Abstract
Like the p16, SMAD4, and RB1 genes, FAM190A (alias CCSER1) lies at a consensus site of homogeneous genomic deletions in human cancer. FAM190A transcripts in 40% of cancers also contain in-frame deletions of evolutionarily conserved exons. Its gene function was unknown. We found an internal deletion of the FAM190A gene in a pancreatic cancer having prominent focal multinuclearity. The experimental knockdown of FAM190A expression by shRNA caused focal cytokinesis defects, multipolar mitosis, and multinuclearity as observed in time-lapse microscopy. FAM190A was localized to the γ-tubulin ring complex of early mitosis and to the midbody in late cytokinesis by immunofluorescence assay and was present in the nuclear fraction of unsynchronized cells by immunoblot. FAM190A interacted with EXOC1 and Ndel1, which function in cytoskeletal organization and the cell division cycle. Levels of FAM190A protein peaked 12 hours after release from thymidine block, corresponding to M-phase. Slower-migrating phosphorylated forms accumulated toward M-phase and disappeared after release from a mitotic block and before cytokinesis. Studies of FAM190A alterations may provide mechanistic insights into mitotic dysregulation and multinuclearity in cancer. We propose that FAM190A is a regulator or structural component required for normal mitosis and that both the rare truncating mutations and common in-frame deletion alteration of FAM190A may contribute to the chromosomal instability of cancer.
Chromosomal instability promotes the development of most cancers. Somatic mutations in mitotic genes occur in some human cancers, including mitotic checkpoint genes and cohesins, but these accounts for <15% of all cancers.1–3 There is thus an unmet need to explain widespread aneuploidy in cancers.
We previously reported a human pancreatic cancer with focal prominent multinuclearity.4 Although the typical mutations of pancreatic cancer would not be able to explain this phenotype, a high-throughput examination of rearrangements revealed the tumor to have an out-of-frame deletion of exons 4 and 5 of the FAM190A gene.5 This mutation was unusual for FAM190A, perhaps representing a more severe gene disruption than typically observed. We previously reported that approximately 40% of the human cancers have internal rearrangements (exon deletions) in FAM190A transcripts,6 generally being in-frame and affecting downstream exons. The FAM190A gene was not silenced (we observed its transcripts) or lost in entirety. FAM190A genomic homozygous deletions were also reported in pancreatic cancer cell lines7 and in an esophageal cancer8 examined by a single-nucleotide array, and a homozygous missense mutation was observed in a non–small cell lung cancer.9 FAM190A was associated with attention-deficit hyperactivity disorder in a genome-wide association study, but after correction for multiple comparisons it was not statistically significant.10
The FAM190A protein was notable in having 23 serines among the first 69 amino acids but had no other defined domains or known function. We induced FAM190A deficiency in cells, creating phenotype of focal multinuclearity and multipolar mitosis, recalling the phenotype of the described pancreatic cancer. The multinuclear phenotype accompanied cellular defects in cytokinesis. We observed phosphorylated forms of FAM190A in cells related to stages of the division cycle. This work may provide a link between FAM190A deletions and genomic abnormalities of cancer.
Materials and Methods
Biological Samples and Supplies
Xenografted human cancers were obtained from our described tissue banks.11 Human tissue was also collected through The Gastrointestinal Cancer Rapid Medical Donation Program at The Johns Hopkins Hospital. Tissues were obtained and used under institutional review board–approved protocols. Selected pancreatic cancer xenografts PX19-R2 and PX188-3 had FAM190A genomic deletions; FAM190A genomic status was unknown for unselected pancreatic xenografts PX120-4A and PX121-3.
All cell lines were from ATCC (Manassas, VA) except AAV-293, which was from Agilent Technologies (Santa Clara, CA). HeLa, DLD1, and AsPC1 cells had wild-type FAM190A transcripts. AAV-293 cells had transcript deletions of exons 7 or 7, 8, and 9. RKO cells had a genomic homozygous deletion of exons 4, 5, 6, and 7. H2126 had a genomic homozygous deletion of exons 9 and 10. HeLa, RKO, H2126, and AAV-293 cells were grown in Dulbecco’s modified Eagle’s medium, and AsPC1 and DLD1 cells in RPMI 1640 medium, all with 10% fetal bovine serum and antibiotics.
Dulbecco’s modified Eagle’s medium, Lipofectamine 2000, Lipofectamine, ProLong Gold antifade reagent with DAPI, Celllight Green fluorescent protein (GFP) histone 2B (H2B), and 4% to 12% Bis-Tris acrylamide gels were from Life Technologies Inc (Carlsbad, CA). Protease inhibitors were from Roche (Basel, Switzerland). Thymidine, nocodazole, demecolcine, N-ethymaleimide, propidium iodide (PI), Protein A-Sepharose, LabTek 8-well glass, and Permanox slides were from Sigma-Aldrich (St. Louis, MO). Calf intestinal phosphatase (CIP) was used in restriction enzyme buffer 3 (New England BioLabs, Ipswich, MA).
Rabbit anti-FAM190A monoclonal antibodies (mAbs) mAb7, mAb62, mAb96, mAb108, and mAb170 were developed by immunizing rabbits with recombinant FAM190A protein (performed by Epitomics Inc, Burlingame, CA). For polyclonal anti-FAM190A antibodies, rabbits were immunized with peptides of exons 7, 8, 9, and 10 of FAM190A (performed by Creative Diagnostics, Shirley, NY). Exon-specific polyclonal antibodies were purified from sera by Protein A-Sepharose. Anti–γ-tubulin (GTU-88) antibody was from Sigma-Aldrich. Anti–α-tubulin (DM1A), anti–securin B, anti–lamin B, anti-actin, and anti-GAPDH antibodies and horseradish peroxidase–tagged secondary antibodies goat anti-mouse, goat anti-rabbit, and donkey anti-goat were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti–centrin 3 was from Abnova (Tapei City, Japan). Anti-FAM190A (AP10022b) was from Abgent (San Diego, CA). Anti–cyclin E was from BD Biosciences (San Jose, CA). Anti-Ndel1, anti-EXOC1, and anti-FAM190A (ab90508) were from Abcam (Cambridge, UK). Horseradish peroxidase–tagged mouse monoclonal anti-rabbit antibody was from Jackson ImmunoResearch Laboratories Inc (West Grove, PA).
Generation of FAM190A-Deficient Cell Lines
Retroviral control, scrambled, and FAM190A-targeting shRNA plasmids were from Origene Technologies (Rockville, MD). FAM190A-targeting sequences were as follows: plasmid 77, 5′-TACCTGCTACAAGTGTGAGCCACTCAGAG-3′; plasmid 78, 5′-GCTCAACCTGGTCACAGCAATATGCAGAA-3′; plasmid 79, 5′-ATGTGCCGCAGTAGTTCTTACTCCTATGG-3′; plasmid 80, 5′-CTCTGCCATTCAGACTGATGTTACAGGAC-3′; plasmid 96A, 5′- TGAACTAGGTTGCATTGCCTTGAAGACTT-3′; and plasmid 96B, 5′- GTTGAAGTACCGTGACATAACACAAGGTG-3′. HeLa, AAV-293, RKO, AsPC1, and DLD1 cells were transfected with shRNA plasmids or infected with retroviral shRNA plasmid. New antibiotic-containing medium was added to cultures 48 hours after transfection or infection. Cells were selected with puromycin; stable clones were established by colony-selection or limiting dilution.
Yeast Two-Hybrid
A yeast two-hybrid screen was performed (Hybrigenics Services SAS, Paris, France) using full-length FAM190A as bait and human fetal brain cDNA RP1 as the prey library. Interactions (67.9 million) were recorded in the presence of 10 mmol/L 3-aminotriazole selection. For each interaction, a predicted biological score was computed to assess the interaction reliability. This score represents the probability of an interaction to be nonspecific: it is an E-value primarily comparing the number of independent prey fragments found for an interaction and the chance of finding them at random (background noise).
Cell Lysis, IB, and IP
Cells were lyzed with detergent (150 mmol/L NaCl, 50 mmol/L Tris-HCl, 1 mmol/L EDTA, 1% Triton-X, 20 mmol/L N-ethymaleimide, pH 7.4) or RIPA solution (150 mmol/L NaCl, 50 mmol/L Tris-HCl, 1 mmol/L EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, pH 7.4). For immunoblotting (IB), clarified cell lysates were quantified by the DC method (Bio-Rad Laboratories, Hercules, CA). Protein in Laemmli buffer (20 to 40 μg per well) was loaded and resolved on 4% to 12% Bis-Tris gels, and transferred to a polyvinylidene difluoride membrane, blocked with 5% milk, incubated with primary antibody and subsequently with horseradish peroxidase–tagged secondary antibody, developed with chemiluminescence [Luminol from ThermoScientific (Rockford, IL) or Luminata Forte from Millipore (Billerica, MA)], and imaged on film. For immunoprecipitation (IP), 1 mg of lysate was incubated with 2 μg of antibodies; complexes were captured by addition of Protein A-Sepharose and bound proteins eluted by denaturation using 1× Laemmli buffer and incubating at 100°C for 5 minutes. Human tissues and xenografts were lyzed in lysis detergent containing 1% SDS. For IP, 2 mg of tissue lysate was incubated with 10 μg of antibodies; complexes were captured by addition of Protein A-Sepharose and bound proteins eluted by denaturation using 1× Laemmli buffer and incubating at 100°C for 5 minutes.
Cell Synchronization
Double-Thymidine Block
HeLa cells were treated with thymidine (5 mmol/L) for 16 hours. Washed cells were grown for 8 to 10 hours without thymidine and then treated again with thymidine for 16 hours; thrice-washed cells were fed with fresh medium and harvested at 0, 8, 10, 12, 14, and 16 hours in cell lysis detergent or prepared for flow cytometry. Protein (20 μg per well) was examined by IB.
Thymidine-Nocodazole Block
HeLa cells were treated with 5 mmol/L thymidine for 16 hours. Washed cells were immediately treated with medium containing 50 ng/mL of nocodazole for 16 hours. Rounded cells were harvested at prometaphase by shake-off and gentle pipetting. Washed cells were resuspended in medium in a culture flask and harvested at 0, 0.5, 1, 2, 4, 6, and 24 hours in cell lysis detergent or for flow cytometry. Protein (20 μg per well) was examined by IB.
Flow Cytometry
Harvested cells were fixed by slow addition of 70% ethanol and stored at −20°C until analysis. PI-stained cells were analyzed in a flow cytometer (FACSCalibur; BD Biosciences, San Jose, CA). Data were analyzed with CellQuest Pro version 5.2.1 software (BD Biosciences).
Immunofluorescence
Cells were grown on 8-well slides, washed with PBS, fixed with −20°C methanol for 10 minutes, incubated with 0.1% Triton X-100 in PBS for 10 minutes, blocked (1% goat serum, 0.05% NP40 in PBS) for 10 minutes, incubated with primary antibody for 2 hours, incubated with fluorophore-tagged secondary antibodies for 1 hour in blocking solution, and mounted with ProLong Gold anti-fade containing DAPI (Life Technologies, Grand Island, NY).
Microscopy
Conventional light and phase-contrast microscopy and fluorescence microscopy used a Zeiss Axiovert microscope (Carl Zeiss AG, Oberkochen, Germany) mounted with incandescent and halogen lamps. Images were acquired and analyzed with MetaMorph software version 6.5 (Molecular Devices, Downington, PA). For confocal microscopy, images were acquired with Zeiss Meta Laser-Scanning Confocal Microscope System (Zeiss AG) and Zen software 2009 (Carl Zeiss Microscopy, LLC, Thornwood, NY). An argon laser exciting at 488 nm and a red HeNe laser at 561 nm were used to obtain optical sections. Narrow-band emission filters were used to eliminate channel cross talk. A 1.0-μm confocal aperture was used to obtain z-plane sections. Slides were imaged with a 63× oil immersion Plan Apochromat objective lens (numerical aperture 1.4) through a Zeiss Axiovert microscope. For time-lapse microscopy, images were acquired every 10 minutes with a Zeiss Axiovert 200 microscope fitted with a Yokogawa CSU 22 spinning disk (Yokogawa Corporation of America, Sugar Land, TX) for time-lapse microscopy and a Photometrics Cascade II camera (Photometrics, Tucson, AZ). Images were analyzed by SlideBook software version 5.0 (Intelligent Imaging Innovations, Inc., Denver, CO). For better visualization of cells, we introduced GFP-tagged H2B using Celllight H2B-GFP (Baculovirus).
IHC and Histology
HeLa cells were washed with PBS, fixed with formalin, collected by scrapping, washed, and mixed with low-melting agarose to form a plug, which was mounted in paraffin. Immunohistochemistry (IHC) on sections used anti-FAM190A antibody (mAb7) and Dako kit (Dako, Carpinteria, CA). Human tissue and xenografts were fixed in formalin and mounted in paraffin. H&E sections were prepared for human tissues, xenografts, and HeLa cell plugs.
Phosphatase Treatment
HeLa cells were harvested in phosphatase lysis solution (restriction enzyme 1× buffer 3 with 1% Triton X; New England Biolab). After IP, washed beads containing captured protein were resuspended in 1× buffer 3. CIP was added in either sample type and incubated at 37°C for 60 minutes. Laemmli buffer was added to stop the reaction and for gel separation.
Results
Natural and Induced Multinuclearity
As reported, focal multinuclearity was a pronounced feature of a particular pancreatic exocrine adenocarcinoma (Figure 1A),4 which in another study was found to have an internal gene rearrangement creating an out-of-frame deletion of exons 4 and 5 in the FAM190A gene.5
Figure 1.
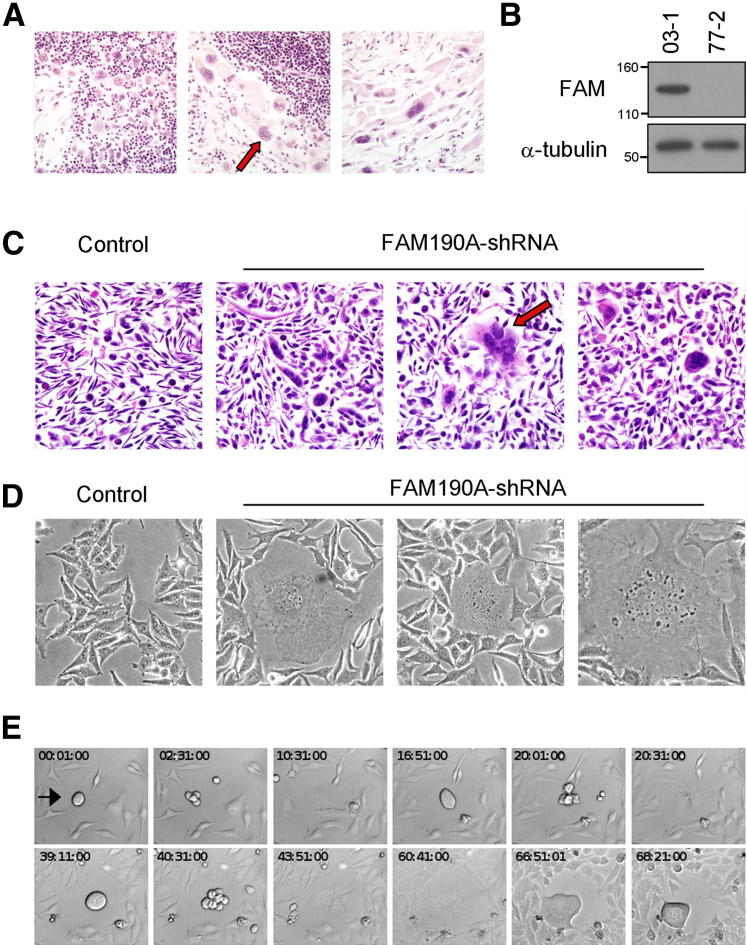
Natural and induced multinuclearity is induced by FAM190A absence. A: H&E-stained section of a pancreatic cancer tissue with known FAM190A exon 4 to 5 deletions. The tissue has focal multinuclear cells (arrow). B: HeLa cell lines were generated by stable transfection of control shRNA plasmid or four different FAM190A-targeting shRNA plasmids. FAM190A-targeting shRNA clone 77-2 had a reduced level of FAM190A protein compared with control shRNA clone 03-1. IB with mAb7 and anti–α-tubulin. C: Agarose plugs were prepared from two shRNA lines from B. Multinuclear cells were present in clone 77-2 by H&E staining (arrow). Control cells lacked multinuclearity. D: Phase-contrast images of FAM190A-knockdown clone 77-2 and control cells. Multinuclear cells were present in clone 77-2. E: Phase-contrast images from a time-lapse movie of FAM190A-knockdown clone 77-2 (Supplemental Video S1). A multinuclear cell underwent multiple rounds of cell division, generating progressively more nuclei in the cell (arrow).
We generated several stable HeLa cell clones by transfection of four different shRNA plasmids targeting the FAM190A transcript. On screening the clones by IB, we identified clone 77-2 (having integrated FAM190A shRNA sequence 77) to have a very low FAM190A protein expression compared with control clone 03-1 (having control shRNA plasmid) (Figure 1B). Focal multinuclear cells were present in the culture of clone 77-2 but not in control clone 03-1. On H&E histologic examination of an agarose plug of clone 77-2 and clone 03-1, multiple isolated multinuclear cells were again seen in the cells of the knockdown clone (Figure 1C).
Phase-contrast microscopy confirmed the presence of focal cells having multiple nuclei in the FAM190A-knockdown clone (Figure 1D). Some multinucleated cells had bizarre nuclear shapes, as seen by DAPI staining (Supplemental Figure S1A). We stained cells with PI and quantified DNA by flow cytometry. FAM190A-knockdown cells had a higher fraction of cells having >2N DNA (percentage of cells, means ± SEM, 4.1 ± 0.72) relative to control cells (1.10 ± 0.05; P < 0.05; Supplemental Figure S1B).
To visualize the process by which these cells became multinuclear, we performed time-lapse microscopy. Cells were imaged every 10 minutes. The resultant movies revealed isolated mitotic cells failing to separate fully from each other at cytokinesis. The unseparated cells sometimes remained adjacent but eventually merged their cytoplasm before synchronous mitosis, in the process forming a fused cell with two or more nuclei. The multinucleated cells underwent multipolar mitosis, failed to separate, and formed cells with yet higher numbers of nuclei (Figure 1E and Supplemental Videos S1, S2, S3, S4, S5, and S6). At times, individual cells appeared to break away from the main mass but became detached or died. Cells with high numbers of nuclei tended to become syncytial rather than retaining a degree of separateness; thus, they resembled the giant cells seen in some human cancers, the osteoclasts of normal bone, and the cells in injured liver (multinucleate hepatocytes). We used baculoviral infection to introduce GFP-tagged H2B into clone 77-2. This permitted fluorescence tracking of the chromosomes and nuclei (Supplemental Videos S1, S2, S3, S4, S5, and S6).
Multinucleate cell morphology was observed in replication studies after stable transfection or infection of the same shRNA plasmid. Multinucleate morphology was also observed in multiple cell lines, including HeLa, RKO, 293-AAV, AsPC-1, and DLD1, after stable transfection or infection of different shRNA plasmids (Supplemental Figure S1C).
Cellular Localization
Immunofluorescence
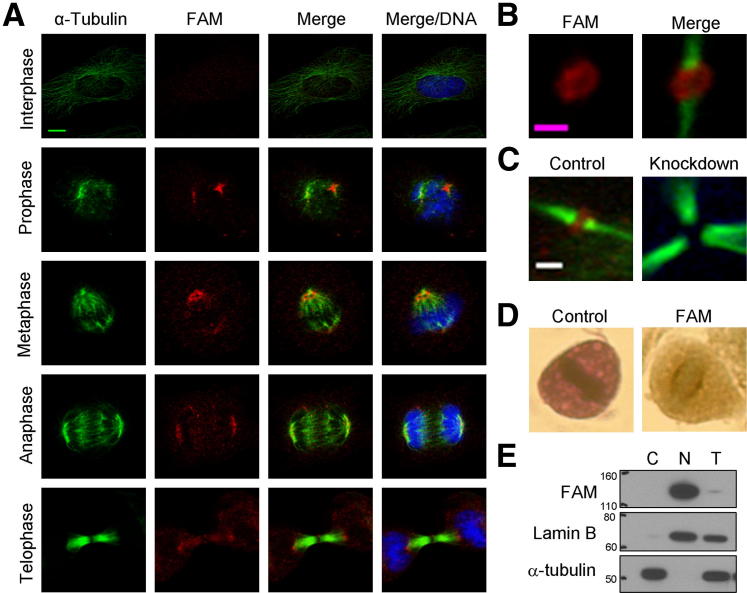
Rabbit monoclonal antibodies localized FAM190A to the mitotic spindle in HeLa cells (Figure 2A). FAM190A appeared in early prophase as a punctuate pattern, later forming two dot-like foci. In metaphase, FAM190A appeared as a purse string–like structure, resembling tethering to microtubules. FAM190A surrounded centrin 3 and γ-tubulin as a pericentriolar material (Supplemental Figure S2, A and B). In anaphase, the FAM190A fluorescence intensity decreased or dispersed, forming an arcuate structure at the mitotic spindle pole. There was faint signal between the planes of separated chromosomes. FAM190A appeared as a ring structure at the midbody (Figure 2B) in the late stage of cytokinesis (Supplemental Figure S2C). Some FAM190A-knockdown cells lacked FAM190A immunofluorescence (IF) signal at the midbody, including in cells undergoing multipolar mitosis (Figure 2C). During interphase, FAM190A signal was low, with fluorescence seen faintly as nuclear speckles. FAM190A similarly localized to the mitotic spindle and midbody in other cell lines, such as AAV-293, RKO, DLD1, and AsPC1 (Supplemental Figure S3A). Besides anti-FAM190A monoclonal antibodies, four FAM190A exon-specific rabbit polyclonal antibodies and two commercially available antibodies also localized FAM190A at the mitotic spindle and midbody (Supplemental Figure S3B).
Figure 2.
FAM190A cellular localization suggests mitotic functions. A: Representative confocal IF images of HeLa cells at various cell cycle stages. Red, FAM190A; green, α-tubulin; blue, DNA. B: Representative IF image of Flemming body of HeLa cells. Red, FAM190A; green, α-tubulin. C: Representative IF image of midbody of FAM190A-knockdown clone 77-2 and control cells. Red, FAM190A; green, α-tubulin. D: Representative images from IHC of HeLa cells using anti-FAM190A antibody (mAb7), cells in mitosis. E: IB comparing nuclear and cytoplasmic fractions of HeLa cells. IB with mAb7, anti–α-tubulin, and anti–Lamin B. C, cytoplasmic; N, nuclear; T, total. Scale bars: 5 μm (A); 1 μm (B and C).
We exogenously overexpressed FAM190A transiently (see below), but GFP-tagged FAM190A produced a nonphysiologic diffuse pattern never observed in native cells. Attempts to generate HeLa and 293 stable lines overexpressing tagged FAM190A were unsuccessful. Long-term, rather than short-term, overexpression of FAM190A might be toxic to the cells.
Immunohistochemistry
In HeLa cells, using an anti-FAM190A rabbit monoclonal antibody, immunopositivity was seen at the spindle location in mitotic cells (Figure 2D). We could determine neither late telophase nor the midbody in these preparations.
Immunoblot
We prepared nuclear and cytoplasmic fractions of HeLa cells. By IB, we found FAM190A exclusively in the nuclear fraction (Figure 2E). In IBs of unfractionated cell lines, only HeLa had an approximately 130-kDa band; this band was depleted by shRNA targeting FAM190A (Supplemental Figure S4A). This larger protein form was also present in normal tissues (Supplemental Figure S4B), but the pancreatic cancer xenografts lacked this band. The band was observed in IB using other mAbs against FAM190A (Supplemental Figures S4, S5, and S6). In histologic sections, xenografts had high nuclear-to-cytoplasmic ratios, prominent nuclear polymorphism, and atypia, and only PX-120 had occasional multinuclear cells. The following experiments thus investigated this protein species in HeLa cells. Exogenously expressed untagged full-length FAM190A in transfected cells was smaller in size than the upper band of native FAM190A of HeLa cells, apparently having been processed nonphysiologically. Moreover, FAM190A seems to be an intrinsically unstructured protein. Intrinsically unstructured proteins bind less SDS and appear 1.2 to 1.8 times higher than calculated molecular weight in SDS-PAGE gels12; FAM190A was seen at approximately 130 kDa than the calculated molecular weight of approximately 100 kDa.
FAM190A Changes During the Cell Cycle
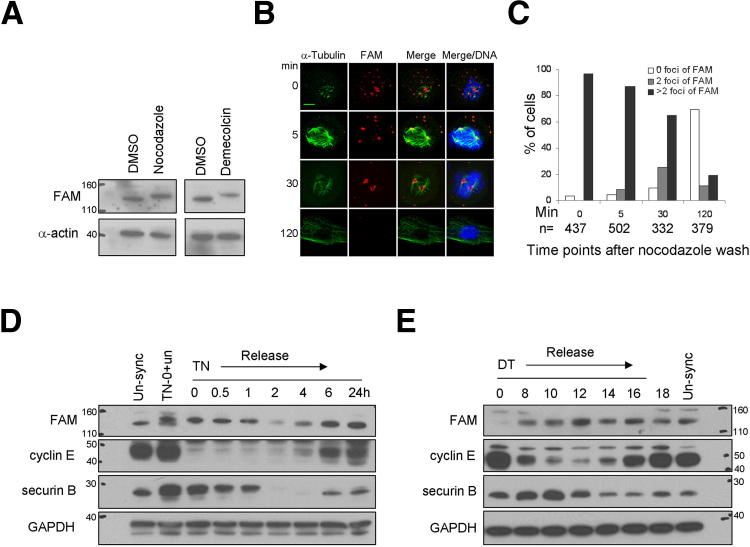
In IBs of HeLa cells treated with the microtubule polymerization inhibitors nocodazole or demecolcine, FAM190A became a slower-migrating isoform (Figure 3A), owing to protein phosphorylation. To determine the subcellular manifestation of this change, we examined cells treated with nocodazole for 16 hours and cells at postrelease time points after nocodazole washout by IF. In arrested cells, FAM190A appeared as a punctuate pattern (multiple foci per cell) with an intense fluorescence. After release, multiple foci became rearranged to form two foci per cell at the spindle poles during mitosis and became nearly unapparent as cells entered interphase (Figure 3B). Cell cycle progression after nocodazole wash was followed by counting FAM190A foci per cell at various time points (Figure 3C). FAM190A was co-localized with microtubules (α-tubulin) at spindle poles from prometaphase to anaphase.
Figure 3.
FAM190A changes during cell cycle. A: HeLa cells were treated with nocodazole for 16 hours or demecolcine for 24 hours. After microtubule depolymerizing drugs, FAM190A migrated slower. IB with mAb7 and anti–α-actin. B and C: HeLa cells were treated with nocodazole for 16 hours. At different times after drug washout, cells were prepared for IF. FAM190A foci were counted at different times. 0 foci, cells at interphase; 2 foci, cells in mitosis; >2 foci, cells under nocodazole effect or ongoing redistribution of FAM190A after drug release. n = foci counted from total number of cells at a time point. D: HeLa cells were synchronized at prometaphase by nocodazole preceded by thymidine block. Cells were harvested at different points after release from nocodazole and examined by IB. IB with mAb170, anti–cyclin E, anti–securin B, and anti-GAPDH. TN, thymidine-nocodazole block. E: HeLa cells were synchronized at G1/S-phase by double thymidine block. Cells were harvested at different times after release from block and examined by IB. IB with mAb170, anti–cyclin E, anti–securin B, and anti-GAPDH. DT, double thymidine block. Scale bar = 5 μm (B and C).
We synchronized HeLa cells at prometaphase by a thymidine-nocodazole block and harvested samples at various time points after drug washout. By IB, we observed the slower-migrating forms of FAM190A during the nocodazole block, but on release it disappeared by approximately 2 hours, when the native (faster-migrating) isoform progressively appeared, returning to the isoform pattern observed in the unsynchronized cells (Figure 3D). The timing of the disappearance of the slower isoform coincided with the timing of degradation of securin B and reappearance of cyclin E in the same cells, which are time posts of cell cycle progression.13,14 The cell cycle progression of blocked and released cells were also determined by flow cytometry (Supplemental Figure S5A). Mixing of samples revealed the two species of FAM190A were indeed distinct (Supplemental Figure S5B).
On synchronizing cells at G1/S-phase by double thymidine block, the levels of FAM190A protein had a pronounced decrease compared with unsynchronized cells. After release, levels of FAM190A protein increased, peaked in M-phase at 12 hours, and subsequently decreased (Figure 3E). Cell cycle progression was determined by flow cytometry (Supplemental Figure S5A). A pronounced decrease in FAM190A protein also occurred by 48 hours after serum starvation, and an increase occurred after the readdition of serum to the culture (Supplemental Figure S5C).
FAM190A Interactome
Several FAM190A-interacting proteins were suggested by the yeast two-hybrid screen (Supplemental Table S1). We verified Ndel1 and EXOC1 as binding partners of FAM190A by IP (Figure 4). The Ndel1 inverse interaction with FAM190A was previously reported using Ndel1 as the yeast two-hybrid bait.15 Ndel1 plays a role in cytoskeletal organization and neuronal migration.16 EXOC1 is a part of the exocyst complex, participates in actin cytoskeletal remodeling and vesicular transport,17 and plays an important role in mediating the cargo transport of proteins to the midbody during cytokinesis.18
Figure 4.
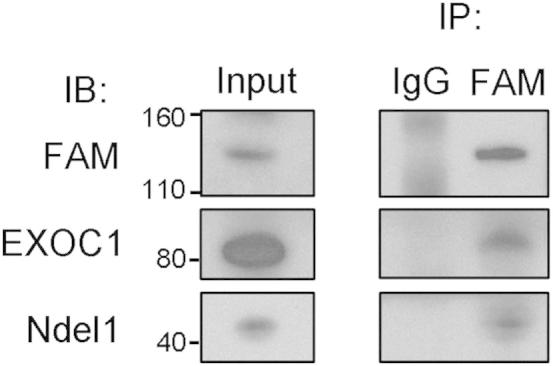
FAM190A interacts with EXOC1 and Ndel1. FAM190A was captured with mAb170 and mAb62. FAM190A, EXOC1, and Ndel1 were identified in captured proteins by IB. IP by rabbit IgG is a control.
FAM190A Isoforms in Mitosis
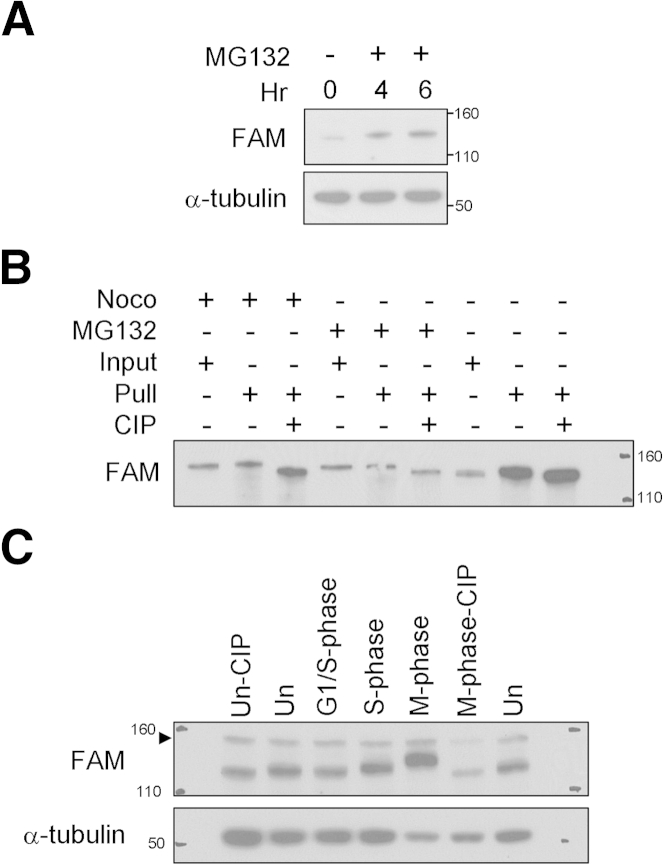
We treated HeLa cells with MG132, which inhibits S26 proteasomal protein degradation. After MG132 treatment, levels of FAM190A protein were modestly increased and the slow-migrating form of FAM190A appeared (Figure 5A). It is possible that the mitotic arrest indirectly induced by MG132 mimicked the arrest earlier observed by nocodazole and demecolcine, preventing the mitotic degradation of FAM190A protein as suggested in earlier experiments.
Figure 5.
FAM190A is phosphorylated in mitosis. A: HeLa cells were incubated with MG132 for 4 hours and 6 hours and harvested in lysis solution. IB with mAb7 and anti–α-tubulin. B: HeLa cells were synchronized at prometaphase by nocodazole or released into MG132 after nocodazole block for 4 hours. FAM190A was captured with mAb170 and mAb62 and incubated with or without CIP for 60 minutes at 37°C. IB using mAb170. C: HeLa cells were synchronized to a different phase of cell cycle by double thymidine block (G1/S-phase), 2-hour release after double thymidine block (S-phase), or thymidine-nocodazole block (M-phase). Lysates were treated with CIP for 60 minutes. IB with mAb170 and mAb62 and α-tubulin. Un, unsynchronized cells. The arrowhead indicates a nonspecific band.
Using IP, we captured FAM190A from HeLa cells synchronized by nocodazole, cells synchronized by nocodazole and treated with MG132 for 4 hours, and unsynchronized cells. Captured protein was treated with CIP. CIP-treated FAM190A converted to a fast-migrating form, indicating that the slower-migrating isoform of M-phase–arrested cells and even the FAM190A isoform in unsynchronized cells were phosphorylated species of FAM190A (Figure 5B).
To examine the difference in the phosphorylation isoforms, we compared unsynchronized cells, cells synchronized by double thymidine block (G0/G1-phase), cells synchronized by double thymidine block and released for 2 hours (S-phase), and cells synchronized by thymidine-nocodazole block (M-phase) by IB. We found progressively slower migration of FAM190A from G0/G1-phase, S-phase, to M-phase (Figure 5C). FAM190A at S-phase was phosphorylated because CIP treatment increased its gel migration (Supplemental Figure S6, A and B). FAM190A was phosphorylated in HeLa cells after release from thymidine block (Supplemental Figure S6A) and after release from serum starvation (Supplemental Figure S6B). Thus, the phosphorylated FAM190A form accumulating before M-phase existed independent of nocodazole, although FAM190A in nocodazole-arrested cells had a slower migration than in S-phase, apparently due to additional phosphorylation.
Discussion
Theodor Boveri proposed that aberrant multipolar mitosis might generate the abnormal chromatin findings of cancers.19 He wrote, “When I published the results of my experiments on the development of double-fertilized sea-urchin eggs in 1902, I added the suggestion that malignant tumors might be the result of a certain abnormal condition of the chromosomes, which may arise from multipolar mitosis. So I have carried on for a long time the kind of experiments I suggested, which are so far without success, but my conviction remains unshaken.” Since his proposal, the overexpression of aurora kinases has been one of the potential mechanisms suggested to form multipolar mitotic division in cancer.20,21
We present novel characteristics of the FAM190A gene, which is frequently altered in human cancers but had an unknown function. A pancreatic cancer with foci of multinuclear cells was previously described to have an unusually early and out-of-frame FAM190A deletion/rearrangement. Human tumors and cancer cell lines often had an absence of the larger protein form that was otherwise observed in normal human tissues (Supplemental Figure S4B) and in FAM190A wild-type HeLa cells. In cellular studies, FAM190A was present in the nuclear fraction of unsynchronized populations, but its form and quantity were modified and potentially regulated by posttranslational modification, during the cell division cycle. At S-phase and in mitotic arrest, transient phosphorylated isoforms appeared. FAM190A localized to early and late mitotic structures, including the Flemming body of the cytokinesis midbody structure.
FAM190A had protein interactions with EXOC1 and Ndel1. EXOC1 is a core component of the exocyst complex, which transports proteins and lipids to cell membrane destinations, including the midbody,22,23 where exocyst function and endosomal trafficking are required for cytokinesis.18 Ndel1 participates in dynein-dependent and dynein-independent centrosomal protein assembly.24
We induced cytokinetic defects and multipolar asynchronous division of multinuclear cells on the depletion of FAM190A. We propose that FAM190A normally plays essential structural or regulatory roles in mitosis. This role is especially likely at the midbody, where interactions of FAM190A with the exocyst complex may be required for normal cytokinesis. In the absence of adequate FAM190A function, abscission fails, eventually yielding multinuclearity and several multipolar mitotic cycles.
We speculate that the focal nature of the multinuclear cells in both the cited pancreatic cancer and in our cellular FAM190A-knockdown model may represent a threshold effect. There might exist a functional level below which a cell experiences negative selective pressures due to a severe FAM190A deficiency (creating a multinuclear phenotype), in turn limiting the tolerated number of generations of aberrant multipolar cell division and causing cell death. Other cells might tolerate having low FAM190A. Some of these will occasionally decrease below the threshold of FAM190A expression level, creating a dynamic equilibrium of multinuclear cells at approximately 4% to 5% of the total cells after FAM190A knockdown. On the basis of our knockdown model, we further speculate that there would be a focal and transient presence of multinuclear cells in human tumors rather than the whole tumor having multinuclear cells.
The predicted formation of truncated and internally deleted proteins requires that we consider the possibility of dominant-negative effects of any altered FAM190A proteins, helping to explain the co-existence in many tumors of a single deleted FAM190A transcript structure with a wild-type transcript.6 We thus also suggest that the more common in-frame homozygous deletions and deletion/rearrangements of FAM190A transcripts in cancers create analogous, yet more subtle, mitotic defects, which contribute to genetic instability during tumorigenesis.
Acknowledgments
We thank the JHU-SOM Institute of Basic Biomedical Sciences Microscope Facility and Barbara Smith, J. Wade Harper (Harvard Medical School, Boston, MA), and Charles J. Sherr (HHMI, St. Jude Children Hospital, Memphis, TN) for helpful suggestions.
Footnotes
Supported by NIH grants P01 CA134292, P50 CA62924, and RO1 CA128920 and by the Everett and Marjorie Kovler Professorship in Pancreas Cancer Research.
Supplemental Data
Natural and induced multinuclearity is created by absence of FAM190A. A: Phase-contrast and corresponding DAPI-stained images of FAM190A-knockdown clone 77-2 and control cells. Multinuclear cells were present in clone 77-2. Some cells had bizarre nuclear shapes. B: After PI staining, flow analysis was done on FAM190A-knockdown clone (77-2) and control cells (03-1). The histogram presents the mean percentage of cells having >2N DNA in both cell types from three independent experiments. Error bars represent SEMs. P = 0.032, Student's t-test. C: Phase-contrast images of FAM190A-knockdown cell lines HeLa, RKO, and DLD1. Clone names are described at the bottom of the images.
FAM190A cellular localization suggests mitotic functions. A: Representative confocal IF images of HeLa cells at various cell cycle stages. Red, FAM190A; green, centrin 3; blue, DNA. B: Representative confocal IF images of HeLa cells at various cell cycle stages. Red, FAM190A; green, gamma-tubulin; blue, DNA. C: FAM190A located at the midbody in late cytokinesis. Representative IF images of HeLa cells. Red, FAM190A; green, alpha-tubulin, blue, DNA. Scale bars: 5 μm (A and B); 2 μm (C).
Similar IF patterns among cell lines and anti-FAM190A antibodies. A: Representative IF images of AAV-293, AsPC1, RKO, and DLD1 cells. Red, FAM190A; green, α-tubulin; blue, DNA. B: FAM190A located at the midbody by various anti-FAM antibodies. Representative IF images of HeLa cells. Red, FAM190A; green, α-tubulin, blue, DNA. The various antibodies produced similar results in IF and IB; however, when tested under similar conditions, differences were seen as expected for independent immunologic reagents. For example, in B, the midbody was detected by antibodies mAb62, mAb96, AP10022b, exon 8, and exons 9 and 10 but not by ab90508 and exon 7 antibodies. Scale bar = 5 μm (A and B).
IBs using anti-FAM190A among cell lines and tissues. A: IB of various cell lines. HeLa cells were transiently transfected to express exogenous FAM190A (HeLa-FAM) as a size reference. One-tenth of the HeLa-FAM lysate was loaded and compared with the neighboring HeLa lane to normalize the chemiluminescence signal. IB used mAb7. B: FAM190A was captured from normal human tissues and pancreatic cancer xenografts by IP. HeLa cells were used as a size reference for endogenous FAM190A. IB used mAb7. The arrowhead indicates a nonspecific band.
Changes in the form and quantity of FAM190A during the cell cycle. A: Flow cytometric cell cycle analysis of samples from Figure 3, D and E. TN, thymidine-nocodazole block; DT, double thymidine block. B: Samples from Figure 3D were mixed and then analyzed by IB using mAb170 and anti-GAPDH. C: HeLa cells were serum starved for 48 hours and harvested at various times after addition of serum. Samples were compared with unstarved samples. IBs used mAb170 and anti–α-actin. Un, untreated cells.
FAM190A is phosphorylated. A: HeLa cells were synchronized by double thymidine block (DT), released for 2 hours (DT2h) after double thymidine block, and harvested in RIPA or phosphatase lysis solution. Sixty minutes after addition of CIP to the appropriate lysate, Laemmli buffer was added. The samples were then equalized by addition of the opposite lysing solution (RIPA or phosphatase lysis solution). IBs used mAb170 and mAb62 and anti–α-actin. B: HeLa cells were serum starved for 48 hours (SS), incubated with serum for 2 hours (SS2h) after serum starvation, and harvested in RIPA or phosphatase lysis solution. Sixty minutes after addition of CIP to appropriate lysate, the reaction was stopped by addition of Laemmli buffer. The samples were then equalized by addition of the opposite lysing solution (RIPA or phosphatase lysis solution). IB using mAb170 and mAb62 and anti–α-actin. The arrowhead indicates FAM190A.
FAM190A knockdown cells (clone 77-2) expressing GFP-tagged H2B to mark chromatin. Images were captured every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) expressing GFP-tagged H2B to mark chromatin. Images were captured every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) expressing GFP-tagged H2B to mark chromatin. Images were captured every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) were imaged every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) were imaged every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) were imaged every 10 minutes. The arrow indicates cells having division defects.
References
- 1.Cahill D.P., Lengauer C., Yu J., Riggins G.J., Willson J.K., Markowitz S.D., Kinzler K.W., Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 2.Solomon D.A., Kim T., Diaz-Martinez L.A., Fair J., Elkahloun A.G., Harris B.T., Toretsky J.A., Rosenberg S.A., Shukla N., Ladanyi M., Samuels Y., James C.D., Yu H.T., Kim J.S., Waldman T. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber T.D., McManus K., Yuen K.W., Reis M., Parmigiani G., Shen D., Barrett I., Nouhi Y., Spencer F., Markowitz S., Velculescu V.E., Kinzler K.W., Vogelstein B., Lengauer C., Hieter P. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iacobuzio-Donahue C.A., Fu B., Yachida S., Luo M., Abe H., Henderson C.M., Vilardell F., Wang Z., Keller J.W., Banerjee P., Herman J.M., Cameron J.L., Yeo C.J., Halushka M.K., Eshleman J.R., Raben M., Klein A.P., Hruban R.H., Hidalgo M., Laheru D. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell P.J., Yachida S., Mudie L.J., Stephens P.J., Pleasance E.D., Stebbings L.A., Morsberger L.A., Latimer C., McLaren S., Lin M.L., McBride D.J., Varela I., Nik-Zainal S.A., Leroy C., Jia M.M., Menzies A., Butler A.P., Teague J.W., Griffin C.A., Burton J., Swerdlow H., Quail M.A., Stratton M.R., Iacobuzio-Donahue C., Futreal P.A. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scrimieri F., Calhoun E.S., Patel K., Gupta R., Huso D.L., Hruban R.H., Kern S.E. FAM190A rearrangements provide a multitude of individualized tumor signatures and neo-antigens in cancer. Oncotarget. 2011;2:69–75. doi: 10.18632/oncotarget.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calhoun E.S., Hucl T., Gallmeier E., West K.M., Arking D.E., Maitra A., Iacobuzio-Donahue C.A., Chakravarti A., Hruban R.H., Kern S.E. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–7928. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 8.Nancarrow D.J., Handoko H.Y., Smithers B.M., Gotley D.C., Drew P.A., Watson D.I., Clouston A.D., Hayward N.K., Whiteman D.C. Genome-wide copy number analysis in esophageal adenocarcinoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2008;68:4163–4172. doi: 10.1158/0008-5472.CAN-07-6710. [DOI] [PubMed] [Google Scholar]
- 9.Manceau G., Letouzé E., Guichard C., Didelot A., Cazes A., Corté H., Fabre E., Pallier K., Imbeaud S., Pimpec-Barthes F.L., Zucman-Rossi J., Laurent-Puig P., Blons H. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma. Int J Cancer. 2013;132:2217–2221. doi: 10.1002/ijc.27900. [DOI] [PubMed] [Google Scholar]
- 10.Lantieri F., Glessner J.T., Hakonarson H., Elia J., Devoto M. Analysis of GWAS top hits in ADHD suggests association to two polymorphisms located in genes expressed in the cerebellum. Am J Med Genet B Neuropsychiatr Genet. 2010;153:1127–1133. doi: 10.1002/ajmg.b.31110. [DOI] [PubMed] [Google Scholar]
- 11.Caldas C., Hahn S.A., da Costa L.T., Redston M.S., Schutte M., Seymour A.B., Weinstein C.L., Hruban R.H., Yeo C.J., Kern S.E. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 12.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 13.Moroy T., Geisen C., Cyclin E. Int J Biochem Cell Biol. 2004;36:1424–1439. doi: 10.1016/j.biocel.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Zhao W.M., Coppinger J.A., Seki A., Cheng X.L., Yates J.R., III, Fang G. RCS1, a substrate of APC/C, controls the metaphase to anaphase transition. Proc Natl Acad Sci U S A. 2008;105:13415–13420. doi: 10.1073/pnas.0709227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camargo L.M., Collura V., Rain J.C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T.P., Whiting P.J., Brandon N.J. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 16.Hippenmeyer S., Youn Y.H., Moon H.M., Miyamichi K., Zong H., Wynshaw-Boris A., Luo L.Q. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68:695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jourdain I., Dooley H.C., Toda T. Fission yeast Sec3 bridges the exocyst complex to the actin cytoskeleton. Traffic. 2012;13:1481–1495. doi: 10.1111/j.1600-0854.2012.01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X.W., Inoue M., Hsu S.C., Saltiel A.R. RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J Biol Chem. 2006;281:38609–38616. doi: 10.1074/jbc.M512847200. [DOI] [PubMed] [Google Scholar]
- 19.Boveri T. Williams & Wilkins Company; Baltimore: 1929. Zur frage der entstehung maligner tumoren. Translation by Marcella Boveri. p 1. [Google Scholar]
- 20.Meraldi P., Honda R., Nigg E.A. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H., Kuang J., Zhong L., Kuo W.L., Gray J.W., Sahin A., Brinkley B.R., Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 22.Heider M.R., Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gromley A., Yeaman C., Rosa J., Redick S., Chen C.T., Mirabelle S., Guha M., Sillibourne J., Doxsey S.J. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol Biol Cell. 2006;17:680–689. doi: 10.1091/mbc.E05-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Natural and induced multinuclearity is created by absence of FAM190A. A: Phase-contrast and corresponding DAPI-stained images of FAM190A-knockdown clone 77-2 and control cells. Multinuclear cells were present in clone 77-2. Some cells had bizarre nuclear shapes. B: After PI staining, flow analysis was done on FAM190A-knockdown clone (77-2) and control cells (03-1). The histogram presents the mean percentage of cells having >2N DNA in both cell types from three independent experiments. Error bars represent SEMs. P = 0.032, Student's t-test. C: Phase-contrast images of FAM190A-knockdown cell lines HeLa, RKO, and DLD1. Clone names are described at the bottom of the images.
FAM190A cellular localization suggests mitotic functions. A: Representative confocal IF images of HeLa cells at various cell cycle stages. Red, FAM190A; green, centrin 3; blue, DNA. B: Representative confocal IF images of HeLa cells at various cell cycle stages. Red, FAM190A; green, gamma-tubulin; blue, DNA. C: FAM190A located at the midbody in late cytokinesis. Representative IF images of HeLa cells. Red, FAM190A; green, alpha-tubulin, blue, DNA. Scale bars: 5 μm (A and B); 2 μm (C).
Similar IF patterns among cell lines and anti-FAM190A antibodies. A: Representative IF images of AAV-293, AsPC1, RKO, and DLD1 cells. Red, FAM190A; green, α-tubulin; blue, DNA. B: FAM190A located at the midbody by various anti-FAM antibodies. Representative IF images of HeLa cells. Red, FAM190A; green, α-tubulin, blue, DNA. The various antibodies produced similar results in IF and IB; however, when tested under similar conditions, differences were seen as expected for independent immunologic reagents. For example, in B, the midbody was detected by antibodies mAb62, mAb96, AP10022b, exon 8, and exons 9 and 10 but not by ab90508 and exon 7 antibodies. Scale bar = 5 μm (A and B).
IBs using anti-FAM190A among cell lines and tissues. A: IB of various cell lines. HeLa cells were transiently transfected to express exogenous FAM190A (HeLa-FAM) as a size reference. One-tenth of the HeLa-FAM lysate was loaded and compared with the neighboring HeLa lane to normalize the chemiluminescence signal. IB used mAb7. B: FAM190A was captured from normal human tissues and pancreatic cancer xenografts by IP. HeLa cells were used as a size reference for endogenous FAM190A. IB used mAb7. The arrowhead indicates a nonspecific band.
Changes in the form and quantity of FAM190A during the cell cycle. A: Flow cytometric cell cycle analysis of samples from Figure 3, D and E. TN, thymidine-nocodazole block; DT, double thymidine block. B: Samples from Figure 3D were mixed and then analyzed by IB using mAb170 and anti-GAPDH. C: HeLa cells were serum starved for 48 hours and harvested at various times after addition of serum. Samples were compared with unstarved samples. IBs used mAb170 and anti–α-actin. Un, untreated cells.
FAM190A is phosphorylated. A: HeLa cells were synchronized by double thymidine block (DT), released for 2 hours (DT2h) after double thymidine block, and harvested in RIPA or phosphatase lysis solution. Sixty minutes after addition of CIP to the appropriate lysate, Laemmli buffer was added. The samples were then equalized by addition of the opposite lysing solution (RIPA or phosphatase lysis solution). IBs used mAb170 and mAb62 and anti–α-actin. B: HeLa cells were serum starved for 48 hours (SS), incubated with serum for 2 hours (SS2h) after serum starvation, and harvested in RIPA or phosphatase lysis solution. Sixty minutes after addition of CIP to appropriate lysate, the reaction was stopped by addition of Laemmli buffer. The samples were then equalized by addition of the opposite lysing solution (RIPA or phosphatase lysis solution). IB using mAb170 and mAb62 and anti–α-actin. The arrowhead indicates FAM190A.
FAM190A knockdown cells (clone 77-2) expressing GFP-tagged H2B to mark chromatin. Images were captured every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) expressing GFP-tagged H2B to mark chromatin. Images were captured every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) expressing GFP-tagged H2B to mark chromatin. Images were captured every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) were imaged every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) were imaged every 10 minutes. The arrow indicates cells having division defects.
FAM190A knockdown cells (clone 77-2) were imaged every 10 minutes. The arrow indicates cells having division defects.