Abstract
Cyclins encode regulatory subunits of holoenzymes that phosphorylate a variety of cellular substrates. Although the classic role of cyclins in cell cycle progression and tumorigenesis has been well characterized, new functions have been identified, including the induction of cellular migration and invasion, enhancement of angiogenesis, inhibition of mitochondrial metabolism, regulation of transcription factor signaling via a DNA-bound form, the induction of chromosomal instability, enhancement of DNA damage sensing and DNA damage repair, and feedback governing expression of the noncoding genome. This review describes the mechanisms of these new functions of cyclin D1.
The family of D-type cyclins (D1, D2, and D3) regulates the G1/S-phase transition, and D cyclins bind and activate cyclin-dependent kinases (Cdk4 and Cdk6) to phosphorylate the retinoblastoma (pRb) protein, and through titration of the Cdk inhibitors p21Cip1 and p27Kip1, the cyclin D–Cdk4/6 complexes activate cyclin E/Cdk2.1 Genetic analysis studies in mice have demonstrated an essential role for cyclin D1 in normal development of the retina, components of the nervous system, and terminal alveolar breast bud development.2,3 Cyclin D1 promotes neural basal progenitors,4 and use of a kinase-deficient cyclin D1 cDNA (K112E) defined an important role for CDK activity in the self-renewal properties of mammary epithelial cell progenitors.5 Cyclin E can partially rescue the retinal and mammary epithelial cell developmental abnormalities likely related to the progenitor pool expansion.6 The kinase-defective cyclin D1 K112E binds p27Kip1, and, in conjunction with prior studies showing p27Kip1, is epistemic to cyclin D1 in the development of the retina and mammary epithelial cell proliferation, raises the possibility of a role for p27Kip1 binding in this stem cell differentiation function.7
Studies of cyclin D1−/− bone marrow macrophages demonstrated an essential role for cyclin D1 in cellular adhesion and migration, a finding common to other cell types, including fibroblasts and mammary epithelial cells.8–10 Cyclin D1b, a common polymorphism at the exon-4 intron-4 boundary of the human cyclin D1 gene, did not enhance cell migration.11 The mechanism by which cyclin D1a promotes cell migration has been examined in detail. Cyclin D1a stabilizes p27Kip1, inhibiting RhoA-inducing Rho-associated protein kinase and myosin light chain kinase.9–11 Cyclin D1 also conveys an indirect effect to inhibit migration by inhibiting epithelial-mesenchymal transition,12 suggesting the effect of cyclin D1 on migration may vary by cell type and differentiation status.12 Mass spectrometry identified protein kinase C and casein kinase substrate in neurons 213 and filamin A13 as additional factors involved in cyclin D1–mediated migration. The effects of cyclin D1 to promote migration were observed in cyclin D1−/− cells that express cyclin D1 and cyclin A3; thus, the function appears to be specific to cyclin D1, which was necessary and sufficient for the promigratory function.
Cyclin D1 Inhibits Mitochondrial Metabolism by Phosphorylating NRF-1
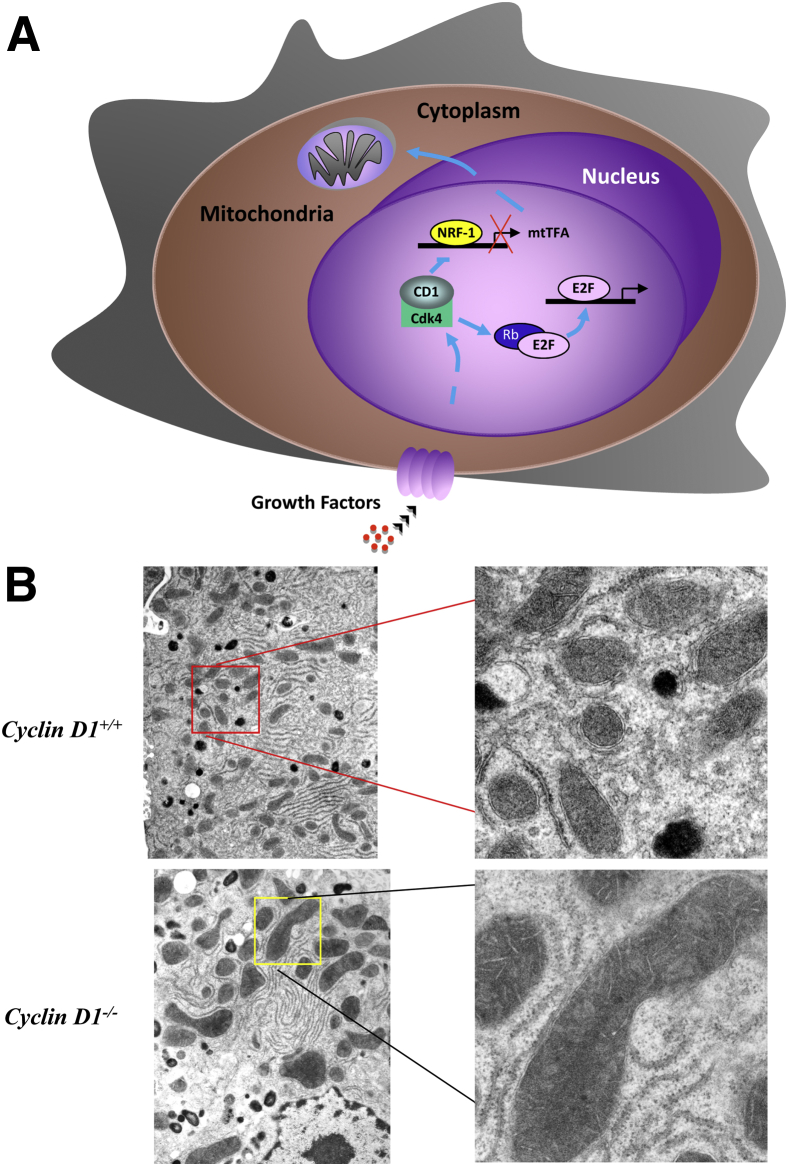
Cyclin D1 deficiency in cells and in vivo is associated with increased mitochondrial size and activity (Figure 1).14,15 Rescue of the cyclin D1−/− cells' mitochondrial phenotype required the Cdk function of cyclin D1. Nuclear respiratory factor 1 (NRF-1), which induces expression of nuclear-encoded mitochondrial genes, was repressed in expression and activity by cyclin D1, which phosphorylated NRF-1 at S47 (Figure 1A).14 By using cyclin D1 antisense mammary epithelial targeted transgenic mice, the role for cyclin D1 as an inhibitor of mitochondrial biogenesis and activity was confirmed in vivo. Endogenous cyclin D1 was shown to inhibit mitochondrial biogenesis and activity by gene expression signaling and in vivo imaging. Consistent with these findings, cyclin D1–induced mammary tumors showed inhibition of mitochondrial activity and aerobic glycolysis with enhanced cytosolic glycolysis.15 In vivo measurements of relative use of amino acids from the tricarboxylic acid cycle using nuclear magnetic resonance in cyclin D1−/− mammary epithelium showed changes in the ratio of glutamate and glutamine/citrate, associated with the induction of mitochondrial metabolism and reduced cytosolic glycolysis, and the glutamate plus glutamine/citrate ratio was increased.15 A similar change in cellular metabolism was observed in the mammary epithelium of ponasterone-inducible cyclin D1 antisense transgenic mice.15 Cyclin D1 was also shown to inhibit mitochondrial function through binding to the mitochondrial voltage-dependent anion channel, thereby competing with hexokinase II.16 The cyclin D1–mediated inhibition of NRF-1 and, thereby mitochondrial transcription factor A, to reduce mitochondrial activity was observed in cells expressing cyclin E.
Figure 1.
Cyclin D1 inhibits mitochondrial biogenesis. A: Schematic representation of the mechanism by which cyclin D1 inhibits mitochondrial function. Cyclin D1–dependent kinase phosphorylates and inhibits NRF-1 and, thereby, mitochondrial transcription factor A and mitochondrial activity. B: Cyclin D1 deficiency enhances mitochondrial size and function. Transmission electron microscopic images of hepatocytes from liver tissue of cyclin D1+/+ (red box, enlarged area) and cyclin D1−/− (yellow box, enlarged area) show increased mitochondria size in cyclin D1−/−. Catalase-positive peroxisomes (dark spherical structures) are evident. Original magnification, ×5000 (right column).
Cyclin E is regulated by mitochondrial activity in that disruption of the mitochondrial electron transport chain activates a G1/S checkpoint that degrades cyclin E.17 Consistent with these findings, pRb was subsequently shown to couple cell cycle exit with mitochondrial biogenesis.18 Inactivation of pRb, or p21 overexpression, also inhibits mitochondrial metabolism, thereby increasing cytosolic glycolysis, suggesting the effect of cyclin D1 is part of a broader role for the cyclin D1/Rb/CDK inhibitor pathway. Collectively, these studies demonstrated that increased abundance of cyclin D1 determines metabolic substrate prioritization toward amino acid synthesis from the tricarboxylic acid cycle, consistent with a known role for cyclin D1 in the induction of DNA synthesis and the known induction of cytosolic glycolysis in tumors. Whether the cyclin D1–dependent inhibition of mitochondrial biogenesis contributes to the glucose avidity of carcinomas remains to be determined.
Cyclin D1 Governs Transcription Factor Activity and Gene Expression by Recruiting Chromatin Remodeling Proteins to the Coding and Noncoding Genome
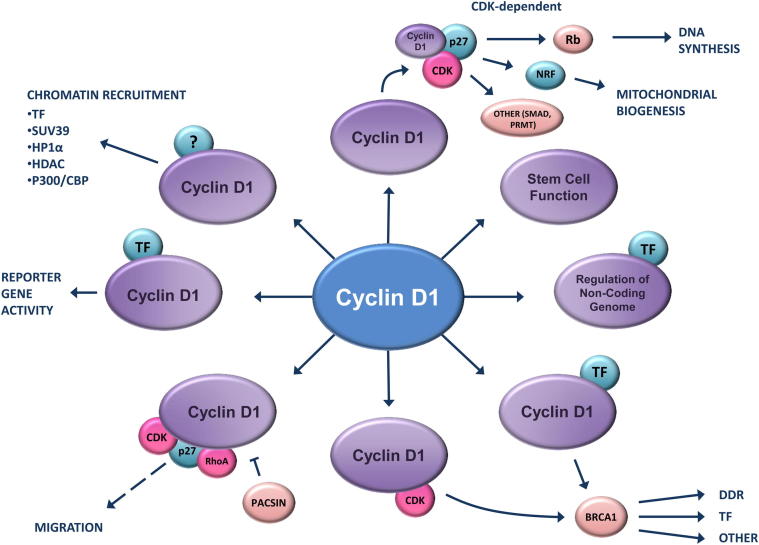
More than 35 distinct transcription factors are regulated by cyclin D1 expression.19 The mechanism appears to involve recruitment to DNA of transcription factors and associated chromatin-modifying enzymes.19 By using cyclin D1−/− mice, it was shown that cyclin D1 plays a critical role in the recruitment of transcription factors in the context of local chromatin to their cognate DNA-binding site.20 Peroxisome proliferator-activated receptor (PPAR)γ recruitment to the murine lipoprotein lipase promoter, characterized in chromatin immunoprecipitation (ChIP) assays, was dependent on the relative abundance of endogenous cyclin D1. These original studies by Hulit et al20 demonstrated the essential role for cyclin D1 in the recruitment of transcription factors in the context of local chromatin. Although it was known that cyclin D1 inhibited PPARγ reporter gene activity in a Cdk-independent manner, and that endogenous cyclin D1 played an important role in fat metabolism in vivo, these studies by Hulit and coworkers,21 were the first to demonstrate the critical role for cyclin D1 in recruiting a transcription factor to target genes in vivo. Subsequent studies using ChIP analysis provided a framework demonstrating that cyclin D1 was recruited in the context of local chromatin to target genes. Fu et al22 conducted ChIP analysis of cyclin D1−/− mouse embryo fibroblasts (MEFs), demonstrating that the recruitment of cyclin D1, in turn, recruited PPARγ, histone deacetylase (HDAC), and HDAC3 in the context of local chromatin. Fu et al22,23 showed that the recruitment of cyclin D1 was associated with the corecruitment of Su (Var) 39H1 and heterochromatin protein 1α (Figure 2) and an induction of dimethylation of H3K9.
Figure 2.
Schematic representation of cyclin D1 interactions. Schematic representation of functions shown to involve CDK binding with other interactions shown as either CDK independent or unknown.
Cyclins E and D regulate transcription factor activity through several mechanisms.24 First, CDK activity regulates the function of a variety of transcription factors, including p53, E2 transcription factor, B lymphoma Mo-MLV insertion region 1 homolog, and inhibitor of DNA-binding protein 2, in a Cdk-dependent manner. In addition, cyclins regulate the activity of the basal transcription apparatus and co-activators. Thus, the RNA polymerase II large subunit contains the essential carboxyl terminal domain, which is phosphorylated by the general transcription factor, TFIIH. The co-activator proteins p300/calcium-binding protein (CBP) undergo phosphorylation during the cell cycle, and the activity of p300 is directly regulated by cyclin D1.23 The repression of p300 by cyclin D1 involved distinct domains from those regulated by p21Cip1. Recent studies demonstrated the association of p300/CBP with cyclin D1 and the co-occupancy of p300 and cyclin D1 in the context of local chromatin using ChIP-ChIP on a −5.5- to 2.5-kb ChIP-ChIP microarray containing approximately 17,000 genes. At the p21Cip1/Waf1 promoter, cyclin D1 reduced the recruitment of CBP,25 whereas cyclin D1 enhanced recruitment of the related p300 to the murine LPL promoter,23 suggesting cyclin D1 conveys gene-specific co-integrator recruitment function. Cyclin D1 inhibits p300-mediated acetylation of histones and autoacetylation, providing an alternative mechanism by which cyclin D1 may regulate gene transcription (Figure 2).23 In addition to binding histone acetyltransferases (p300, CBP, and p300/CBP-associated factor)26,27 to regulate histone acetylation and histone deacetylases,28 cyclin D1 also recruits the estrogen receptor (ER)α co-activator, AIB1, to enhance ERα activity at a synthetic estrogen-responsive element.29,30
Cyclin D1 phosphorylates methylosome protein 50 and thereby increases activity of PRMT5, a histone arginine methyltransferase.31 Recent studies identified approximately 132 cyclin D1–interactive proteins,32 which included several previously identified cyclin D1–binding proteins (RAD51, HDAC, and p27), but did not identify other recently identified proteins, PRMT533 or C/EBP,34 suggesting cell type–specific effects. BRCA proteins were shown to bind cyclin D1,32 consistent with prior studies. In prior studies, BRCA1 colocalized in nuclear foci with RAD51 and RAD50, and cyclin D1 and BRCA1 formed distinct complexes with ERα in breast cancer cells.35 The well documented inhibition of ERα activity by BRCA136–39 was reversed by cyclin D1, requiring an HLH-like region between residues 141 and 178 (Figure 2).20 Cyclin D1 expression enhanced ERα recruitment in the context of local chromatin to an estrogen-responsive element in ChIP assays, and cyclin D1 interacted with ERα and BRCA1 at the pS2 gene promoter.
ChIP-ChIP and ChIP-Sequencing analyses have revealed the complexity of genomic binding by the DNA-associated form of cyclin D1. ChIP of cyclin D1, followed by ChIP-Sequencing, mapped at high resolution the entire genomic region bound by cyclin D1. The genome-wide distribution of binding sites in relation to the transcriptional start site showed that peak values of active regions within the promoter are comparable to those at 10 kb and beyond, suggesting that cyclin D1 localizes to both promoter-proximal elements and distant elements (Figure 2). The transcription factor–binding sites enriched in the cyclin D1 peak interval sequences of ChIP-ChIP identified several top hit transcription factors that correlated in function with the previous studies by Sicinski using promoter ChIP analysis, including mitochondrial metabolism, cellular division, and RNA processing (Figure 2). The cis elements included Ctcf, Zfx, Sp1, Mizf, Esr1, ERα, E2f1, and Creb1. Subsequent studies identified cyclin D3 in ChIP at PPARγ target promoters, much like cyclin D1.40 The ability of cyclins to regulate transcription factor activity is not restricted to cyclin D1, although the mechanisms appear to differ; thus, cyclin E/CDK2 phosphorylates TF (NFKB, inhibitor of DNA-binding protein 2, and AR) and coactivators (CBP and p220NPAT) to regulate their activity.41
Cyclin D1 Rapidly Induces CIN and Binds in ChIP to a Cluster of Genes Governing CIN
Functional pathway analysis of the gene-regulatory elements bound by cyclin D1 uncovered enrichment for genes that govern chromosomal stability and demonstrated, by ChIP, occupancy of cyclin D1 at the regulatory regions of several genes involved in chromosomal stability.42 The DNA sequences associated with cyclin D1 enrichment included Ctcf, Zfx, Sp1, Mizf, Esr1, and E2F1. The enrichment for E2f1 was significantly less, and the P value for E2f1 was two orders of magnitude less than that for Esr1 (ERα) and three orders of magnitude less than that for Ctcf. These findings suggest that cyclin D1–dependent regulation of E2f1 signaling, which is Cdk dependent,43 provides only a modest contribution to this signaling activity. Chromosomal instability (CIN) in tumors is characterized by chromosomal abnormalities and an altered gene expression signature. CIN in tumors is characterized by an elevated rate of gain or loss of whole chromosomes (aneuploidy) and/or structural chromosomal aberrations.44–46 One of the most striking differences between cancer and normal cells is aneuploidy. The molecular mechanisms inducing CIN in tumors are poorly understood.47,48 Cell cycle–associated factors have been implicated in CIN, including cyclin E.49 The relative enrichment of a molecular genetic signature of CIN-related genes has been used to quantitate a CIN score,50 including AURKB, TOP2A, CENPP, MLF1IP, ZW10, and CKAP2.46 Interrogation of gene expression from 2254 breast tumors identified cyclin D1 expression correlating with CIN in luminal B breast cancer (Figure 2).42 Cyclin D1 rescue of cyclin D1−/− MEFs induced CIN gene expression and CIN gene-regulatory region occupancy by the DNA-bound form of cyclin D1. Mammary gland–targeted cyclin D1 expression induced tumors with CIN, and short-term transgenic expression of cyclin D1 induced CIN in vivo.
Previous studies had characterized a function for cyclin E in regulating the centrosome. The in vivo analysis of E-type cyclins identified a key role for cyclin E in re-entry to the cell cycle from the quiescent state, and in cultured cells, centrosomal localization of cyclin E was required for S-phase entry in a Cdk2-independent manner.51,52 Cyclin E overexpression also induces CIN in tumors thought to be related to the induction of double-standard DNA breaks.53
Cyclin D1 Governs DNA Damage Repair through Recruiting DNA Repair Complexes
Cell type–dependent radiation–induced sensitivity is determined by the relative abundance of cyclin D1. Gamma irradiation–induced apoptosis was enhanced in cyclin D1−/− MEFs,54 and cyclin D1 expression inhibited UV-induced apoptosis in a p300-dependent manner. In contrast, breast cancer cell lines show enhanced apoptosis in response to γ radiation when cyclin D1 is overexpressed.55,56 Cyclin D1 is important in G1 cell arrest in response to gamma irradiation–induced DNA damage because interference in cyclin D1 degradation prevents both G1 arrest and G2-M arrest induced by gamma irradiation.57 Intriguingly, lymphoid compartment–targeted transgenic expression of a cyclin D1 mutant (D1T286A) that is nuclear in expression during S phase induced aneuploidy lymphoid tumors in which a DNA damage response (DDR) was triggered.31
Because a physical tethering of DNA repair factors to chromatin induces the DDR signaling cascade, and cyclin D1 is recruited to local chromatin, as evidenced by ChIP assays, Li et al61 examined the role of cyclin D1 in regulating the DNA damage signaling response. By using the comet assay, in which intact DNA composes the head, and the tail consists of damaged DNA at a neutral pH, cyclin D1+/+ had a 14-fold increase in comet tail formation compared with cyclin D1−/− cells. H2AX phosphorylation on serine 139 (γH2A), a sensitive mark of double-strand breaks, was increased with doxorubicin treatment in cyclin D1+/+ compared with cyclin D1−/− cells. Cyclin D1 siRNA mediated reduction in endogenous cyclin D1 and reduced 5-fluorouracil–induced γH2A phosphorylation. The enhancement of the DDR by cyclin D1 occurred rapidly (15 minutes), preceding the effect on DNA synthesis (>6 hours). Cyclin D1a enhanced the DDR induced by doxorubicin but did not enhance S-phase entry in the absence of serum, providing corroborative evidence that the induction of the DDR by cyclin D1a can be uncoupled from the induction of DNA synthesis. Cyclin D1 was shown to bind p21Cip1 and RAD51 using a cherry red fluorescent LacR reporter system and immune precipitation. By using p21−/− MEFs, cyclin D1 induction of the DDR was shown to require p21Cip1. Cyclin D1a recapitulated the recruitment of H2AX at the LacO site in a similar manner as the DNA damage repair signaling pathway proteins ATM, NBS1, or MDC1. Mutational analysis demonstrated a requirement for the cyclin D1 carboxyl terminus in the recruitment to H2AX foci. Cyclin D1a was shown to recruit RAD51 in the context of local chromatin in response to the DNA damage.
The induction of RAD51 abundance by cyclin D1a, its binding to cyclin D1a, and recruitment by cyclin D1 in the context of chromatin to sites of DNA damage support the notion that cyclin D1 enhances the DDR, but does not formally determine its role in DNA damage repair. Cyclin D1a induces genes involved in DNA replication and/or the DNA damage checkpoint in fibroblasts and in the mammary epithelium.15,22 A protein interaction analysis of cyclin D1–interacting proteins identified RAD51.32 By using a homologous recombination repair reporter system, endogenous cyclin D1 was shown to increase the homologous recombination rate. BRCA2 was also identified as a cyclin D1–interacting protein, and BRCA2 knockdown reduced cyclin D1 recruitment to DNA damage sites. Cyclin D1 depletion did not affect BRCA2 recruitment. However, cyclin D1 depletion reduced RAD51 recruitment to DNA damage sites.32 Again, the regulation of DNA replication and/or DNA damage checkpoints is not unique to cyclin D1. Cyclin D1 induces the expression of minichromosome maintenance-deficient (MCM) 2, MCM3, and MCM4,15,23 whereas cyclin E/CDK regulates loading of MCM onto chromatin52 through both kinase-dependent and kinase-independent mechanisms.58
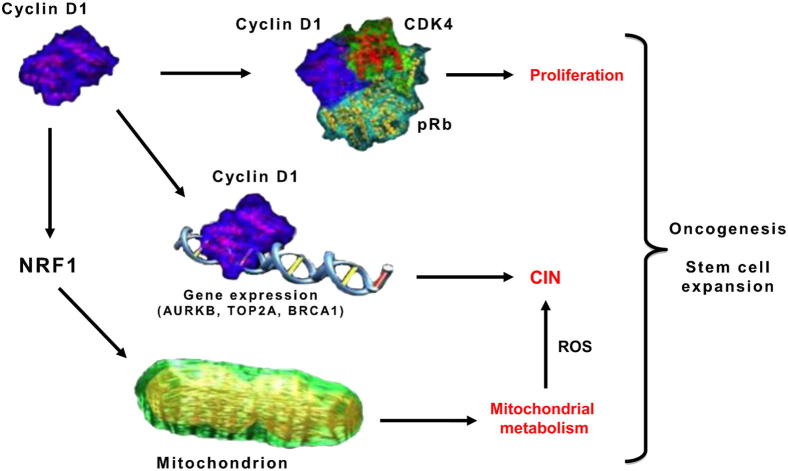
Collectively, these studies illustrate the diverse roles that cyclin D1 plays, including the inhibition of mitochondrial biogenesis through phosphorylation of NRF-1, inhibition of G1/S-phase transition through phosphorylation of pRb, and the regulation of gene expression, at least in part through a DNA-bound form of cyclin D1 that interacts with transcription factors in the context of chromatin (Figure 3). The mechanisms by which each of these functions contributes to oncogenesis and stem cell function remain to be fully understood.
Figure 3.
Hypothetical functional interaction between cyclin D1 as a kinase complex, which phosphorylates pRb and NRF1, and the DNA-bound form, which binds promoters of genes regulating CIN, to promote oncogenesis.
Future Directions: Cyclin D1 Regulates the Noncoding Genome
At this time, relatively little is known of the mechanisms by which the cell cycle and cyclins regulate the noncoding genome. miRNAs are 21- to 22-nucleotide molecules that regulate cellular phenotype via regulation of translational efficiency or the stability of targeted mRNAs. Compelling evidence has demonstrated the importance of the noncoding genome and the miRNA biogenesis apparatus in tumorigenesis. Analysis of miRNAs regulated in cyclin D1–induced transgenic mammary tumors and those reciprocally regulated in cyclin D1 antisense or knockout mice identified the miR-17/20 cluster as a cyclin D1–induced regulator of mammary tumor growth. miR-17/20 repressed cyclin D1 expression via its 3′ untranslated region binding site.59 In ChIP assays, cyclin D1 associated with the miR-17/20 regulatory region between nucleotides −1050 and −1200. To our knowledge, these studies were the first to demonstrate cyclin-dependent regulation of noncoding RNA via binding to the regulatory region of the noncoding miRNA cluster. Yu et al60 demonstrated that miR-17/20 expression regulated the secretion of cytokines and plasminogen activator via expression of α-enolase and cytokeratin 8. The inhibition of plasminogen activator by miR-17/20 required cyclin D1, indicating that complex regulatory loops between the noncoding and coding genome govern cellular migration of breast cancer cells.60
At this time, published studies suggest cyclin E is regulated by several different miRNAs, including miR-223, miR-161, and miR-195. However, there is no evidence that cyclin E binds to regulatory regions of the noncoding genome to coordinate miRNA expression. As yet, there is no clear mechanism by which the cell cycle coordinates signaling of noncoding RNA biogenesis. Given the disruption of noncoding genome precursors in many tumors, it will be of particular interest to determine the potential role for cyclins in regulating broader aspects of noncoding RNA biogenesis.
Footnotes
Supported in part by NIH grants R01CA70896, R01CA75503, R01CA86072, R01CA137494, and R01CA132115; the Kimmel Cancer Center was supported by NIH Cancer Center Core grant P30CA56036; and this project is supported in part by the Dr. Ralph and Marian C. Falk Medical Research Trust, the Breast Cancer Research Foundation, and a Pennsylvania Department of Health grant.
The Pennsylvania Department of Health specifically disclaims responsibility for analyses, interpretations, or conclusions.
References
- 1.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 2.Fantl V., Stamp G., Andrews A., Rosewell I., Dickson C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- 3.Sicinski P., Donaher J.L., Parker S.B., Li T., Fazeli A., Gardner H., Haslam S.Z., Bronson R.T., Elledge S.J., Weinberg R.A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 4.Lange C., Huttner W.B., Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Jeselsohn R., Brown N.E., Arendt L., Klebba I., Hu M.G., Kuperwasser C., Hinds P.W. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geng Y., Whoriskey W., Park M.Y., Bronson R.T., Medema R.H., Li T., Weinberg R.A., Sicinski P. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell. 1999;97:767–777. doi: 10.1016/s0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
- 7.Geng Y., Yu Q., Sicinska E., Das M., Bronson R.T., Sicinski P. Deletion of the p27Kip1 gene restores normal development in cyclin D1-deficient mice. Proc Natl Acad Sci U S A. 2001;98:194–199. doi: 10.1073/pnas.011522998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumeister P., Pixley F.J., Xiong Y., Xie H., Wu K., Ashton A., Cammer M., Chan A., Symons M., Stanley E.R., Pestell R.G. Cyclin D1 governs adhesion and motility of macrophages. Mol Biol Cell. 2003;14:2005–2015. doi: 10.1091/mbc.02-07-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Wang C., Jiao X., Lu Y., Fu M., Quong A.A., Dye C., Yang J., Dai M., Ju X., Zhang X., Li A., Burbelo P., Stanley E.R., Pestell R.G. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006;26:4240–4256. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z., Jiao X., Wang C., Ju X., Lu Y., Yuan L., Lisanti M.P., Katiyar S., Pestell R.G. Cyclin D1 induction of cellular migration requires p27(KIP1) Cancer Res. 2006;66:9986–9994. doi: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- 11.Li Z., Wang C., Jiao X., Katiyar S., Casimiro M.C., Prendergast G.C., Powell M.J., Pestell R.G. Alternate cyclin D1 mRNA splicing modulates p27KIP1 binding and cell migration. J Biol Chem. 2008;283:7007–7015. doi: 10.1074/jbc.M706992200. [DOI] [PubMed] [Google Scholar]
- 12.Tobin N.P., Sims A.H., Lundgren K.L., Lehn S., Landberg G. Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer. 2011;11:417. doi: 10.1186/1471-2407-11-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng H., Tian L., Zhou J., Li Z., Jiao X., Li W.W., Plomann M., Xu Z., Lisanti M.P., Wang C., Pestell R.G. PACSIN 2 represses cellular migration through direct association with cyclin D1 but not its alternate splice form cyclin D1b. Cell Cycle. 2011;10:73–81. doi: 10.4161/cc.10.1.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C., Li Z., Lu Y., Du R., Katiyar S., Yang J., Fu M., Leader J.E., Quong A., Novikoff P.M., Pestell R.G. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakamaki T., Casimiro M.C., Ju X., Quong A.A., Katiyar S., Liu M., Jiao X., Li A., Zhang X., Lu Y., Wang C., Byers S., Nicholson R., Link T., Shemluck M., Yang J., Fricke S.T., Novikoff P.M., Papanikolaou A., Arnold A., Albanese C., Pestell R. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26:5449–5469. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tchakarska G., Roussel M., Troussard X., Sola B. Cyclin D1 inhibits mitochondrial activity in B cells. Cancer Res. 2011;71:1690–1699. doi: 10.1158/0008-5472.CAN-10-2564. [DOI] [PubMed] [Google Scholar]
- 17.Mandal S., Freije W.A., Guptan P., Banerjee U. Metabolic control of G1-S transition: cyclin E degradation by p53-induced activation of the ubiquitin-proteasome system. J Cell Biol. 2010;188:473–479. doi: 10.1083/jcb.200912024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sankaran V.G., Orkin S.H., Walkley C.R. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 2008;22:463–475. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu M., Wang C., Li Z., Sakamaki T., Pestell R.G. Minireview: cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 20.Hulit J., Wang C., Li Z., Albanese C., Rao M., Di Vizio D., Shah S., Byers S.W., Mahmood R., Augenlicht L.H., Russell R., Pestell R.G. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol Cell Biol. 2004;24:7598–7611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C., Pattabiraman N., Zhou J.N., Fu M., Sakamaki T., Albanese C., Li Z., Wu K., Hulit J., Neumeister P., Novikoff P.M., Brownlee M., Scherer P.E., Jones J.G., Whitney K.D., Donehower L.A., Harris E.L., Rohan T., Johns D.C., Pestell R.G. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol Cell Biol. 2003;23:6159–6173. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu M., Rao M., Bouras T., Wang C., Wu K., Zhang X., Li Z., Yao T.P., Pestell R.G. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280:16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 23.Fu M., Wang C., Rao M., Wu X., Bouras T., Zhang X., Li Z., Jiao X., Yang J., Li A., Perkins N.D., Thimmapaya B., Kung A.L., Munoz A., Giordano A., Lisanti M.P., Pestell R.G. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280:29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 24.Pestell R.G., Albanese C., Reutens A.T., Segall J.E., Lee R.J., Arnold A. The cyclins and cyclin-dependent kinase inhibitors in hormonal regulation of proliferation and differentiation. Endocr Rev. 1999;20:501–534. doi: 10.1210/edrv.20.4.0373. [DOI] [PubMed] [Google Scholar]
- 25.Bienvenu F., Barre B., Giraud S., Avril S., Coqueret O. Transcriptional regulation by a DNA-associated form of cyclin D1. Mol Biol Cell. 2005;16:1850–1858. doi: 10.1091/mbc.E04-08-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reutens A.T., Fu M., Wang C., Albanese C., McPhaul M.J., Sun Z., Balk S.P., Janne O.A., Palvimo J.J., Pestell R.G. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 27.McMahon C., Suthiphongchai T., DiRenzo J., Ewen M.E. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc Natl Acad Sci U S A. 1999;96:5382–5387. doi: 10.1073/pnas.96.10.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin H.M., Zhao L., Cheng S.Y. Cyclin D1 is a ligand-independent co-repressor for thyroid hormone receptors. J Biol Chem. 2002;277:28733–28741. doi: 10.1074/jbc.M203380200. [DOI] [PubMed] [Google Scholar]
- 29.Zwijsen R.M., Wientjens E., Klompmaker R., van der Sman J., Bernards R., Michalides R.J. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 30.Neuman E., Ladha M.H., Lin N., Upton T.M., Miller S.J., DiRenzo J., Pestell R.G., Hinds P.W., Dowdy S.F., Brown M., Ewen M.E. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aggarwal P., Lessie M.D., Lin D.I., Pontano L., Gladden A.B., Nuskey B., Goradia A., Wasik M.A., Klein-Szanto A.J., Rustgi A.K., Bassing C.H., Diehl J.A. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jirawatnotai S., Hu Y., Michowski W., Elias J.E., Becks L., Bienvenu F., Zagozdzon A., Goswami T., Wang Y.E., Clark A.B., Kunkel T.A., van Harn T., Xia B., Correll M., Quackenbush J., Livingston D.M., Gygi S.P., Sicinski P. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal P., Vaites L.P., Kim J.K., Mellert H., Gurung B., Nakagawa H., Herlyn M., Hua X., Rustgi A.K., McMahon S.B., Diehl J.A. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18:329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb J., Ramaswamy S., Ford H.L., Contreras B., Martinez R.V., Kittrell F.S., Zahnow C.A., Patterson N., Golub T.R., Ewen M.E. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 35.Wang C., Fan S., Li Z., Fu M., Rao M., Ma Y., Lisanti M.P., Albanese C., Katzenellenbogen B.S., Kushner P.J., Weber B., Rosen E.M., Pestell R.G. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65:6557–6567. doi: 10.1158/0008-5472.CAN-05-0486. [DOI] [PubMed] [Google Scholar]
- 36.Fan S., Wang J., Yuan R., Ma Y., Meng Q., Erdos M.R., Pestell R.G., Yuan F., Auborn K.J., Goldberg I.D., Rosen E.M. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 37.Ma Y., Fan S., Hu C., Meng Q., Fuqua S.A., Pestell R.G., Tomita Y.A., Rosen E.M. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol Endocrinol. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan S., Ma Y.X., Wang C., Yuan R.Q., Meng Q., Wang J.A., Erdos M., Goldberg I.D., Webb P., Kushner P.J., Pestell R.G., Rosen E.M. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene. 2001;20:77–87. doi: 10.1038/sj.onc.1204073. [DOI] [PubMed] [Google Scholar]
- 39.Fan S., Ma Y.X., Wang C., Yuan R.Q., Meng Q., Wang J.A., Erdos M., Goldberg I.D., Webb P., Kushner P.J., Pestell R.G., Rosen E.M. p300 Modulates the BRCA1 inhibition of estrogen receptor activity. Cancer Res. 2002;62:141–151. [PubMed] [Google Scholar]
- 40.Sarruf D.A., Iankova I., Abella A., Assou S., Miard S., Fajas L. Cyclin D3 promotes adipogenesis through activation of peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2005;25:9985–9995. doi: 10.1128/MCB.25.22.9985-9995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma T., Van Tine B.A., Wei Y., Garrett M.D., Nelson D., Adams P.D., Wang J., Qin J., Chow L.T., Harper J.W. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casimiro M.C., Crosariol M., Loro E., Ertel A., Yu Z., Dampier W., Saria E.A., Papanikolaou A., Stanek T.J., Li Z., Wang C., Fortina P., Addya S., Tozeren A., Knudsen E.S., Arnold A., Pestell R.G. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. J Clin Invest. 2012;122:833–843. doi: 10.1172/JCI60256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe G., Albanese C., Lee R.J., Reutens A., Vairo G., Henglein B., Pestell R.G. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 45.Lengauer C., Kinzler K.W., Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 46.Thompson S.L., Bakhoum S.F., Compton D.A. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gollin S.M. Mechanisms leading to chromosomal instability. Semin Cancer Biol. 2005;15:33–42. doi: 10.1016/j.semcancer.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Draviam V.M., Xie S., Sorger P.K. Chromosome segregation and genomic stability. Curr Opin Genet Dev. 2004;14:120–125. doi: 10.1016/j.gde.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Spruck C.H., Won K.A., Reed S.I. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 50.Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto Y., Maller J.L. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 2004;306:885–888. doi: 10.1126/science.1103544. [DOI] [PubMed] [Google Scholar]
- 52.Geng Y., Lee Y.M., Welcker M., Swanger J., Zagozdzon A., Winer J.D., Roberts J.M., Kaldis P., Clurman B.E., Sicinski P. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–139. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 53.Loeb K.R., Kostner H., Firpo E., Norwood T., D Tsuchiya K., Clurman B.E., Roberts J.M. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell. 2005;8:35–47. doi: 10.1016/j.ccr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Albanese C., D'Amico M., Reutens A.T., Fu M., Watanabe G., Lee R.J., Kitsis R.N., Henglein B., Avantaggiati M., Somasundaram K., Thimmapaya B., Pestell R.G. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274:34186–34195. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- 55.Coco Martin J.M., Balkenende A., Verschoor T., Lallemand F., Michalides R. Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res. 1999;59:1134–1140. [PubMed] [Google Scholar]
- 56.Zhou Q., Fukushima P., DeGraff W., Mitchell J.B., Stetler Stevenson M., Ashkenazi A., Steeg P.S. Radiation and the Apo2L/TRAIL apoptotic pathway preferentially inhibit the colonization of premalignant human breast cells overexpressing cyclin D1. Cancer Res. 2000;60:2611–2615. [PubMed] [Google Scholar]
- 57.Agami R., Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 58.Coverley D., Laman H., Laskey R.A. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat Cell Biol. 2002;4:523–528. doi: 10.1038/ncb813. [DOI] [PubMed] [Google Scholar]
- 59.Yu Z., Wang C., Wang M., Li Z., Casimiro M.C., Liu M., Wu K., Whittle J., Ju X., Hyslop T., McCue P., Pestell R.G. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Z., Willmarth N.E., Zhou J., Katiyar S., Wang M., Liu Y., McCue P.A., Quong A.A., Lisanti M.P., Pestell R.G. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A. 2010;107:8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]