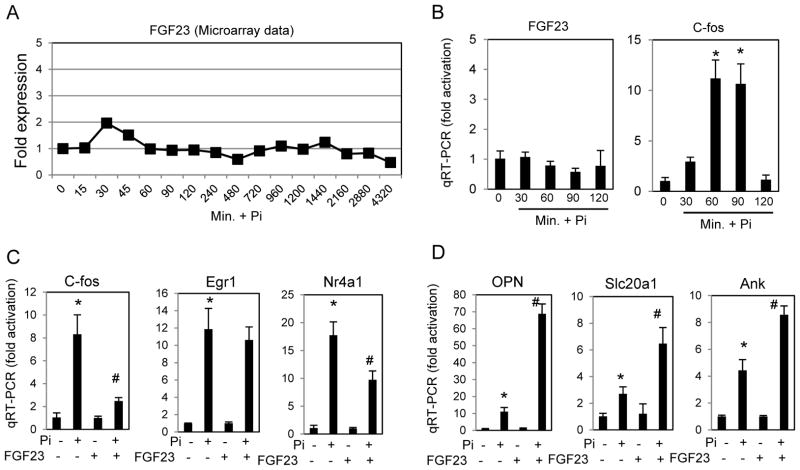
FGF23 is not regulated by Pi in MC3T3-E1 cells and is modestly synergistic for late Pi-induced gene expression
To date FGF23 is the most closely linked FGF to Pi regulation and we therefore determined if this particular FGF was regulated by Pi. Our transcriptomic study detected expression of FGF23 RNA however our clustering analysis did not detect a significant change in FGF23 levels (Fig. 4A) and this lack of response was confirmed using qRT-PCR, with c-fos induced expression acting as a positive control for the Pi-response (Fig. 4B). To determine if FGF23 was involved in the Pi response we added FGF23 peptide in combination with Klotho and heparin alone or with elevated Pi and analyzed gene expression at early and late time points. Although FGF23 alone stimulated gene expression and provided an additive effect on baseline (not shown) when results were calculated as fold increase to Pi (FGF23 relative to FGF23 + Pi), the early genes were not synergistically activated or were modestly decreased (Fig. 4C), however the late genes analyzed demonstrated a synergistic effect in response to added Pi (Fig. 4D). The results suggest that under these in vitro conditions FGF23 acts to enhance the late Pi-response.
Figure 4. FGF23 RNA levels are not regulated by Pi but the peptide synergistically enhances late gene expression.
(A) FGF23 levels in response to a time course of Pi treatment as measured by microarray. (B) The lack of FGF23 response to elevated Pi was confirmed by qRT-PCR in MC3T3-E1 cells treated for indicated times. C-fos is shown as a positive control. (C) Cells were serum starved overnight and treated with FGF23 (500ng/ml) with Klotho (500ng/ml) and Heparin (5ug) with or without Pi as indicated for 60 minutes and RNA analyzed by qRT-PCR. Results are calculated as fold increase relative to the non-treated control for each pair. (D) Cells were treated for 24 hours in growth medium with FGF23 and Pi as in (C) and RNA analyzed by qRT-PCR. Results were calculated as in (C) for each pair. (n=3) *p<0.01 compared to control (1mM), #p<0.01 compared to Pi-stimulated (student T test).
Pi increases ATP production through oxidative phosphorylation
To predict changes in cell function, we used four different bioinformatics approaches as described for the signaling analysis above. As predicted from the above results many of the predicted changes in cell function were related to proliferation (Table 3). The analyses also predicted a change in oxidative phosphorylation (OxPhos). A consequence of increased OxPhos is an increase in ATP levels. Exposure of MC3T3-E1 cells to elevated Pi for 24 h resulted in an approximate 20% increase in cellular ATP relative to control (Fig. 5A). Pretreatment with the electron transport chain inhibitor Rotenone completely inhibited the Pi-induced increase in ATP. Additionally, inhibition of MEK-ERK also completely blocked this increase suggesting that ERK1/2 signaling is required event upstream of the response (Fig. 5A). To determine if OxPhos is a required upstream event for the expression of late Pi-responsive genes, cells were pretreated with the inhibitor Rotenone and RNA was analyzed by Northern blotting. Results suggest that a functional electron transport chain is required for Pi induced expression of all of the late genes examined (Fig. 5B). Pi-induced expression of Egr1 and c-fos was not inhibited by Rotenone (Fig. 5C), defining the requirement of OxPhos as a critical intermediate between early and late responses.
Table 3.
Predicted phosphate induced changes in cell functions
| Early | Intermediate | ||
| Role of MAL in Rho activation of SRF | B | Cell cycle, G2, M checkpoint | B, GB, InP, InM |
| Negative regulation of transport | GB | Purine/Pyrimidine synthesis and metabol. | GB, InM |
| Ectoderm development | GB | DNA metabolism/replication/repair/mod. | GB, InM |
| Gliogenesis | GB | Basic sumoylation & ubiquitination | B, InM |
| Cell cycle/cell growth/proliferation | InP | Glycolysis/gluconeogenesis | InP, InM |
| Apoptosis | InP | Oxidative phosphorylation | InM |
| Cellular metabolism | InP | Fatty acid metabolism | InP |
| Parkinson’s signaling | InM | Citrate cycle | InM |
| Delayed Early | Late | ||
| Cell cycle, G2, M checkpoint | B, GB, IP, InM | Regulation of transcription | GB, InM |
| Positive regulation of cell death | GB, InP, InM | Negative regulation of proliferation | GB, InP |
| Negative regulation of apoptosis | GB, InP | Connective tissue dev. and function | InP, InM |
| Cardiovasc. system dev. and function | InP, InM | Phosphorous metabolism | GB |
| Influence of rho and ras on G1 to S | B | Ion homeostasis | GB |
| Oxidative phosphorylation | InP | Inhibition of matrix metalloproteinases | B |
| Reg. of lipid and fatty acid metabol. | GB | Apoptosis signaling | GB |
| Protein biosynthesis | GB | Reg. of ossification and mineralization | GB |
| Cell adhesion | GB | Myeloid/monocyte/cell differentiation | GB |
| Anagen/hair follicle maturation | GB | Mesoderm development/Gastrulation | GB |
| Odontogenesis | GB | Function of SLRP in bone * | B |
B; Biocarta, InM; Ingenuity for microarrays, InP; Ingenuity for proteomics, GB; gene ontology/biological processes database.
=down regulated.
Figure 5. Elevated Pi increases oxidative phosphorylation.
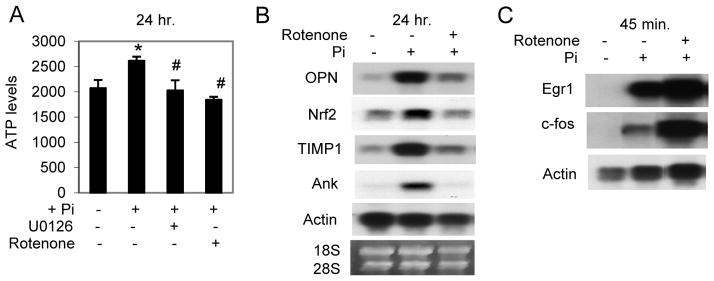
(A) MC3T3-E1 were cultured in growth medium and pretreated with the MEK-ERK1/2 inhibitor U0126 (30μM) or the OxPhos inhibitor Rotenone (1μM) as indicated followed by addition of Pi (10mM) for 24 h. Cells were harvested and ATP quantified. (B) Northern Blot analysis of cells cultured in growth medium and pretreated with Rotenone (1μM) before addition of Pi for 24 hours. (C) Northern Blot analysis of serum starved cells pretreated with Rotenone (1μM) before addition of Pi for 45 min. *p<0.01 compared to control (1mM), #p<0.01 compared to Pi-stimulated (student T test).
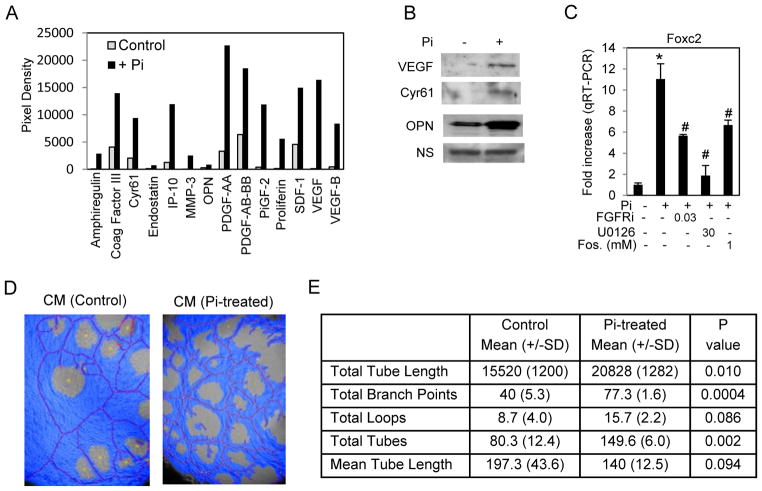
Elevated Pi promotes angiogenesis
Our transcriptomic analysis revealed the strong induction of Vascular Endothelial Growth Factor alpha (Vegfα) and OPN, genes/proteins associated with vascularization as well as the key transcriptional regulator Foxc2. To determine if elevated Pi stimulated an angiogenic response we performed an angiogenesis protein array using conditioned medium from Pi-treated cells. Results suggested a number of angiogenic proteins with increased levels in the medium following long-term treatment (96 hours) with elevated Pi (Figure 6A,B). To determine if our defined Pi signaling pathways were required for the response we chose to analyze Foxc2 a Forkhead transcription factor demonstrated to be important in the angiogenesis (Kume, 2008) (Table S1). Analysis of RNA after 2 h of Pi treatment revealed that activation of FGFR, ERK1/2, and Pi-transport are all required events for pi-induced Foxc2 expression (Figure 6C). To determine if in fact the secretion of these proteins alters angiogenesis, the HUVEC tube formation assay was used. Tubes were generated in differentiation medium supplemented with either control conditioned medium from untreated MC3T3-E1 cells or conditioned medium from MC3T3-E1 cells treated with Pi for 7 days. The resulting images were analyzed and quantified using WIMASIS software. Results revealed that conditioned medium from Pi treated pre-osteoblast cells increased tube formation (Figure 6D,E) suggesting that Pi is capable of stimulating neovasculargenesis.
Figure 6. Elevated Pi stimulates angiogenesis.
(A) MC3T3-E1 cells were cultured in growth medium (1mM Pi) (control) or with 10mM Pi added for 96 hours and medium changed to serum free medium for 20 h. The conditioned medium was concentrated by filter centrifugation and used for angiogenesis protein arrays. The resulting blots were densitometrically quantitated and proteins with a > 3fold change in response to Pi are shown. (B) A Western blot of samples derived as in (A) was probed as indicated. NS=non-specific (C) MC3T3-E1 cells were serum starved overnight and pretreated with inhibitors as indicated (μM) (Fos in mM). RNA was analyzed by qRT-PCR for Foxc2 expression *p<0.01 compared to control (1mM), #p<0.01 compared to Pi-stimulated (student T test). (D) Conditioned medium from control or Pi-treated E1 cells was generated as in (A) added to the HUVEC tube formation assay. Results were quantified using Wimasis software, a representative image is shown (D) and quantification data presented in (E) (n=3).
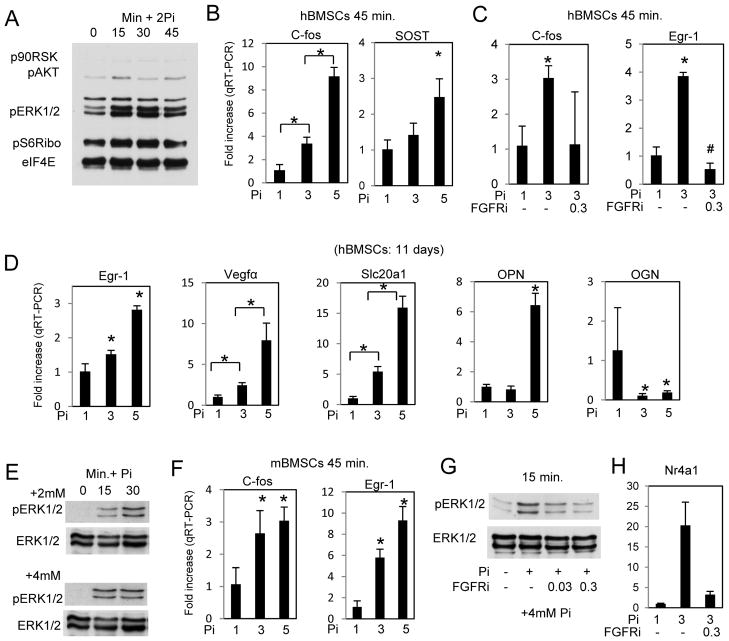
Pi induced signaling occurs in mouse and human bone marrow stromal cells
To determine if the response defined above was applicable to other cells types we tested primary bone marrow stromal cells from both human (hBMSCs) and mouse (mBMSCs). A number of representative endpoints were used. hBMSCs were serum starved overnight (1mM Pi) and treated with 2mM Pi (3mM final). Western blotting revealed a similar increase in ERK1/2 and pAKT as demonstrated in the MC3T3-E1 cells (Fig. 7A) and qRT-PCR detected increased expression of the early marker genes c-fos and SOST (Fig. 7B). Pre-treatment with the FGFRi (PD173074) inhibited the induction of c-fos and Egr1 (Fig. 7C). Our previous results have described an inverse relationship between the concentration of extracellular Pi and time in regards to increased expression of “late” genes (Beck, 2003). To determine the effects of relatively small changes in extracellular Pi that may be more relevant to BMSCs than osteoblasts, RNA was obtained from hBMSCs and cultured in 1, 3, or 5mM Pi for 11 days. Results revealed similar changes in gene expression as our studies in MC3T3-E1 cells above (Fig. 7D). Representative studies using mBMSCs also revealed a similar response in both signaling and RNA expression to elevated Pi (Fig. 7E,F,G,H). These results confirm that the Pi-stimulated responses we have identified in the MC3T3-E1 cells are applicable to primary cultures with a range of concentrations of Pi.
Figure 7. Similar effects of elevated Pi in human and mouse bone marrow stromal cells.
(A) Human bone marrow stromal cells were serum starved overnight followed by addition of Pi (2mM) for times as indicated. The resulting cell lysate was used for Western blotting and probed as indicated (Rep. of 2 patients). (B) hBMSCs were serum starved overnight and treated with Pi as indicated for 45 min. and cells harvested for RNA analysis by qRT-PCR (Rep. of 3 patients). (C) Serum starved hBMSCs were pretreated with FGFR inhibitor (300nM) followed by Pi as indicated for 45 min. and harvested for RNA analysis by qRT-PCR. (D) hBMSCs were treated with the indicated concentrations of Pi and cells harvested for qRT-PCR after 11 days (Rep. of 3 patients). (E) Mouse BMSCs were serum starved overnight followed by addition of Pi (2mM or 4mM) for times as indicated and the resulting cell lysate was used for Western blotting. (F) Serum starved mBMSCs were treated with Pi as indicated for 45 min. and analyzed by qRT-PCR. (G) Serum starved mBMSCs were pretreated with FGFR inhibitor (300nM) followed by Pi as indicated for 15 min. and harvested for Western blotting analysis. (H) mBMSCs were pretreated with FGFR inhibitor 300nM followed by addition of 2mM Pi for 45 min and resulting RNA analyzed by qRT-PCR. *p<0.05 compared to control or as indicated (student T test).
DISCUSSION
We used an integrative biology approach to identify novel cellular and molecular links in the Pi-response pathway and functionally connect these with previously identified Pi-events. The results provide a a more complete understanding of upstream signaling events and pathways required for the initiation of proliferation and identify Pi as a novel regulator of immediate early genes (Cochran et al., 1983; Greenberg et al., 1985; Lau and Nathans, 1985). Many of the immediate early genes were initially identified as TPA inducible genes (TIS) (Lim et al., 1987) and were subsequently demonstrated to be induced by other factors including epidermal growth factor (EGF) and platelet derived growth factor (PDGF) among others (Arenander et al., 1989). Several immediate early genes are regulated by SRF and the promoters uniformly contain a SRE in addition to other regulatory elements. (Treisman, 1986; Treisman, 1987; Treisman et al., 1992). The c-fos promoter was used to determine that elevated Pi functions like a traditional mitogen to potently induce the immediate and delayed early genes requiring at least SRE and AP-1 elements. The stimulation of growth responsive pathways provides additional insight into the results from two recent studies which have linked elevated serum Pi to increased tumorigenesis (Camalier et al., 2010; Jin et al., 2009). The propensity of high Pi to push cells to a proliferative state even under normal growth conditions may influence the outcome of a transforming event relative to a low Pi environment, tipping the balance towards a cancer phenotype.
A number of the genes/proteins identified herein may be relevant to both physiological matrix mineralization as well as pathological calcification. These genes are generally clustered with the “late” time points and represent the long-term cellular response to elevated Pi which was predicted to be involved in matrix mineralization and Pi metabolism (Fig. S5). The most responsive Pi-induced genes/proteins such as OPN, DMP1, and Ank have been previously described to be both Pi responsive and to regulate mineralization. Newly identified Pi-responsive genes/proteins such as calreticulin and calgizzarin (S100A11) are intracellular and known to bind calcium at a high capacity and emphasize the possible role of buffering against calcification within the cell. Annexin A1 and Scramblase 3 are calcium and phospholipid binding proteins that have been linked to membrane regulation and matrix vesicle formation in osteoblasts (Golub, 2011; Xiao et al., 2007). A number of Pi-downregulated genes are involved in controlling events in the extracellular matrix including members of the small leucine-rich proteoglycan family such as decorin, asporin, fibromodulin, osteoglycin, and osteoadherin as well as matrix assembly proteins periostin, collagen, thrombospondin, and nephronectin, all of which were downregulated at the late time points. Changes in serum Pi have been demonstrated to influence cells/tissues beyond functionally mineralizing cells such as kidney and vascular smooth muscle (Giachelli, 2009) and the dysfunction of these genes/proteins could lead to non-intended calcification. Furthermore, the identification of genes more commonly associated with regulation of these cell types such as nephronectin, periostin, serpine1 (PAI-1), alpha-2-macroglobulin, gremlin, serum/glucocorticoid regulated kinase-1 (Sgk1), among others, as Pi-responsive suggest that our results may have implications beyond functionally mineralizing cells to pathological calcification.
We also identified a number of novel Pi-responsive genes/proteins that show Pi’s role as a signaling molecule in bone metabolism. Colony stimulating factor (csf), necessary for the differentiation of osteoclasts, was upregulated at all time points however its receptor (csfr1) was downregulated. Another gene defined as Pi-responsive in this study is SOST (Sclerostin), an osteoblast/osteocyte secreted Wnt signaling regulator known to inhibit mineralization (Winkler et al., 2003). The expression of this gene again may represent a proactive response of the pre-osteoblast preventing the cell from differentiating too early in the matrix mineralization process or it may represent a systemic response limiting pathological calcification. Additionally, Pi-induced proteins such as HMGB1, CD40 ligand, Slamf6, epiregulin (Ereg), HB-EGF stimulate osteoclast resorption or modulate immune system function which may represent a communication link between mineralizing cells and surrounding cell types, particularly in the bone marrow microenvironment. Interestingly, most of these proteins were only identified as regulated by our proteomics analysis emphasizing the increased insight provided by combining technologies.
Our analysis identified the specific requirement of FGFR signaling for both the early and late Pi-response. The family of fibroblast growth factors (FGFs) functions by binding to one of four different receptors (FGFR 1-4) and mediate a range of cellular functions in developing organisms and adults (Ornitz and Itoh, 2001) including tissue repair, wound healing, and tumor angiogenesis (Eswarakumar et al., 2005). FGF signaling and Pi-transport were first linked in a study that demonstrated FGF2 stimulated Pi-transport (Suzuki et al., 2000). A recent study has also linked the activation of FGFR1 signaling by FGF23 to Pi transport, specifically Slc20a1, in kidney cells (Yamazaki et al., 2010) and another has demonstrated the requirement of Pi-transport for the Pi-induced phosphorylation of FRS2α in chondrocytes (Kimata et al., 2010). Our results agree with these findings and additionally identify GTP related signaling for the Pi-induced phosphorylation of FRS2α. The results suggest that a number of membrane events may be required for the Pi-induced signaling response and these membrane events converge on specific signaling pathways. Understanding the mechanisms(s) by which Pi alters cell function may provide novel therapeutic targets in the control of high Pi altered cell function related to functional mineralization and pathological calcification.
FGFs and FGFR signaling have been implicated in numerous disease processes (Beenken and Mohammadi, 2009) as well as skeletal development and maintenance (Du et al., 2012). FGF23 is widely accepted as a key phosphatonin which regulates systemic serum Pi levels (Quarles, 2012) and changes in Pi are known to increase expression of FGF23 from bone (Mirams et al., 2004; Sitara et al., 2004). We however did not detect a significant change in FGF23 RNA levels either by our microarray analysis or qRT-PCR. MC3T3-E1 cells represent an osteoblast precursor and it has been suggested that FGF23 is mainly regulated by mature osteocytes and therefore our data support this notion. Although FGF23 levels were not altered by elevated Pi we did identify a synergistic effect of added FGF23 peptide with Pi-induced gene expression. Interestingly, this response was specific for the late genes such as OPN and Ank with no synergism detected for the early genes. FGF23 is an endocrine factor and therefore Pi-induced expression by osteocytes and possibly the kidney (Ito et al., 2005) could represent a physiological response to either sustained elevated serum Pi or local changes in the bone microenvironment.
The identification of N-Ras activation in response to elevated Pi represents a novel potential regulator of osteoblast function. There are four Ras isoforms that are associated with cell growth signaling ((Malumbres and Pellicer, 1998) however, much less is known about potential roles in differentiation and osteoblasts in particular. H-ras has been linked to cell adhesion (Tanaka et al., 2002) and mechanical strain (Rubin et al., 2006) in osteoblasts but other Ras isoforms have not been reported. Our results suggest a role for N-Ras in osteoblast function and mineralization. The Pi-driven increase in proliferation may be relevant to the bone-remodeling microenvironment. Osteoblasts originate from pluripotent mesenchymal stromal cells that reside in the bone marrow. Elevated Pi generated from the byproduct of osteoclast resorption or activity of the ecto-enzyme alkaline phosphatase in the early stages of osteoblast differentiation may be required to drive proliferation of precursor cells necessary to generate sufficient osteoblasts for bone formation.
We identified a number of transcription factors that are coordinately regulated in response to Pi including AP-1 family members. Genetic manipulation of AP-1 genes through either transgenic or knockout mice have revealed that most AP-1 family members play important roles in osteoblast development and bone metabolism (reviewed in (Wagner, 2002). AP-1 proteins are differentially regulated during osteoblast differentiation (McCabe et al., 1996) and numerous in vivo, in vitro and ex vivo studies have linked AP-1 transcriptional regulation with various functional changes in osteoblast phenotype (Lian et al., 1991; Owen et al., 1990). Although many of the AP-1 genes/proteins have been individually linked to bone metabolism, less is known about the forces that regulate this family of transcription factors during bone metabolism. Our results identify elevated extracellular Pi as a regulator of AP-1 transcriptional regulation during osteoblast cell growth and differentiation.
The requirement of mitochondrial OxPhos for the Pi-induced expression of late genes, such as OPN (spp1) and Ank, identifies an important and novel functional link in the Pi-response pathway. OxPhos is the process by which respiratory enzymes in the mitochondria synthesize ATP from ADP and Pi during the oxidation of NADH by molecular oxygen as part of the electron transport chain. The products of increased metabolism include ATP, inorganic pyrophosphate (PPi), and reactive oxygen species (ROS). We have previously demonstrated the requirement of Pi-transport for expression of late response genes (Beck et al., 2003) and the additional requirement of mitochondrial function identified here suggests a dual regulatory mechanism. Whether this response is at the transcriptional or signal transduction level has yet to be determined. One candidate is the generation of PPi which has been demonstrated in a number of tissues to be an important inhibitor of mineralization and to stimulate OPN expression (Addison et al., 2007; Johnson et al., 2003). It will be interesting to determine if PPi represents a link between the OxPhos and Pi generated signaling in the regulation of late genes.
The increase in angiogenic related genes and proteins suggest a potentially important physiological and/or pathological consequence of elevated extracellular Pi. Neovascularization is required by a number of processes associated with normal tissue development and repair as well as pathological states such as tumorigenesis (Goel et al., 2011). A number of the proteins such as VEGFα, Cyr61, F3 (coagulation factor III), and OPN were detected as strongly induced at the RNA level, however, others such as PDGF, proliferin, and SDF-1 were not, suggesting these proteins are regulated at the level of protein stability and/or post-translational modification by elevated Pi. We also identified the strong induction of Nr4a1 (nuclear receptor subfamily 4 group A member 1 and Nur77, NGFI-B) an orphan receptor that has been linked to angiogenesis (Zeng et al., 2006) and osteopontin expression (Lammi et al., 2004). These gene/proteins represent the early and late response to Pi indicating a coordination of both signaling phases in the ultimate functional cell response to elevated Pi emphasized by the requirement of FGFR, Pi-transport, and ERK1/2 signaling for FoxC2 and OPN expression. New vessel formation is also a critical component to fracture repair (Hankenson et al., 2011) and the stimulation of angiogenesis by high Pi could have implications for bone development and fracture repair. In this case, the high Pi environment created during osteoblast mineralization of a fracture would represent a signal for subsequent required vascularization. These results are also relevant to the cancer studies in which mice on a high Pi diet had increased tumor number and size (Camalier et al., 2010; Jin et al., 2009). The coordinated increase in angiogenesis in response to elevated Pi would provide the growing cell mass with required blood flow. In fact, a high phosphate diet was demonstrated to increase markers of angiogenesis such as MMP2, FGF2, CD31 and CD34 in the liver and lung of mice (Jin et al., 2007; Xu et al., 2008). A study describing a knockout of the sodium dependent Pi-transporter Slc20A1 (Pit-1) identified yolk sacs that appear anemic and lack mature vasculature relative to wild type (Festing et al., 2009) supporting the relevance of Pi-signaling to angiogenesis in vivo.
Taken together, these results describe the temporally coordinated response of extracellular ion sensing and growth factor receptor (GFR) signaling with cell growth, changes in the matrix environment, angiogenesis, and mineralization/calcification. The results also suggest that a short exposure to elevated Pi is sufficient to drive proliferation however a sustained exposure is required for the stimulation of osteo-regulatory genes. Recent studies have suggested that serum Pi levels may influence initiation and/or progression of a pathological states associated with cardiovascular disease, bone metabolism, kidney function, and cancer and results presented herein provide insight into cell autonomous and autocrine mechanisms of the cellular response to high Pi and identify potential novel therapeutic targets to modulate the Pi-response.
Supplementary Material
Acknowledgments
This project has been funded by grants from the National Cancer Institute CA84573, CA136059, CA136716 and Emory University-URC1433 (CEC, KAC, YJL, YL, LMG, and GRB Jr.). YJL is also supported by a research grant from Jeju National University, Korea (2010). This project has also been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
Footnotes
The authors have no conflicts of interest to declare.
References
- Addison WN, Azari F, Sorensen ES, Kaartinen MT, McKee MD. Pyrophosphate inhibits mineralization of osteoblast cultures by binding to mineral, up-regulating osteopontin, and inhibiting alkaline phosphatase activity. The Journal of biological chemistry. 2007;282(21):15872–15883. doi: 10.1074/jbc.M701116200. [DOI] [PubMed] [Google Scholar]
- Arenander AT, Lim RW, Varnum BC, Cole R, de Vellis J, Herschman HR. TIS gene expression in cultured rat astrocytes: multiple pathways of induction by mitogens. J Neurosci Res. 1989;23(3):257–265. doi: 10.1002/jnr.490230303. [DOI] [PubMed] [Google Scholar]
- Barsh GS, Greenberg DB, Cunningham DD. Phosphate uptake and control of fibroblasts growth. Journal of cellular physiology. 1977;92(1):115–128. doi: 10.1002/jcp.1040920114. [DOI] [PubMed] [Google Scholar]
- Beck GR., Jr Inorganic phosphate as a signaling molecule in osteoblast differentiation. Journal of cellular biochemistry. 2003;90(2):234–243. doi: 10.1002/jcb.10622. [DOI] [PubMed] [Google Scholar]
- Beck GR, Jr, Khazai NB, Bouloux GF, Camalier CE, Lin Y, Garneys LM, Siqueira J, Peng L, Pasquel F, Umpierrez D, Smiley D, Umpierrez GE. The effects of thiazolidinediones on human bone marrow stromal cell differentiation in vitro and in thiazolidinedione-treated patients with type 2 diabetes. Translational research: the journal of laboratory and clinical medicine. 2012 doi: 10.1016/j.trsl.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck GR, Jr, Knecht N. Osteopontin regulation by inorganic phosphate is ERK1/2-, protein kinase C-, and proteasome-dependent. The Journal of biological chemistry. 2003;278(43):41921–41929. doi: 10.1074/jbc.M304470200. [DOI] [PubMed] [Google Scholar]
- Beck GR, Jr, Moran E, Knecht N. Inorganic phosphate regulates multiple genes during osteoblast differentiation, including Nrf2. Experimental cell research. 2003;288(2):288–300. doi: 10.1016/s0014-4827(03)00213-1. [DOI] [PubMed] [Google Scholar]
- Beck GR, Jr, Sullivan EC, Moran E, Zerler B. Relationship between alkaline phosphatase levels, osteopontin expression, and mineralization in differentiating MC3T3-E1 osteoblasts. Journal of cellular biochemistry. 1998;68(2):269–280. doi: 10.1002/(sici)1097-4644(19980201)68:2<269::aid-jcb13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Beck L, Leroy C, Salaun C, Margall-Ducos G, Desdouets C, Friedlander G. Identification of a novel function of PiT1 critical for cell proliferation and independent of its phosphate transport activity. The Journal of biological chemistry. 2009;284(45):31363–31374. doi: 10.1074/jbc.M109.053132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nature reviews Drug discovery. 2009;8(3):235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergwitz C, Rasmussen MD, DeRobertis C, Wee MJ, Sinha S, Chen HH, Huang J, Perrimon N. Roles of major facilitator superfamily transporters in phosphate response in Drosophila. PloS one. 2012;7(2):e31730. doi: 10.1371/journal.pone.0031730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R, Burckhardt J, Curran T, Muller R. Expression of c-fos in NIH3T3 cells is very low but inducible throughout the cell cycle. EMBO J. 1986;5(4):695–700. doi: 10.1002/j.1460-2075.1986.tb04269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byskov K, Jensen N, Kongsfelt IB, Wielsoe M, Pedersen LE, Haldrup C, Pedersen L. Regulation of cell proliferation and cell density by the inorganic phosphate transporter PiT1. Cell division. 2012;7(1):7. doi: 10.1186/1747-1028-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camalier CE, Young MR, Bobe G, Perella CM, Colburn NH, Beck GR., Jr Elevated phosphate activates N-ras and promotes cell transformation and skin tumorigenesis. Cancer Prev Res (Phila Pa) 2010;3(3):359–370. doi: 10.1158/1940-6207.CAPR-09-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil DL, Rose DM, Terkeltaub R, Liu-Bryan R. Role of interleukin-8 in PiT-1 expression and CXCR1-mediated inorganic phosphate uptake in chondrocytes. Arthritis Rheum. 2005;52(1):144–154. doi: 10.1002/art.20748. [DOI] [PubMed] [Google Scholar]
- Chang SH, Yu KN, Lee YS, An GH, Beck GR, Jr, Colburn NH, Lee KH, Cho MH. Elevated inorganic phosphate stimulates Akt-ERK1/2-Mnk1 signaling in human lung cells. American journal of respiratory cell and molecular biology. 2006;35(5):528–539. doi: 10.1165/rcmb.2005-0477OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran BH, Reffel AC, Stiles CD. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983;33(3):939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- Collins JF, Bai L, Ghishan FK. The SLC20 family of proteins: dual functions as sodium-phosphate cotransporters and viral receptors. Pflugers Arch. 2004;447(5):647–652. doi: 10.1007/s00424-003-1088-x. [DOI] [PubMed] [Google Scholar]
- Conrads KA, Yi M, Simpson KA, Lucas DA, Camalier CE, Yu LR, Veenstra TD, Stephens RM, Conrads TP, Beck GR., Jr A Combined Proteome and Microarray Investigation of Inorganic Phosphate-induced Pre-osteoblast Cells. Mol Cell Proteomics. 2005;4(9):1284–1296. doi: 10.1074/mcp.M500082-MCP200. [DOI] [PubMed] [Google Scholar]
- Conrads KA, Yu LR, Lucas DA, Zhou M, Chan KC, Simpson KA, Schaefer CF, Issaq HJ, Veenstra TD, Beck GR, Jr, Conrads TP. Quantitative proteomic analysis of inorganic phosphate-induced murine MC3T3-E1 osteoblast cells. Electrophoresis. 2004;25(9):1342–1352. doi: 10.1002/elps.200405892. [DOI] [PubMed] [Google Scholar]
- Cunningham DD, Pardee AB. Transport changes rapidly initiated by serum addition to “contact inhibited” 3T3 cells. Proceedings of the National Academy of Sciences of the United States of America. 1969;64(3):1049–1056. doi: 10.1073/pnas.64.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55(3):395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- de Asua LJ, Rozengurt E, Dulbecco R. Kinetics of early changes in phosphate and uridine transport and cyclic AMP levels stimulated by serum in density-inhibited 3T3 cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71(1):96–98. doi: 10.1073/pnas.71.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra R, Sullivan LM, Fox CS, Wang TJ, D’Agostino RB, Sr, Gaziano JM, Vasan RS. Relations of serum phosphorus and calcium levels to the incidence of cardiovascular disease in the community. Arch Intern Med. 2007;167(9):879–885. doi: 10.1001/archinte.167.9.879. [DOI] [PubMed] [Google Scholar]
- Du X, Xie Y, Xian CJ, Chen L. Role of FGFs/FGFRs in skeletal development and bone regeneration. Journal of cellular physiology. 2012;227(12):3731–3743. doi: 10.1002/jcp.24083. [DOI] [PubMed] [Google Scholar]
- Engstrom W, Zetterberg A. Phosphate and the regulation of DNA replication in normal and virus-transformed 3T3 cells. The Biochemical journal. 1983;214(3):695–702. doi: 10.1042/bj2140695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16(2):139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ferro CJ, Chue CD, Steeds RP, Townend JN. Is lowering phosphate exposure the key to preventing arterial stiffening with age? Heart. 2009;95(21):1770–1772. doi: 10.1136/hrt.2008.162594. [DOI] [PubMed] [Google Scholar]
- Festing MH, Speer MY, Yang HY, Giachelli CM. Generation of mouse conditional and null alleles of the type III sodium-dependent phosphate cotransporter PiT-1. Genesis. 2009;47(12):858–863. doi: 10.1002/dvg.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LW, Stubbs JT, 3rd, Young MF. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl. 1995;266:61–65. [PubMed] [Google Scholar]
- Foster BL, Nociti FH, Jr, Swanson EC, Matsa-Dunn D, Berry JE, Cupp CJ, Zhang P, Somerman MJ. Regulation of cementoblast gene expression by inorganic phosphate in vitro. Calcif Tissue Int. 2006;78(2):103–112. doi: 10.1007/s00223-005-0184-7. [DOI] [PubMed] [Google Scholar]
- Fujita T, Meguro T, Izumo N, Yasutomi C, Fukuyama R, Nakamuta H, Koida M. Phosphate stimulates differentiation and mineralization of the chondroprogenitor clone ATDC5. Jpn J Pharmacol. 2001;85(3):278–281. doi: 10.1254/jjp.85.278. [DOI] [PubMed] [Google Scholar]
- Giachelli CM. Vascular calcification: in vitro evidence for the role of inorganic phosphate. Journal of the American Society of Nephrology: JASN. 2003;14(9 Suppl 4):S300–304. doi: 10.1097/01.asn.0000081663.52165.66. [DOI] [PubMed] [Google Scholar]
- Giachelli CM. The emerging role of phosphate in vascular calcification. Kidney international. 2009;75(9):890–897. doi: 10.1038/ki.2008.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girnita A, Girnita L, del Prete F, Bartolazzi A, Larsson O, Axelson M. Cyclolignans as inhibitors of the insulin-like growth factor-1 receptor and malignant cell growth. Cancer Res. 2004;64(1):236–242. doi: 10.1158/0008-5472.can-03-2522. [DOI] [PubMed] [Google Scholar]
- Goel S, Duda DG, Xu L, Munn LL, Boucher Y, Fukumura D, Jain RK. Normalization of the vasculature for treatment of cancer and other diseases. Physiological reviews. 2011;91(3):1071–1121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub EE. Biomineralization and matrix vesicles in biology and pathology. Seminars in immunopathology. 2011;33(5):409–417. doi: 10.1007/s00281-010-0230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer science. 2008;99(7):1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Greene LA, Ziff EB. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. The Journal of biological chemistry. 1985;260(26):14101–14110. [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Hankenson KD, Dishowitz M, Gray C, Schenker M. Angiogenesis in bone regeneration. Injury. 2011;42(6):556–561. doi: 10.1016/j.injury.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Hilborn DA. Serum stimulation of phosphate uptake into 3T3 cells. Journal of cellular physiology. 1976;87(1):111–121. doi: 10.1002/jcp.1040870114. [DOI] [PubMed] [Google Scholar]
- Holley RW, Kiernan JA. Control of the initiation of DNA synthesis in 3T3 cells: low-molecular weight nutrients. Proceedings of the National Academy of Sciences of the United States of America. 1974;71(8):2942–2945. doi: 10.1073/pnas.71.8.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen MM, Pietila PE, Viljakainen HT, Lamberg-Allardt CJ. Prolonged increase in dietary phosphate intake alters bone mineralization in adult male rats. The Journal of nutritional biochemistry. 2006;17(7):479–484. doi: 10.1016/j.jnutbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Huttunen MM, Tillman I, Viljakainen HT, Tuukkanen J, Peng Z, Pekkinen M, Lamberg-Allardt CJ. High dietary phosphate intake reduces bone strength in the growing rat skeleton. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2007;22(1):83–92. doi: 10.1359/jbmr.061009. [DOI] [PubMed] [Google Scholar]
- Ito M, Sakai Y, Furumoto M, Segawa H, Haito S, Yamanaka S, Nakamura R, Kuwahata M, Miyamoto K. Vitamin D and phosphate regulate fibroblast growth factor-23 in K-562 cells. American journal of physiology Endocrinology and metabolism. 2005;288(6):E1101–1109. doi: 10.1152/ajpendo.00502.2004. [DOI] [PubMed] [Google Scholar]
- Ji QS, Mulvihill MJ, Rosenfeld-Franklin M, Cooke A, Feng L, Mak G, O’Connor M, Yao Y, Pirritt C, Buck E, Eyzaguirre A, Arnold LD, Gibson NW, Pachter JA. A novel, potent, and selective insulin-like growth factor-I receptor kinase inhibitor blocks insulin-like growth factor-I receptor signaling in vitro and inhibits insulin-like growth factor-I receptor dependent tumor growth in vivo. Mol Cancer Ther. 2007;6(8):2158–2167. doi: 10.1158/1535-7163.MCT-07-0070. [DOI] [PubMed] [Google Scholar]
- Jin H, Chang SH, Xu CX, Shin JY, Chung YS, Park SJ, Lee YS, An GH, Lee KH, Cho MH. High dietary inorganic phosphate affects lung through altering protein translation, cell cycle, and angiogenesis in developing mice. Toxicological sciences: an official journal of the Society of Toxicology. 2007;100(1):215–223. doi: 10.1093/toxsci/kfm202. [DOI] [PubMed] [Google Scholar]
- Jin H, Hwang SK, Yu K, Anderson HK, Lee YS, Lee KH, Prats AC, Morello D, Beck GR, Jr, Cho MH. A high inorganic phosphate diet perturbs brain growth, alters Akt-ERK signaling, and results in changes in cap-dependent translation. Toxicological sciences: an official journal of the Society of Toxicology. 2006;90(1):221–229. doi: 10.1093/toxsci/kfj066. [DOI] [PubMed] [Google Scholar]
- Jin H, Xu CX, Lim HT, Park SJ, Shin JY, Chung YS, Park SC, Chang SH, Youn HJ, Lee KH, Lee YS, Ha YC, Chae CH, Beck GR, Jr, Cho MH. High dietary inorganic phosphate increases lung tumorigenesis and alters Akt signaling. Am J Respir Crit Care Med. 2009;179(1):59–68. doi: 10.1164/rccm.200802-306OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Goding J, Van Etten D, Sali A, Hu SI, Farley D, Krug H, Hessle L, Millan JL, Terkeltaub R. Linked deficiencies in extracellular PP(i) and osteopontin mediate pathologic calcification associated with defective PC-1 and ANK expression. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2003;18(6):994–1004. doi: 10.1359/jbmr.2003.18.6.994. [DOI] [PubMed] [Google Scholar]
- Jono S, McKee MD, Murry CE, Shioi A, Nishizawa Y, Mori K, Morii H, Giachelli CM. Phosphate regulation of vascular smooth muscle cell calcification. Circulation research. 2000;87(7):E10–17. doi: 10.1161/01.res.87.7.e10. [DOI] [PubMed] [Google Scholar]
- Julien M, Magne D, Masson M, Rolli-Derkinderen M, Chassande O, Cario-Toumaniantz C, Cherel Y, Weiss P, Guicheux J. Phosphate stimulates matrix Gla protein expression in chondrocytes through the extracellular signal regulated kinase signaling pathway. Endocrinology. 2007;148(2):530–537. doi: 10.1210/en.2006-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatani M, Sugimoto T, Kano J, Kanzawa M, Chihara K. Effect of high phosphate concentration on osteoclast differentiation as well as bone-resorbing activity. Journal of cellular physiology. 2003;196(1):180–189. doi: 10.1002/jcp.10270. [DOI] [PubMed] [Google Scholar]
- Kido S, Miyamoto K, Mizobuchi H, Taketani Y, Ohkido I, Ogawa N, Kaneko Y, Harashima S, Takeda E. Identification of regulatory sequences and binding proteins in the type II sodium/phosphate cotransporter NPT2 gene responsive to dietary phosphate. The Journal of biological chemistry. 1999;274(40):28256–28263. doi: 10.1074/jbc.274.40.28256. [DOI] [PubMed] [Google Scholar]
- Kim DW, Cheriyath V, Roy AL, Cochran BH. TFII-I enhances activation of the c-fos promoter through interactions with upstream elements. Molecular and cellular biology. 1998;18(6):3310–3320. doi: 10.1128/mcb.18.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata M, Michigami T, Tachikawa K, Okada T, Koshimizu T, Yamazaki M, Kogo M, Ozono K. Signaling of extracellular inorganic phosphate up-regulates cyclin D1 expression in proliferating chondrocytes via the Na+/Pi cotransporter Pit-1 and Raf/MEK/ERK pathway. Bone. 2010;47(5):938–947. doi: 10.1016/j.bone.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Koshihara M, Katsumata S, Uehara M, Suzuki K. Effects of dietary phosphorus intake on bone mineralization and calcium absorption in adult female rats. Biosci Biotechnol Biochem. 2005a;69(5):1025–1028. doi: 10.1271/bbb.69.1025. [DOI] [PubMed] [Google Scholar]
- Koshihara M, Masuyama R, Uehara M, Suzuki K. Reduction in dietary calcium/phosphorus ratio reduces bone mass and strength in ovariectomized rats enhancing bone turnover. Biosci Biotechnol Biochem. 2005b;69(10):1970–1973. doi: 10.1271/bbb.69.1970. [DOI] [PubMed] [Google Scholar]
- Kume T. Foxc2 transcription factor: a newly described regulator of angiogenesis. Trends in cardiovascular medicine. 2008;18(6):224–228. doi: 10.1016/j.tcm.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi J, Huppunen J, Aarnisalo P. Regulation of the osteopontin gene by the orphan nuclear receptor NURR1 in osteoblasts. Molecular endocrinology. 2004;18(6):1546–1557. doi: 10.1210/me.2003-0247. [DOI] [PubMed] [Google Scholar]
- Lau LF, Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. Embo J. 1985;4(12):3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang HY, Giachelli CM. Role of the sodium-dependent phosphate cotransporter, Pit-1, in vascular smooth muscle cell calcification. Circulation research. 2006;98(7):905–912. doi: 10.1161/01.RES.0000216409.20863.e7. [DOI] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Bortell R, Owen TA. Phenotype suppression: a postulated molecular mechanism for mediating the relationship of proliferation and differentiation by Fos/Jun interactions at AP-1 sites in steroid responsive promoter elements of tissue-specific genes. Journal of cellular biochemistry. 1991;45(1):9–14. doi: 10.1002/jcb.240450106. [DOI] [PubMed] [Google Scholar]
- Lim RW, Varnum BC, Herschman HR. Cloning of tetradecanoyl phorbol ester-induced ‘primary response’ sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1(3):263–270. [PubMed] [Google Scholar]
- Lundquist P, Murer H, Biber J. Type II Na+-Pi cotransporters in osteoblast mineral formation: regulation by inorganic phosphate. Cell Physiol Biochem. 2007;19(1–4):43–56. doi: 10.1159/000099191. [DOI] [PubMed] [Google Scholar]
- Lundquist P, Ritchie HH, Moore K, Lundgren T, Linde A. Phosphate and calcium uptake by rat odontoblast-like MRPC-1 cells concomitant with mineralization. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2002;17(10):1801–1813. doi: 10.1359/jbmr.2002.17.10.1801. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Pellicer A. RAS pathways to cell cycle control and cell transformation. Front Biosci. 1998;3:d887–912. doi: 10.2741/a331. [DOI] [PubMed] [Google Scholar]
- Mansfield K, Teixeira CC, Adams CS, Shapiro IM. Phosphate ions mediate chondrocyte apoptosis through a plasma membrane transporter mechanism. Bone. 2001;28(1):1–8. doi: 10.1016/s8756-3282(00)00409-9. [DOI] [PubMed] [Google Scholar]
- Mathew S, Tustison KS, Sugatani T, Chaudhary LR, Rifas L, Hruska KA. The mechanism of phosphorus as a cardiovascular risk factor in CKD. Journal of the American Society of Nephrology: JASN. 2008;19(6):1092–1105. doi: 10.1681/ASN.2007070760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe LR, Banerjee C, Kundu R, Harrison RJ, Dobner PR, Stein JL, Lian JB, Stein GS. Developmental expression and activities of specific fos and jun proteins are functionally related to osteoblast maturation: role of Fra-2 and Jun D during differentiation. Endocrinology. 1996;137(10):4398–4408. doi: 10.1210/endo.137.10.8828501. [DOI] [PubMed] [Google Scholar]
- Meng Z, Camalier CE, Lucas DA, Veenstra TD, Beck GR, Jr, Conrads TP. Probing early growth response 1 interacting proteins at the active promoter in osteoblast cells using oligoprecipitation and mass spectrometry. J Proteome Res. 2006;5(8):1931–1939. doi: 10.1021/pr060009l. [DOI] [PubMed] [Google Scholar]
- Mirams M, Robinson BG, Mason RS, Nelson AE. Bone as a source of FGF23: regulation by phosphate? Bone. 2004;35(5):1192–1199. doi: 10.1016/j.bone.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Mozar A, Haren N, Chasseraud M, Louvet L, Maziere C, Wattel A, Mentaverri R, Morliere P, Kamel S, Brazier M, Maziere JC, Massy ZA. High extracellular inorganic phosphate concentration inhibits RANK-RANKL signaling in osteoclast-like cells. J Cell Physiol. 2007 doi: 10.1002/jcp.21283. [DOI] [PubMed] [Google Scholar]
- Mulvihill MJ, Ji QS, Coate HR, Cooke A, Dong H, Feng L, Foreman K, Rosenfeld-Franklin M, Honda A, Mak G, Mulvihill KM, Nigro AI, O’Connor M, Pirrit C, Steinig AG, Siu K, Stolz KM, Sun Y, Tavares PA, Yao Y, Gibson NW. Novel 2-phenylquinolin-7-yl-derived imidazo[1,5-a]pyrazines as potent insulin-like growth factor-I receptor (IGF-IR) inhibitors. Bioorg Med Chem. 2008;16(3):1359–1375. doi: 10.1016/j.bmc.2007.10.061. [DOI] [PubMed] [Google Scholar]
- Murai K, Treisman R. Interaction of serum response factor (SRF) with the Elk-1 B box inhibits RhoA-actin signaling to SRF and potentiates transcriptional activation by Elk-1. Molecular and cellular biology. 2002;22(20):7083–7092. doi: 10.1128/MCB.22.20.7083-7092.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Taguchi E, Miura T, Yamamoto A, Takahashi K, Bichat F, Guilbaud N, Hasegawa K, Kubo K, Fujiwara Y, Suzuki R, Kubo K, Shibuya M, Isae T. KRN951, a highly potent inhibitor of vascular endothelial growth factor receptor tyrosine kinases, has antitumor activities and affects functional vascular properties. Cancer Res. 2006;66(18):9134–9142. doi: 10.1158/0008-5472.CAN-05-4290. [DOI] [PubMed] [Google Scholar]
- Onufrak SJ, Bellasi A, Shaw LJ, Herzog CA, Cardarelli F, Wilson PW, Vaccarino V, Raggi P. Phosphorus levels are associated with subclinical atherosclerosis in the general population. Atherosclerosis. 2008;199(2):424–431. doi: 10.1016/j.atherosclerosis.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Orfanidou T, Malizos KN, Varitimidis S, Tsezou A. 1,25-Dihydroxyvitamin D(3) and extracellular inorganic phosphate activate mitogen-activated protein kinase pathway through fibroblast growth factor 23 contributing to hypertrophy and mineralization in osteoarthritic chondrocytes. Experimental biology and medicine. 2012;237(3):241–253. doi: 10.1258/ebm.2011.011301. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2(3):REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen TA, Bortell R, Yocum SA, Smock SL, Zhang M, Abate C, Shalhoub V, Aronin N, Wright KL, van Wijnen AJ, et al. Coordinate occupancy of AP-1 sites in the vitamin D-responsive and CCAAT box elements by Fos-Jun in the osteocalcin gene: model for phenotype suppression of transcription. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(24):9990–9994. doi: 10.1073/pnas.87.24.9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek RL, Lu GH, Dahring TK, Batley BL, Connolly C, Hamby JM, Brown KJ. In vitro biological characterization and antiangiogenic effects of PD 166866, a selective inhibitor of the FGF-1 receptor tyrosine kinase. J Pharmacol Exp Ther. 1998;286(1):569–577. [PubMed] [Google Scholar]
- Quarles LD. Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nature reviews Endocrinology. 2012;8(5):276–286. doi: 10.1038/nrendo.2011.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M, Flavell RA. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. Embo J. 1994;13(18):4370–4381. doi: 10.1002/j.1460-2075.1994.tb06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussanne MC, Lieberherr M, Souberbielle JC, Sarfati E, Drueke T, Bourdeau A. Human parathyroid cell proliferation in response to calcium, NPS R-467, calcitriol and phosphate. Eur J Clin Invest. 2001;31(7):610–616. doi: 10.1046/j.1365-2362.2001.00809.x. [DOI] [PubMed] [Google Scholar]
- Rubin H, Chu BM. Solute concentration effects on the expression of cellular heterogeneity of anchorage-independent growth among spontaneously transformed BALB/c3T3 cells. In Vitro. 1984;20(7):585–596. doi: 10.1007/BF02639774. [DOI] [PubMed] [Google Scholar]
- Rubin H, Sanui H. Complexes of inorganic pyrophosphate, orthophosphate, and calcium as stimulants of 3T3 cell multiplication. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(11):5026–5030. doi: 10.1073/pnas.74.11.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J, Murphy TC, Rahnert J, Song H, Nanes MS, Greenfield EM, Jo H, Fan X. Mechanical inhibition of RANKL expression is regulated by H-Ras-GTPase. The Journal of biological chemistry. 2006;281(3):1412–1418. doi: 10.1074/jbc.M508639200. [DOI] [PubMed] [Google Scholar]
- Sitara D, Razzaque MS, Hesse M, Yoganathan S, Taguchi T, Erben RG, Juppner H, Lanske B. Homozygous ablation of fibroblast growth factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix biology: journal of the International Society for Matrix Biology. 2004;23(7):421–432. doi: 10.1016/j.matbio.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromberg T, Ekman S, Girnita L, Dimberg LY, Larsson O, Axelson M, Lennartsson J, Hellman U, Carlson K, Osterborg A, Vanderkerken K, Nilsson K, Jernberg-Wiklund H. IGF-1 receptor tyrosine kinase inhibition by the cyclolignan PPP induces G2/M-phase accumulation and apoptosis in multiple myeloma cells. Blood. 2006;107(2):669–678. doi: 10.1182/blood-2005-01-0306. [DOI] [PubMed] [Google Scholar]
- Sudo H, Kodama HA, Amagai Y, Yamamoto S, Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J Cell Biol. 1983;96(1):191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzukawa K, Weber TJ, Colburn NH. AP-1, NF-kappa-B, and ERK activation thresholds for promotion of neoplastic transformation in the mouse epidermal JB6 model. Environ Health Perspect. 2002;110(9):865–870. doi: 10.1289/ehp.02110865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Ghayor C, Guicheux J, Magne D, Quillard S, Kakita A, Ono Y, Miura Y, Oiso Y, Itoh M, Caverzasio J. Enhanced expression of the inorganic phosphate transporter Pit-1 is involved in BMP-2-induced matrix mineralization in osteoblast-like cells. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2006;21(5):674–683. doi: 10.1359/jbmr.020603. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Palmer G, Bonjour JP, Caverzasio J. Stimulation of sodium-dependent phosphate transport and signaling mechanisms induced by basic fibroblast growth factor in MC3T3-E1 osteoblast-like cells. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2000;15(1):95–102. doi: 10.1359/jbmr.2000.15.1.95. [DOI] [PubMed] [Google Scholar]
- Takeyama S, Yoshimura Y, Deyama Y, Sugawara Y, Fukuda H, Matsumoto A. Phosphate decreases osteoclastogenesis in coculture of osteoblast and bone marrow. Biochemical and biophysical research communications. 2001;282(3):798–802. doi: 10.1006/bbrc.2001.4652. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nakayamada S, Fujimoto H, Okada Y, Umehara H, Kataoka T, Minami Y. H-Ras/mitogen-activated protein kinase pathway inhibits integrin-mediated adhesion and induces apoptosis in osteoblasts. The Journal of biological chemistry. 2002;277(24):21446–21452. doi: 10.1074/jbc.M202238200. [DOI] [PubMed] [Google Scholar]
- Teixeira CE, Teixeira SA, Antunes E, De Nucci G. The role of nitric oxide on the relaxations of rabbit corpus cavernosum induced by Androctonus australis and Buthotus judaicus scorpion venoms. Toxicon. 2001;39(5):633–639. doi: 10.1016/s0041-0101(00)00187-2. [DOI] [PubMed] [Google Scholar]
- Tenenhouse HS. Regulation of phosphorus homeostasis by the type iia na/phosphate cotransporter. Annu Rev Nutr. 2005;25:197–214. doi: 10.1146/annurev.nutr.25.050304.092642. [DOI] [PubMed] [Google Scholar]
- Tenenhouse HS. Phosphate transport: molecular basis, regulation and pathophysiology. J Steroid Biochem Mol Biol. 2007;103(3–5):572–577. doi: 10.1016/j.jsbmb.2006.12.090. [DOI] [PubMed] [Google Scholar]
- Tonelli M, Sacks F, Pfeffer M, Gao Z, Curhan G. Relation between serum phosphate level and cardiovascular event rate in people with coronary disease. Circulation. 2005;112(17):2627–2633. doi: 10.1161/CIRCULATIONAHA.105.553198. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46(4):567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. Embo J. 1987;6(9):2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. Embo J. 1992;11(12):4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Mizuno M, Komori T, Tamura M. Extracellular inorganic phosphate regulates gibbon ape leukemia virus receptor-2/phosphate transporter mRNA expression in rat bone marrow stromal cells. Journal of cellular physiology. 2004;198(1):40–47. doi: 10.1002/jcp.10383. [DOI] [PubMed] [Google Scholar]
- Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Annals of the rheumatic diseases. 2002;61(Suppl 2):ii40–42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MJ, Edlin G. Phosphate transport, nucleotide pools, and ribonucleic acid synthesis in growing and in density-inhibited 3T3 cells. The Journal of biological chemistry. 1971;246(6):1828–1833. [PubMed] [Google Scholar]
- Werner A, Dehmelt L, Nalbant P. Na+-dependent phosphate cotransporters: the NaPi protein families. J Exp Biol. 1998;201(Pt 23):3135–3142. doi: 10.1242/jeb.201.23.3135. [DOI] [PubMed] [Google Scholar]
- Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, Appleby M, Brunkow ME, Latham JA. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22(23):6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Camalier CE, Nagashima K, Chan KC, Lucas DA, de la Cruz MJ, Gignac M, Lockett S, Issaq HJ, Veenstra TD, Conrads TP, Beck GR., Jr Analysis of the extracellular matrix vesicle proteome in mineralizing osteoblasts. Journal of cellular physiology. 2007;210(2):325–335. doi: 10.1002/jcp.20826. [DOI] [PubMed] [Google Scholar]
- Xu CX, Jin H, Lim HT, Kim JE, Shin JY, Lee ES, Chung YS, Lee YS, Beck G, Jr, Lee KH, Cho MH. High dietary inorganic phosphate enhances cap-dependent protein translation, cell-cycle progression, and angiogenesis in the livers of young mice. Am J Physiol Gastrointest Liver Physiol. 2008;295(4):G654–663. doi: 10.1152/ajpgi.90213.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Ozono K, Okada T, Tachikawa K, Kondou H, Ohata Y, Michigami T. Both FGF23 and extracellular phosphate activate Raf/MEK/ERK pathway via FGF receptors in HEK293 cells. Journal of cellular biochemistry. 2010;111(5):1210–1221. doi: 10.1002/jcb.22842. [DOI] [PubMed] [Google Scholar]
- Yates AJ, Oreffo RO, Mayor K, Mundy GR. Inhibition of bone resorption by inorganic phosphate is mediated by both reduced osteoclast formation and decreased activity of mature osteoclasts. J Bone Miner Res. 1991;6(5):473–478. doi: 10.1002/jbmr.5650060508. [DOI] [PubMed] [Google Scholar]
- Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Mudunuri U, Che A, Stephens RM. Seeking unique and common biological themes in multiple gene lists or datasets: pathway pattern extraction pipeline for pathway-level comparative analysis. BMC Bioinformatics. 2009;10:200. doi: 10.1186/1471-2105-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiko Y, Candeliere GA, Maeda N, Aubin JE. Osteoblast autonomous Pi regulation via Pit1 plays a role in bone mineralization. Molecular and cellular biology. 2007;27(12):4465–4474. doi: 10.1128/MCB.00104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Qin L, Zhao D, Tan X, Manseau EJ, Van Hoang M, Senger DR, Brown LF, Nagy JA, Dvorak HF. Orphan nuclear receptor TR3/Nur77 regulates VEGF-A-induced angiogenesis through its transcriptional activity. The Journal of experimental medicine. 2006;203(3):719–729. doi: 10.1084/jem.20051523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Carney DH, Jo H, Boyan BD, Schwartz Z. Inorganic Phosphate Induces Mammalian Growth Plate Chondrocyte Apoptosis in a Mitochondrial Pathway Involving Nitric Oxide and JNK MAP Kinase. Calcif Tissue Int. 2010 doi: 10.1007/s00223-010-9433-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.